Differential Interactions of Molecular Chaperones and Yeast Prions
Abstract
1. Introduction
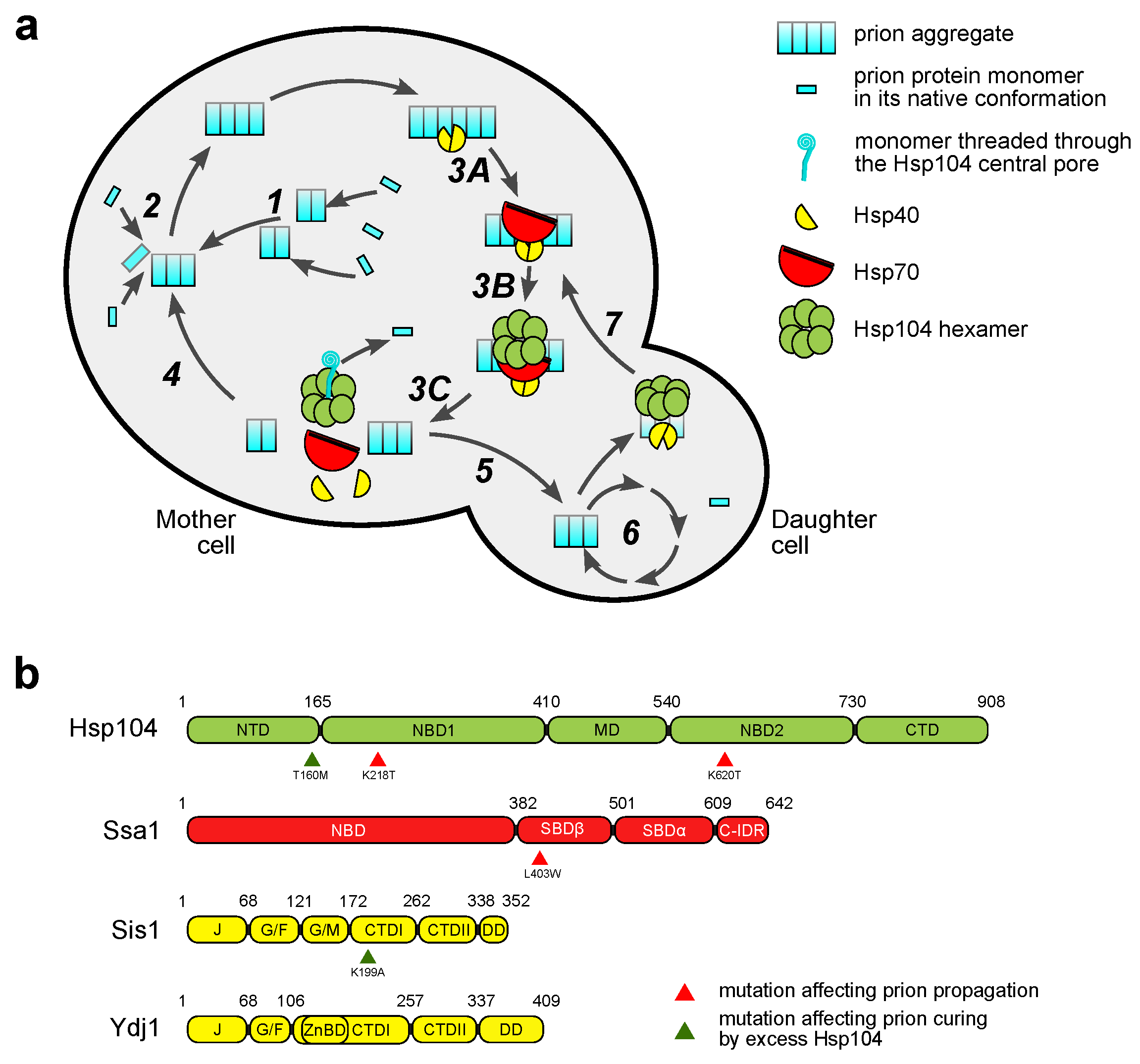
2. Yeast Prions and Their Life Cycle
3. Major Molecular Chaperones Involved in Prion Propagation
3.1. Hsp104
3.2. The Hsp70/Hsp40 Chaperone System
3.3. Other Proteins
4. Differential Effects of Chaperones and Protein-Sorting Factors on Yeast Prion Propagation
4.1. Hsp104 and Its Variants
4.2. Hsp70
4.3. Sis1 and Its Domain Activity
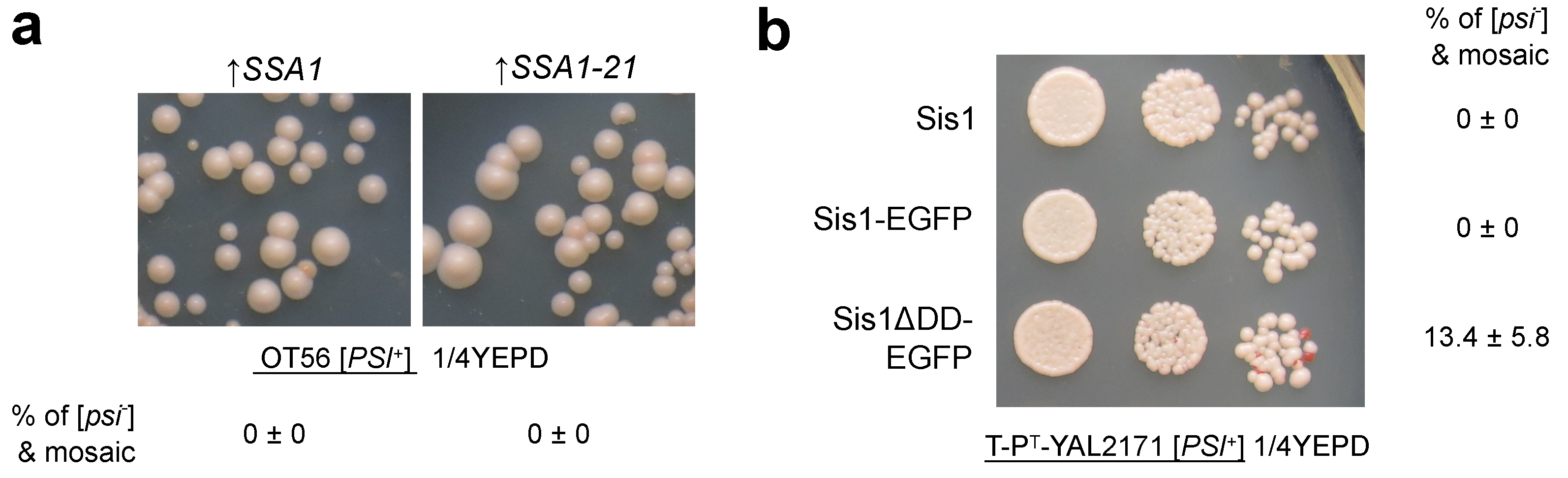
4.4. Other J-Proteins
4.5. Protein-Sorting Factors
5. Mechanistic Basis for the Differential Effects of Chaperones on Yeast Prions
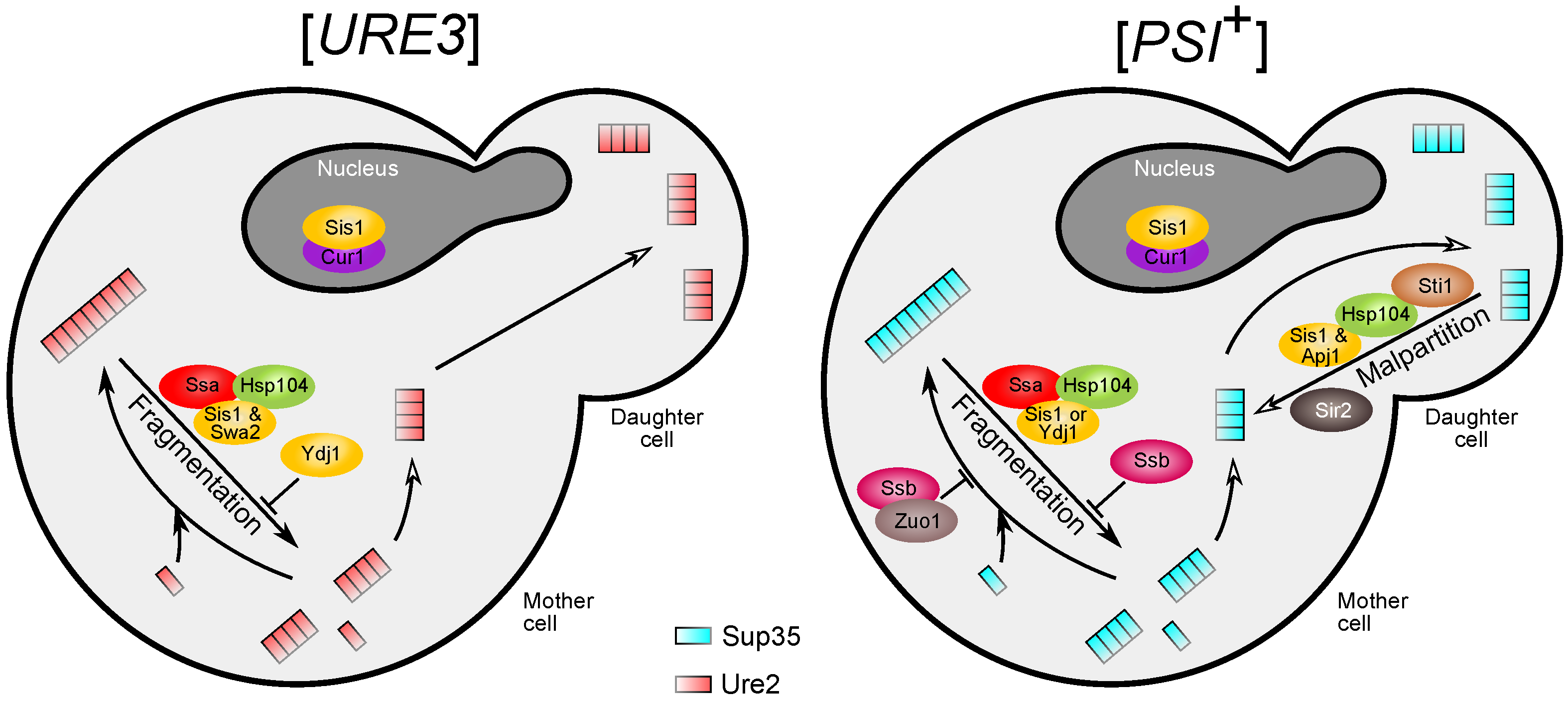
5.1. The Central Role of Sis1 in Protein Aggregation and Disaggregation
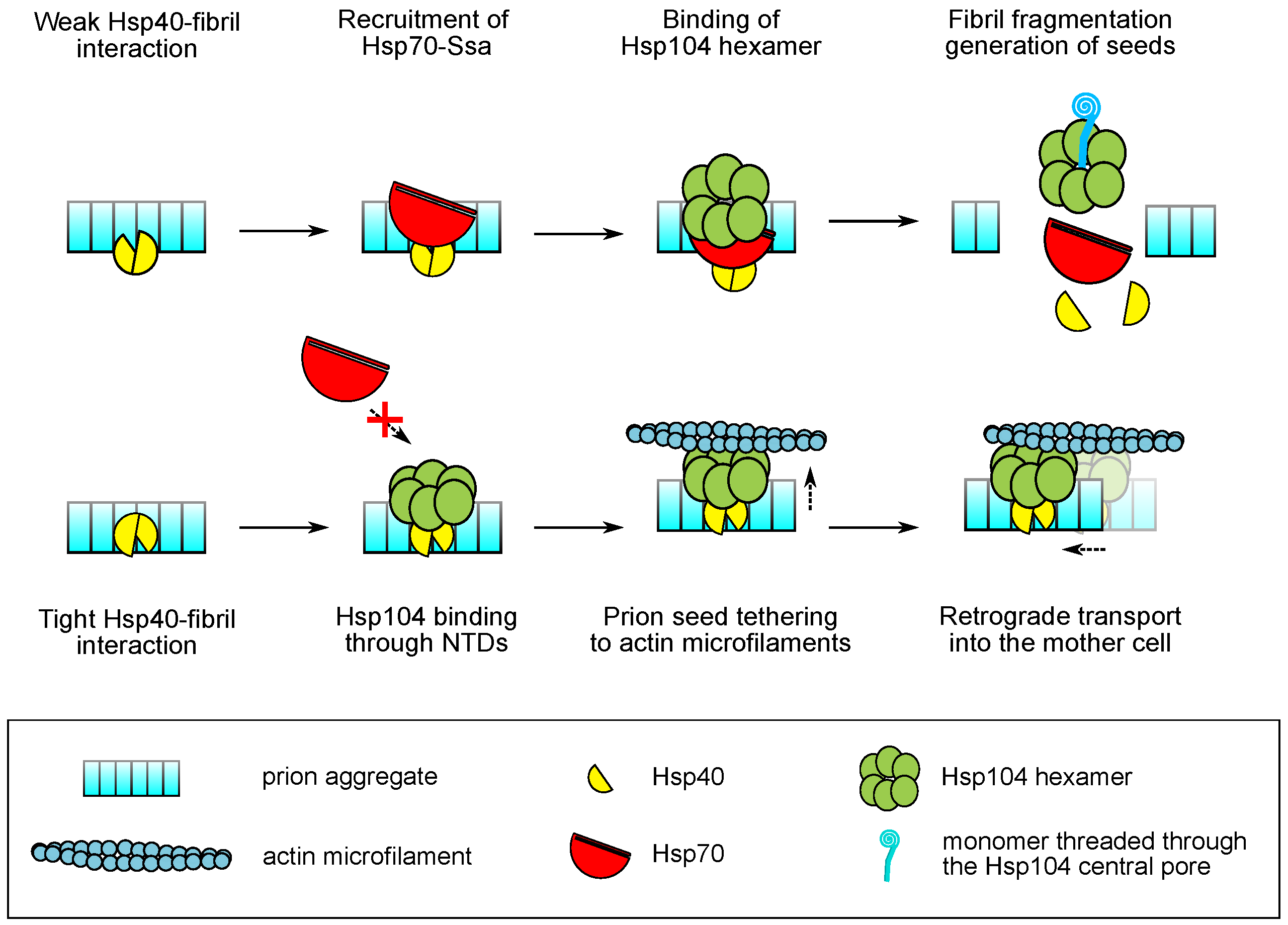
5.2. Modes of Hsp104 Function and the Mechanisms of Malpartition
6. Conclusions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Sample Availability
Abbreviations
| CTD | C-terminal domain |
| NBD | Nucleotide-binding domain |
| NTD | N-terminal domain |
| OE | Overexpression/overproduction |
| PQC | Protein Quality Control |
| PrD | Prion domain |
| RAC | Ribosome-associated complex |
References
- Uversky, V.N. Mysterious oligomerization of the amyloidogenic proteins. FEBS J. 2010, 277, 2940–2953. [Google Scholar] [CrossRef] [PubMed]
- Prusiner, S.B. Prions. Proc. Natl. Acad. Sci. USA 1998, 95, 13363–13383. [Google Scholar] [CrossRef] [PubMed]
- Jaunmuktane, Z.; Brandner, S. Invited Review: The role of prion-like mechanisms in neurodegenerative diseases. Neuropathol. Appl. Neurobiol. 2020, 46, 522–545. [Google Scholar] [CrossRef] [PubMed]
- Saibil, H. Chaperone machines for protein folding, unfolding and disaggregation. Nat. Rev. Mol. Cell Biol. 2013, 14, 630–642. [Google Scholar] [CrossRef] [PubMed]
- Kaganovich, D.; Kopito, R.; Frydman, J. Misfolded proteins partition between two distinct quality control compartments. Nature 2008, 454, 1088–1095. [Google Scholar] [CrossRef] [PubMed]
- Specht, S.; Miller, S.B.M.; Mogk, A.; Bukau, B. Hsp42 is required for sequestration of protein aggregates into deposition sites in Saccharomyces cerevisiae. J. Cell Biol. 2011, 195, 617–629. [Google Scholar] [CrossRef]
- Miller, S.B.M.; Ho, C.T.; Winkler, J.; Khokhrina, M.; Neuner, A.; Mohamed, M.Y.H.; Guilbride, D.L.; Richter, K.; Lisby, M.; Schiebel, E.; et al. Compartment-specific aggregases direct distinct nuclear and cytoplasmic aggregate deposition. EMBO J. 2015, 34, 778–797. [Google Scholar] [CrossRef]
- Sarto-Jackson, I.; Tomaska, L. How to bake a brain: Yeast as a model neuron. Curr. Genet. 2016, 62, 347–370. [Google Scholar] [CrossRef]
- Liebman, S.W.; Chernoff, Y.O. Prions in yeast. Genetics 2012, 191, 1041–1072. [Google Scholar] [CrossRef]
- Tanaka, M.; Weissman, J.S. An efficient protein transformation protocol for introducing prions into yeast. Methods Enzymol. 2006, 412, 185–200. [Google Scholar] [CrossRef]
- Cox, B.S. Ψ, A cytoplasmic suppressor of super-suppressor in yeast. Heredity 1965, 20, 505–521. [Google Scholar] [CrossRef]
- True, H.L.; Berlin, I.; Lindquist, S.L. Epigenetic regulation of translation reveals hidden genetic variation to produce complex traits. Nature 2004, 431, 184–187. [Google Scholar] [CrossRef] [PubMed]
- Halfmann, R.; Jarosz, D.F.; Jones, S.K.; Chang, A.; Lancaster, A.K.; Lindquist, S. Prions are a common mechanism for phenotypic inheritance in wild yeasts. Nature 2012, 482, 363–368. [Google Scholar] [CrossRef] [PubMed]
- Matiiv, A.B.; Trubitsina, N.P.; Matveenko, A.G.; Barbitoff, Y.A.; Zhouravleva, G.A.; Bondarev, S.A. Amyloid and amyloid-like aggregates: Diversity and the term crisis. Biochemistry 2020, 85, 1011–1034. [Google Scholar] [CrossRef] [PubMed]
- Wickner, R.B.; Edskes, H.K.; Engel, A. Yeast prions: Evolution of the prion concept. Prion 2007, 1, 94–100. [Google Scholar] [CrossRef] [PubMed]
- Wickner, R.B. [URE3] as an altered URE2 protein: Evidence for a prion analog in Saccharomyces cerevisiae. Science 1994, 264, 566–569. [Google Scholar] [CrossRef] [PubMed]
- Derkatch, I.L.; Bradley, M.E.; Zhou, P.; Chernoff, Y.O.; Liebman, S.W. Genetic and environmental factors affecting the de novo appearance of the [PSI+] prion in Saccharomyces cerevisiae. Genetics 1997, 147, 507–519. [Google Scholar] [CrossRef]
- Baxa, U.; Taylor, K.L.; Wall, J.S.; Simon, M.N.; Cheng, N.; Wickner, R.B.; Steven, A.C. Architecture of Ure2p prion filaments: The N-terminal domains form a central core fiber. J. Biol. Chem. 2003, 278, 43717–43727. [Google Scholar] [CrossRef]
- Wickner, R.B.; Dyda, F.; Tycko, R. Amyloid of Rnq1p, the basis of the [PIN+] prion, has a parallel in-register β-sheet structure. Proc. Natl. Acad. Sci. USA 2008, 105, 2403–2408. [Google Scholar] [CrossRef]
- Bagriantsev, S.N.; Gracheva, E.O.; Richmond, J.E.; Liebman, S.W. Variant-specific [PSI+] infection is transmitted by Sup35 polymers within [PSI+] aggregates with heterogeneous protein composition. Mol. Biol. Cell 2008, 19, 2433–2443. [Google Scholar] [CrossRef]
- Sergeeva, A.V.; Belashova, T.A.; Bondarev, S.A.; Velizhanina, M.E.; Barbitoff, Y.A.; Matveenko, A.G.; Valina, A.A.; Simanova, A.L.; Zhouravleva, G.A.; Galkin, A.P. Direct proof of the amyloid nature of yeast prions [PSI+] and [PIN+] by the method of immunoprecipitation of native fibrils. FEMS Yeast Res. 2021, 21, foab046. [Google Scholar] [CrossRef] [PubMed]
- Stansfield, I.; Jones, K.M.; Kushnirov, V.V.; Dagkesamanskaya, A.R.; Poznyakovski, A.I.; Paushkin, S.V.; Nierras, C.R.; Cox, B.S.; Ter-Avanesyan, M.D.; Tuite, M.F. The products of the SUP45 (eRF1) and SUP35 genes interact to mediate translation termination in Saccharomyces cerevisiae. EMBO J. 1995, 14, 4365–4373. [Google Scholar] [CrossRef] [PubMed]
- Zhouravleva, G.; Frolova, L.; Le Goff, X.; Le Guellec, R.; Inge-Vechtomov, S.; Kisselev, L.; Philippe, M. Termination of translation in eukaryotes is governed by two interacting polypeptide chain release factors, eRF1 and eRF3. EMBO J. 1995, 14, 4065–4072. [Google Scholar] [CrossRef] [PubMed]
- Serio, T.R.; Cashikar, A.G.; Kowal, A.S.; Sawicki, G.J.; Moslehi, J.J.; Serpell, L.; Arnsdorf, M.F.; Lindquist, S.L. Nucleated conformational conversion and the replication of conformational information by a prion determinant. Science 2000, 289, 1317–1321. [Google Scholar] [CrossRef]
- Lacroute, F. Non-Mendelian mutation allowing ureidosuccinic acid uptake in yeast. J. Bacteriol. 1971, 106, 519–522. [Google Scholar] [CrossRef]
- Derkatch, I.L.; Bradley, M.E.; Hong, J.Y.; Liebman, S.W. Prions affect the appearance of other prions: The story of [PIN+]. Cell 2001, 106, 171–182. [Google Scholar] [CrossRef]
- Du, Z.; Park, K.w.; Yu, H.; Fan, Q.; Li, L. Newly identified prion linked to the chromatin-remodeling factor Swi1 in Saccharomyces cerevisiae. Nat. Genet. 2008, 40, 460–465. [Google Scholar] [CrossRef]
- Nizhnikov, A.A.; Ryzhova, T.A.; Volkov, K.V.; Zadorsky, S.P.; Sopova, J.V.; Inge-Vechtomov, S.G.; Galkin, A.P. Interaction of prions causes heritable traits in Saccharomyces cerevisiae. PLoS Genet. 2016, 12, e1006504. [Google Scholar] [CrossRef]
- Wickner, R.B.; Shewmaker, F.P.; Bateman, D.a.; Edskes, H.K.; Gorkovskiy, A.; Dayani, Y.; Bezsonov, E.E. Yeast prions: Structure, biology, and prion-handling systems. Microbiol. Mol. Biol. Rev. 2015, 79, 1–17. [Google Scholar] [CrossRef]
- Kushnirov, V.V.; Dergalev, A.A.; Alexandrov, A.I. Amyloid fragmentation and disaggregation in yeast and animals. Biomolecules 2021, 11, 1884. [Google Scholar] [CrossRef]
- Tuite, M.F. Yeast prions: Paramutation at the protein level? Semin. Cell Dev. Biol. 2015, 44, 51–61. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, M.; Chien, P.; Naber, N.; Cooke, R.; Weissman, J.S. Conformational variations in an infectious protein determine prion strain differences. Nature 2004, 428, 323–328. [Google Scholar] [CrossRef] [PubMed]
- Drozdova, P.B.; Barbitoff, Y.A.; Belousov, M.V.; Skitchenko, R.K.; Rogoza, T.M.; Leclercq, J.Y.; Kajava, A.V.; Matveenko, A.G.; Zhouravleva, G.A.; Bondarev, S.A. Estimation of amyloid aggregate sizes with semi-denaturing detergent agarose gel electrophoresis and its limitations. Prion 2020, 14, 118–128. [Google Scholar] [CrossRef] [PubMed]
- Inoue, Y. Life cycle of yeast prions: Propagation mediated by amyloid fibrils. Protein Pept. Lett. 2009, 16, 271–276. [Google Scholar] [CrossRef] [PubMed]
- Cox, B.; Tuite, M. The life of [PSI]. Curr. Genet. 2018, 64, 1–8. [Google Scholar] [CrossRef]
- Shorter, J.; Lindquist, S. Destruction or potentiation of different prions catalyzed by similar Hsp104 remodeling activities. Mol. Cell 2006, 23, 425–438. [Google Scholar] [CrossRef]
- Krzewska, J.; Melki, R. Molecular chaperones and the assembly of the prion Sup35p, an in vitro study. EMBO J. 2006, 25, 822–833. [Google Scholar] [CrossRef]
- Savistchenko, J.; Krzewska, J.; Fay, N.; Melki, R. Molecular chaperones and the assembly of the prion Ure2p in vitro. J. Biol. Chem. 2008, 283, 15732–15739. [Google Scholar] [CrossRef]
- Ness, F.; Cox, B.S.; Wongwigkarn, J.; Naeimi, W.R.; Tuite, M.F. Over-expression of the molecular chaperone Hsp104 in Saccharomyces cerevisiae results in the malpartitioning of [PSI+] propagons. Mol. Microbiol. 2017, 104, 125–142. [Google Scholar] [CrossRef]
- Howie, R.L.; Jay-Garcia, L.M.; Kiktev, D.A.; Faber, Q.L.; Murphy, M.; Rees, K.A.; Sachwani, N.; Chernoff, Y.O. Role of the cell asymmetry apparatus and ribosome-associated chaperones in the destabilization of a Saccharomyces cerevisiae prion by heat shock. Genetics 2019, 212, 757–771. [Google Scholar] [CrossRef]
- Drozdova, P.B.; Matveenko, A.G. Prions in yeast: Thinking outside of the brain. In The Prion Phenomena in Neurodegenerative Diseases: New Frontiers in Neuroscience; Legname, G., Giachin, G., Eds.; Nova Science Publishers: Hauppauge, NY, USA, 2015; Chapter 10; pp. 209–236. [Google Scholar]
- Hung, G.C.; Masison, D.C. N-Terminal domain of yeast Hsp104 chaperone is dispensable for thermotolerance and prion propagation but necessary for curing prions by Hsp104 overexpression. Genetics 2006, 173, 611–620. [Google Scholar] [CrossRef] [PubMed]
- Gong, W.; Hu, W.; Xu, L.; Wu, H.; Wu, S.; Zhang, H.; Wang, J.; Jones, G.W.; Perrett, S. The C-terminal GGAP motif of Hsp70 mediates substrate recognition and stress response in yeast. J. Biol. Chem. 2018, 293, 17663–17675. [Google Scholar] [CrossRef] [PubMed]
- Kirkland, P.A.; Reidy, M.; Masison, D.C. Functions of yeast Hsp40 chaperone Sis1p dispensable for prion propagation but important for prion curing and protection from prion toxicity. Genetics 2011, 188, 565–577. [Google Scholar] [CrossRef] [PubMed]
- Borges, J.C.; Seraphim, T.V.; Mokry, D.Z.; Almeida, F.C.; Cyr, D.M.; Ramos, C.H. Identification of regions involved in substrate binding and dimer stabilization within the central domains of yeast Hsp40 Sis1. PLoS ONE 2012, 7, e50927. [Google Scholar] [CrossRef] [PubMed]
- Glover, J.R.; Lindquist, S. Hsp104, Hsp70, and Hsp40: A novel chaperone system that rescues previously aggregated proteins. Cell 1998, 94, 73–82. [Google Scholar] [CrossRef]
- Shorter, J.; Lindquist, S. Hsp104 catalyzes formation and elimination of self-replicating Sup35 prion conformers. Science 2004, 304, 1793–1797. [Google Scholar] [CrossRef]
- Levkovich, S.A.; Rencus-Lazar, S.; Gazit, E.; Laor Bar-Yosef, D. Microbial Prions: Dawn of a New Era. Trends Biochem. Sci. 2021, 46, 391–405. [Google Scholar] [CrossRef]
- Chernoff, Y.O.; Lindquist, S.L.; Ono, B.; Inge-Vechtomov, S.G.; Liebman, S.W. Role of the chaperone protein Hsp104 in propagation of the yeast prion-like factor [psi+]. Science 1995, 268, 880–884. [Google Scholar] [CrossRef]
- Wegrzyn, R.D.; Bapat, K.; Newnam, G.P.; Zink, a.D.; Chernoff, Y.O. Mechanism of prion loss after Hsp104 inactivation in yeast. Mol. Cell. Biol. 2001, 21, 4656–4669. [Google Scholar] [CrossRef]
- Moriyama, H.; Edskes, H.K.; Wickner, R.B. [URE3] prion propagation in Saccharomyces cerevisiae: Requirement for chaperone Hsp104 and curing by overexpressed chaperone Ydj1p. Mol. Cell. Biol. 2000, 20, 8916–8922. [Google Scholar] [CrossRef]
- Alberti, S.; Halfmann, R.; King, O.; Kapila, A.; Lindquist, S. A systematic survey identifies prions and illuminates sequence features of prionogenic proteins. Cell 2009, 137, 146–158. [Google Scholar] [CrossRef] [PubMed]
- Kushnirov, V.V.; Ter-Avanesyan, M.D. Structure and replication of yeast prions. Cell 1998, 94, 13–16. [Google Scholar] [CrossRef]
- Winkler, J.; Tyedmers, J.; Bukau, B.; Mogk, A. Hsp70 targets Hsp100 chaperones to substrates for protein disaggregation and prion fragmentation. J. Cell Biol. 2012, 198, 387–404. [Google Scholar] [CrossRef] [PubMed]
- Chernova, T.A.; Wilkinson, K.D.; Chernoff, Y.O. Prions, chaperones, and proteostasis in yeast. Cold Spring Harb. Perspect. Biol. 2017, 9, a023663. [Google Scholar] [CrossRef]
- Duran, E.C.; Weaver, C.L.; Lucius, A.L. Comparative analysis of the structure and function of AAA+ motors ClpA, ClpB, and Hsp104: Common threads and disparate functions. Front. Mol. Biosci. 2017, 4, 54. [Google Scholar] [CrossRef]
- Shorter, J.; Southworth, D.R. Spiraling in control: Structures and mechanisms of the Hsp104 disaggregase. Cold Spring Harb. Perspect. Biol. 2019, 11, a034033. [Google Scholar] [CrossRef]
- Gates, S.N.; Yokom, A.L.; Lin, J.B.; Jackrel, M.E.; Rizo, A.N.; Kendsersky, N.M.; Buell, C.E.; Sweeny, E.A.; Mack, K.L.; Chuang, E.; et al. Ratchet-like polypeptide translocation mechanism of the AAA+ disaggregase Hsp104. Science 2017, 357, 273–279. [Google Scholar] [CrossRef]
- Deville, C.; Carroni, M.; Franke, K.B.; Topf, M.; Bukau, B.; Mogk, A.; Saibil, H.R. Structural pathway of regulated substrate transfer and threading through an Hsp100 disaggregase. Sci. Adv. 2017, 3, e1701726. [Google Scholar] [CrossRef]
- Lee, J.; Sung, N.; Mercado, J.M.; Hryc, C.F.; Chang, C.; Lee, S.; Tsai, F.T. Overlapping and specific functions of the Hsp104 N-domain define its role in protein disaggregation. Sci. Rep. 2017, 7, 11184. [Google Scholar] [CrossRef]
- Helsen, C.W.; Glover, J.R. Insight into molecular basis of curing of [PSI+] prion by overexpression of 104-kDa heat shock protein (Hsp104). J. Biol. Chem. 2012, 287, 542–556. [Google Scholar] [CrossRef]
- Matveenko, A.G.; Barbitoff, Y.A.; Jay-Garcia, L.M.; Chernoff, Y.O.; Zhouravleva, G.A. Differential effects of chaperones on yeast prions: CURrent view. Curr. Genet. 2018, 64, 317–325. [Google Scholar] [CrossRef] [PubMed]
- Sharma, D.; Masison, D.C. Functionally redundant isoforms of a yeast Hsp70 chaperone subfamily have different antiprion effects. Genetics 2008, 179, 1301–1311. [Google Scholar] [CrossRef] [PubMed]
- Park, Y.N.; Morales, D.; Rubinson, E.H.; Masison, D.; Eisenberg, E.; Greene, L.E. Differences in the curing of [PSI+] prion by various methods of Hsp104 inactivation. PLoS ONE 2012, 7, e37692. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Gorkovskiy, A.; Reidy, M.; Masison, D.C.; Wickner, R.B. Hsp104 disaggregase at normal levels cures many [PSI+] prion variants in a process promoted by Sti1p, Hsp90, and Sis1p. Proc. Natl. Acad. Sci. USA 2017, 114, E4193–E4202. [Google Scholar] [CrossRef]
- Mayer, M.P.; Gierasch, L.M. Recent advances in the structural and mechanistic aspects of Hsp70 molecular chaperones. J. Biol. Chem. 2019, 294, 2085–2097. [Google Scholar] [CrossRef]
- Kampinga, H.H.; Craig, E.A. The HSP70 chaperone machinery: J proteins as drivers of functional specificity. Nat. Rev. Mol. Cell Biol. 2010, 11, 579–592. [Google Scholar] [CrossRef]
- Craig, E.A.; Marszalek, J. How do J-proteins get Hsp70 to do so many different things? Trends Biochem. Sci. 2017, 42, 355–368. [Google Scholar] [CrossRef]
- Zhu, X.; Zhao, X.; Burkholder, W.F.; Gragerov, A.; Ogata, C.M.; Gottesman, M.E.; Hendrickson, W.A. Structural analysis of substrate binding by the molecular chaperone DnaK. Science 1996, 272, 1606–1614. [Google Scholar] [CrossRef]
- Boorstein, W.R.; Ziegelhoffer, T.; Craig, E.A. Molecular evolution of the HSP70 multigene family. J. Mol. Evol. 1994, 38, 1–17. [Google Scholar] [CrossRef]
- Lotz, S.K.; Knighton, L.E.; Nitika; Jones, G.W.; Truman, A.W. Not quite the SSAme: Unique roles for the yeast cytosolic Hsp70s. Curr. Genet. 2019, 65, 1127–1134. [Google Scholar] [CrossRef]
- Nelson, R.J.; Ziegelhoffer, T.; Nicolet, C.; Werner-Washburne, M.; Craig, E.A. The translation machinery and 70 kd heat shock protein cooperate in protein synthesis. Cell 1992, 71, 97–105. [Google Scholar] [CrossRef]
- Newnam, G.P.; Wegrzyn, R.D.; Lindquist, S.L.; Chernoff, Y.O. Antagonistic interactions between yeast chaperones Hsp104 and Hsp70 in prion curing. Mol. Cell. Biol. 1999, 19, 1325–1333. [Google Scholar] [CrossRef] [PubMed]
- Jung, G.; Jones, G.; Wegrzyn, R.D.; Masison, D.C. A role for cytosolic hsp70 in yeast [PSI+] prion propagation and [PSI+] as a cellular stress. Genetics 2000, 156, 559–570. [Google Scholar] [CrossRef] [PubMed]
- Jones, G.W.; Masison, D.C. Saccharomyces cerevisiae Hsp70 mutations affect [PSI+] prion propagation and cell growth differently and implicate Hsp40 and tetratricopeptide repeat cochaperones in impairment of [PSI+]. Genetics 2003, 163, 495–506. [Google Scholar] [CrossRef]
- Needham, P.G.; Masison, D.C. Prion-impairing mutations in Hsp70 chaperone Ssa1: Effects on ATPase and chaperone activities. Arch. Biochem. Biophys. 2008, 478, 167–174. [Google Scholar] [CrossRef]
- Chernoff, Y.O.; Newnam, G.P.; Kumar, J.; Allen, K.; Zink, A.D. Evidence for a protein mutator in yeast: Role of the Hsp70-related chaperone ssb in formation, stability, and toxicity of the [PSI] prion. Mol. Cell. Biol. 1999, 19, 8103–8112. [Google Scholar] [CrossRef]
- Kushnirov, V.V.; Kryndushkin, D.S.; Boguta, M.; Smirnov, V.N.; Ter-Avanesyan, M.D. Chaperones that cure yeast artificial [PSI+] and their prion-specific effects. Curr. Biol. 2000, 10, 1443–1446. [Google Scholar] [CrossRef]
- Allen, K.D.; Wegrzyn, R.D.; Chernova, T.A.; Müller, S.; Newnam, G.P.; Winslett, P.A.; Wittich, K.B.; Wilkinson, K.D.; Chernoff, Y.O. Hsp70 chaperones as modulators of prion life cycle: Novel effects of Ssa and Ssb on the Saccharomyces cerevisiae prion [PSI+]. Genetics 2005, 169, 1227–1242. [Google Scholar] [CrossRef]
- Kiktev, D.A.; Melomed, M.M.; Lu, C.D.; Newnam, G.P.; Chernoff, Y.O. Feedback control of prion formation and propagation by the ribosome-associated chaperone complex. Mol. Microbiol. 2015, 96, 621–632. [Google Scholar] [CrossRef]
- Walsh, P.; Bursać, D.; Law, Y.C.; Cyr, D.; Lithgow, T. The J-protein family: Modulating protein assembly, disassembly and translocation. EMBO Rep. 2004, 5, 567–571. [Google Scholar] [CrossRef]
- Cyr, D.M.; Lu, X.; Douglas, M.G. Regulation of Hsp70 function by a eukaryotic DnaJ homolog. J. Biol. Chem. 1992, 267, 20927–20931. [Google Scholar] [CrossRef]
- Li, J.; Wu, Y.; Qian, X.; Sha, B. Crystal structure of yeast Sis1 peptide-binding fragment and Hsp70 Ssa1 C-terminal complex. Biochem. J. 2006, 398, 353–360. [Google Scholar] [CrossRef]
- Ghaemmaghami, S.; Huh, W.K.; Bower, K.; Howson, R.W.; Belle, A.; Dephoure, N.; O’Shea, E.K.; Weissman, J.S. Global analysis of protein expression in yeast. Nature 2003, 425, 737–741. [Google Scholar] [CrossRef]
- Sondheimer, N.; Lopez, N.; Craig, E.A.; Lindquist, S. The role of Sis1 in the maintenance of the [RNQ+] prion. EMBO J. 2001, 20, 2435–2442. [Google Scholar] [CrossRef]
- Higurashi, T.; Hines, J.K.; Sahi, C.; Aron, R.; Craig, E.A. Specificity of the J-protein Sis1 in the propagation of 3 yeast prions. Proc. Natl. Acad. Sci. USA 2008, 105, 16596–16601. [Google Scholar] [CrossRef]
- Tipton, K.A.; Verges, K.J.; Weissman, J.S. In vivo monitoring of the prion replication cycle reveals a critical role for Sis1 in delivering substrates to Hsp104. Mol. Cell 2008, 32, 584–591. [Google Scholar] [CrossRef]
- Harris, J.M.; Nguyen, P.P.; Patel, M.J.; Sporn, Z.a.; Hines, J.K. Functional diversification of Hsp40: Distinct J-protein functional requirements for two prions allow for chaperone-dependent prion selection. PLoS Genet. 2014, 10, e1004510. [Google Scholar] [CrossRef][Green Version]
- Reidy, M.; Sharma, R.; Shastry, S.; Roberts, B.L.; Albino-Flores, I.; Wickner, S.; Masison, D.C. Hsp40s specify functions of Hsp104 and Hsp90 protein chaperone machines. PLoS Genet. 2014, 10, e1004720. [Google Scholar] [CrossRef]
- Hines, J.K.; Li, X.; Du, Z.; Higurashi, T.; Li, L.; Craig, E.A. [SWI+], the prion formed by the chromatin remodeling factor Swi1, is highly sensitive to alterations in Hsp70 chaperone system activity. PLoS Genet. 2011, 7, e1001309. [Google Scholar] [CrossRef]
- Killian, A.N.; Miller, S.C.; Hines, J.K. Impact of amyloid polymorphism on prion-chaperone interactions in yeast. Viruses 2019, 11, 349. [Google Scholar] [CrossRef]
- Scior, A.; Buntru, A.; Arnsburg, K.; Ast, A.; Iburg, M.; Juenemann, K.; Pigazzini, M.L.; Mlody, B.; Puchkov, D.; Priller, J.; et al. Complete suppression of Htt fibrilization and disaggregation of Htt fibrils by a trimeric chaperone complex. EMBO J. 2018, 37, 282–299. [Google Scholar] [CrossRef] [PubMed]
- Kryndushkin, D.S.; Engel, A.; Edskes, H.; Wickner, R.B. Molecular chaperone Hsp104 can promote yeast prion generation. Genetics 2011, 188, 339–348. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Astor, M.T.; Kamiya, E.; Sporn, Z.A.; Berger, S.E.; Hines, J.K. Variant-specific and reciprocal Hsp40 functions in Hsp104-mediated prion elimination. Mol. Microbiol. 2018, 109, 41–62. [Google Scholar] [CrossRef] [PubMed]
- Reidy, M.; Masison, D.C. Sti1 regulation of Hsp70 and Hsp90 is critical for curing of Saccharomyces cerevisiae [PSI+] prions by Hsp104. Mol. Cell. Biol. 2010, 30, 3542–3552. [Google Scholar] [CrossRef] [PubMed]
- Moosavi, B.; Wongwigkarn, J.; Tuite, M.F. Hsp70/Hsp90 co-chaperones are required for efficient Hsp104-mediated elimination of the yeast [PSI+] prion but not for prion propagation. Yeast 2010, 27, 167–179. [Google Scholar] [CrossRef]
- Jones, G.; Song, Y.; Chung, S.; Masison, D.C. Propagation of Saccharomyces cerevisiae [PSI+] prion is impaired by factors that regulate Hsp70 substrate binding. Mol. Cell. Biol. 2004, 24, 3928–3937. [Google Scholar] [CrossRef][Green Version]
- Kryndushkin, D.S.; Shewmaker, F.; Wickner, R.B. Curing of the [URE3] prion by Btn2p, a Batten disease-related protein. EMBO J. 2008, 27, 2725–2735. [Google Scholar] [CrossRef]
- Malinovska, L.; Kroschwald, S.; Munder, M.C.; Richter, D.; Alberti, S. Molecular chaperones and stress-inducible protein-sorting factors coordinate the spatiotemporal distribution of protein aggregates. Mol. Biol. Cell 2012, 23, 3041–3056. [Google Scholar] [CrossRef]
- Barbitoff, Y.A.; Matveenko, A.G.; Moskalenko, S.E.; Zemlyanko, O.M.; Newnam, G.P.; Patel, A.; Chernova, T.A.; Chernoff, Y.O.; Zhouravleva, G.A. To CURe or not to CURe? Differential effects of the chaperone sorting factor Cur1 on yeast prions are mediated by the chaperone Sis1. Mol. Microbiol. 2017, 105, 242–257. [Google Scholar] [CrossRef]
- Wickner, R.B.; Kelly, A.C.; Bezsonov, E.E.; Edskes, H.K. [PSI+] prion propagation is controlled by inositol polyphosphates. Proc. Natl. Acad. Sci. USA 2017, 114, E8402–E8410. [Google Scholar] [CrossRef]
- Edskes, H.K.; Stroobant, E.E.; DeWilde, M.P.; Bezsonov, E.E.; Wickner, R.B. Proteasome control of [URE3] prion propagation by degradation of anti-prion proteins Cur1 and Btn2 in Saccharomyces cerevisiae. Genetics 2021, 218, iyab037. [Google Scholar] [CrossRef] [PubMed]
- Sondheimer, N.; Lindquist, S. Rnq1: An epigenetic modifier of protein function in yeast. Mol. Cell 2000, 5, 163–172. [Google Scholar] [CrossRef]
- Schwimmer, C.; Masison, D.C. Antagonistic interactions between yeast [PSI+] and [URE3] prions and curing of [URE3] by Hsp70 protein chaperone Ssa1p but not by Ssa2p. Mol. Cell. Biol. 2002, 22, 3590–3598. [Google Scholar] [CrossRef]
- Stein, K.C.; True, H.L. Structural variants of yeast prions show conformer-specific requirements for chaperone activity. Mol. Microbiol. 2014, 93, 1156–1171. [Google Scholar] [CrossRef]
- Kryndushkin, D.S.; Smirnov, V.N.; Ter-Avanesyan, M.D.; Kushnirov, V.V. Increased expression of Hsp40 chaperones, transcriptional factors, and ribosomal protein Rpp0 can cure yeast prions. J. Biol. Chem. 2002, 277, 23702–23708. [Google Scholar] [CrossRef]
- Sharma, D.; Stanley, R.F.; Masison, D.C. Curing of yeast [URE3] prion by the Hsp40 cochaperone Ydj1p is mediated by Hsp70. Genetics 2009, 181, 129–137. [Google Scholar] [CrossRef]
- Aron, R.; Higurashi, T.; Sahi, C.; Craig, E.A. J-protein co-chaperone Sis1 required for generation of [RNQ+] seeds necessary for prion propagation. EMBO J. 2007, 26, 3794–3803. [Google Scholar] [CrossRef]
- Zhao, X.; Lanz, J.; Steinberg, D.; Pease, T.; Ahearn, J.M.; Bezsonov, E.E.; Staguhn, E.D.; Eisenberg, E.; Masison, D.C.; Greene, L.E. Real-time imaging of yeast cells reveals several distinct mechanisms of curing of the [URE3] prion. J. Biol. Chem. 2018, 293, 3104–3117. [Google Scholar] [CrossRef]
- Troisi, E.M.; Rockman, M.E.; Nguyen, P.P.; Oliver, E.E.; Hines, J.K. Swa2, the yeast homolog of mammalian auxilin, is specifically required for the propagation of the prion variant [URE3-1]. Mol. Microbiol. 2015, 97, 926–941. [Google Scholar] [CrossRef]
- Lian, H.Y.; Zhang, H.; Zhang, Z.R.; Loovers, H.M.; Jones, G.W.; Rowling, P.J.; Itzhaki, L.S.; Zhou, J.M.; Perrett, S. Hsp40 interacts directly with the native state of the yeast prion protein Ure2 and inhibits formation of amyloid-like fibrils. J. Biol. Chem. 2007, 282, 11931–11940. [Google Scholar] [CrossRef]
- Amor, A.J.; Castanzo, D.T.; Delany, S.P.; Selechnik, D.M.; van Ooy, A.; Cameron, D.M. The ribosome-associated complex antagonizes prion formation in yeast. Prion 2015, 9, 144–164. [Google Scholar] [CrossRef] [PubMed]
- Wickner, R.B.; Bezsonov, E.; Bateman, D.A. Normal levels of the antiprion proteins Btn2 and Cur1 cure most newly formed [URE3] prion variants. Proc. Natl. Acad. Sci. USA 2014, 111, E2711–E2720. [Google Scholar] [CrossRef] [PubMed]
- Duennwald, M.L.; Echeverria, A.; Shorter, J. Small heat shock proteins potentiate amyloid dissolution by protein disaggregases from yeast and humans. PLoS Biol. 2012, 10, e1001346. [Google Scholar] [CrossRef] [PubMed]
- Reidy, M.; Miot, M.; Masison, D.C. Prokaryotic chaperones support yeast prions and thermotolerance and define disaggregation machinery interactions. Genetics 2012, 192, 185–193. [Google Scholar] [CrossRef] [PubMed]
- Zhao, X.; Rodriguez, R.; Silberman, R.E.; Ahearn, J.M.; Saidha, S.; Cummins, K.C.; Eisenberg, E.; Greene, L.E. Heat shock protein 104 (Hsp104)-mediated curing of [PSI+] yeast prions depends on both [PSI+] conformation and the properties of the Hsp104 homologs. J. Biol. Chem. 2017, 292, 8630–8641. [Google Scholar] [CrossRef] [PubMed]
- Sharma, D.; Masison, D.C. Single methyl group determines prion propagation and protein degradation activities of yeast heat shock protein (Hsp)-70 chaperones Ssa1p and Ssa2p. Proc. Natl. Acad. Sci. USA 2011, 108, 13665–13670. [Google Scholar] [CrossRef]
- Sharma, D.; Martineau, C.N.; Le Dall, M.T.; Reidy, M.; Masison, D.C.; Kabani, M. Function of SSA subfamily of Hsp70 within and across species varies widely in complementing Saccharomyces cerevisiae cell growth and prion propagation. PLoS ONE 2009, 4, e6644. [Google Scholar] [CrossRef]
- Yu, C.i.; King, C.y. Forms and abundance of chaperone proteins influence yeast prion variant competition. Mol. Microbiol. 2019, 111, 798–810. [Google Scholar] [CrossRef]
- James, P.; Pfund, C.; Craig, E.A. Functional specificity among Hsp70 molecular chaperones. Science 1997, 275, 387–389. [Google Scholar] [CrossRef]
- Eaglestone, S.S.; Ruddock, L.W.; Cox, B.S.; Tuite, M.F. Guanidine hydrochloride blocks a critical step in the propagation of the prion-like determinant [PSI+] of Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 2000, 97, 240–244. [Google Scholar] [CrossRef]
- Lee, S.; Fan, C.Y.; Michael Younger, J.; Ren, H. Identification of essential residues in the type II Hsp40 Sis1 that function in polypeptide binding. J. Biol. Chem. 2002, 277, 21675–21682. [Google Scholar] [CrossRef] [PubMed]
- Kumar, J.; Reidy, M.; Masison, D.C. Yeast J-protein Sis1 prevents prion toxicity by moderating depletion of prion protein. Genetics 2021, 219, iyab129. [Google Scholar] [CrossRef]
- Oliver, E.E.; Troisi, E.M.; Hines, J.K. Prion-specific Hsp40 function: The role of the auxilin homolog Swa2. Prion 2017, 11, 174–185. [Google Scholar] [CrossRef]
- Berger, S.E.; Nolte, A.M.; Kamiya, E.; Hines, J.K. Three J-proteins impact Hsp104-mediated variant-specific prion elimination: A new critical role for a low-complexity domain. Curr. Genet. 2020, 66, 51–58. [Google Scholar] [CrossRef]
- Faust, O.; Abayev-Avraham, M.; Wentink, A.S.; Maurer, M.; Nillegoda, N.B.; London, N.; Bukau, B.; Rosenzweig, R. HSP40 proteins use class-specific regulation to drive HSP70 functional diversity. Nature 2020, 587, 489–494. [Google Scholar] [CrossRef]
- Kumar, S.; Dine, E.A.; Paddock, E.; Steinberg, D.N.; Greene, L.E.; Masison, D.C. Mutations outside the Ure2 amyloid-forming region disrupt [URE3] prion propagation and alter interactions with protein quality control factors. Mol. Cell. Biol. 2020, 40, e00294-20. [Google Scholar] [CrossRef]
- Wickner, R.B. Anti-prion systems in yeast. J. Biol. Chem. 2019, 294, 1729–1738. [Google Scholar] [CrossRef]
- Wickner, R.B.; Edskes, H.K.; Son, M.; Wu, S.; Niznikiewicz, M. Innate immunity to prions: Anti-prion systems turn a tsunami of prions into a slow drip. Curr. Genet. 2021, 67, 833–847. [Google Scholar] [CrossRef]
- Shorter, J.; Lindquist, S. Hsp104, Hsp70 and Hsp40 interplay regulates formation, growth and elimination of Sup35 prions. EMBO J. 2008, 27, 2712–2724. [Google Scholar] [CrossRef]
- Park, S.H.; Kukushkin, Y.; Gupta, R.; Chen, T.; Konagai, A.; Hipp, M.; Hayer-Hartl, M.; Hartl, F. PolyQ proteins interfere with nuclear degradation of cytosolic proteins by sequestering the Sis1p chaperone. Cell 2013, 154, 134–145. [Google Scholar] [CrossRef]
- Walters, R.W.; Muhlrad, D.; Garcia, J.; Parker, R. Differential effects of Ydj1 and Sis1 on Hsp70-mediated clearance of stress granules in Saccharomyces cerevisiae. RNA 2015, 21, 1660–1671. [Google Scholar] [CrossRef] [PubMed]
- Gokhale, K.C.; Newnam, G.P.; Sherman, M.Y.; Chernoff, Y.O. Modulation of prion-dependent polyglutamine aggregation and toxicity by chaperone proteins in the yeast model. J. Biol. Chem. 2005, 280, 22809–22818. [Google Scholar] [CrossRef] [PubMed]
- Douglas, P.M.; Treusch, S.; Ren, H.Y.; Halfmann, R.; Duennwald, M.L.; Lindquist, S.; Cyr, D.M. Chaperone-dependent amyloid assembly protects cells from prion toxicity. Proc. Natl. Acad. Sci. USA 2008, 105, 7206–7211. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.; Stone, D.E.; Liebman, S.W. Prion-promoted phosphorylation of heterologous amyloid is coupled with ubiquitin-proteasome system inhibition and toxicity. Mol. Microbiol. 2014, 93, 1043–1056. [Google Scholar] [CrossRef]
- Matveenko, A.G.; Drozdova, P.B.; Belousov, M.V.; Moskalenko, S.E.; Bondarev, S.A.; Barbitoff, Y.A.; Nizhnikov, A.A.; Zhouravleva, G.A. SFP1-mediated prion-dependent lethality is caused by increased Sup35 aggregation and alleviated by Sis1. Genes Cells 2016, 21, 1290–1308. [Google Scholar] [CrossRef] [PubMed]
- Klaips, C.L.; Gropp, M.H.M.; Hipp, M.S.; Hartl, F.U. Sis1 potentiates the stress response to protein aggregation and elevated temperature. Nat. Commun. 2020, 11, 6271. [Google Scholar] [CrossRef] [PubMed]
- Wentink, A.S.; Nillegoda, N.B.; Feufel, J.; Ubartaitė, G.; Schneider, C.P.; De Los Rios, P.; Hennig, J.; Barducci, A.; Bukau, B. Molecular dissection of amyloid disaggregation by human HSP70. Nature 2020, 587, 483–488. [Google Scholar] [CrossRef]
- Barbitoff, Y.A.; Matveenko, A.G.; Bondarev, S.A.; Maksiutenko, E.M.; Kulikova, A.V.; Zhouravleva, G.A. Quantitative assessment of chaperone binding to amyloid aggregates identifies specificity of Hsp40 interaction with yeast prion fibrils. FEMS Yeast Res. 2020, 20, foaa025. [Google Scholar] [CrossRef]
- Kroschwald, S.; Maharana, S.; Mateju, D.; Malinovska, L.; Elisabeth, N.; Poser, I.; Richter, D.; Alberti, S. Promiscuous interactions and protein disaggregases determine the material state of stress-inducible RNP granules. eLife 2015, 4, e06807. [Google Scholar] [CrossRef]
- Kroschwald, S.; Munder, M.C.; Maharana, S.; Franzmann, T.M.; Richter, D.; Ruer, M.; Hyman, A.A.; Alberti, S. Different material states of Pub1 condensates define distinct modes of stress adaptation and recovery. Cell Rep. 2018, 23, 3327–3339. [Google Scholar] [CrossRef]
- Saarikangas, J.; Barral, Y. Protein aggregates are associated with replicative aging without compromising protein quality control. eLife 2015, 4, e06197. [Google Scholar] [CrossRef] [PubMed]
- Saarikangas, J.; Caudron, F.; Prasad, R.; Moreno, D.F.; Bolognesi, A.; Aldea, M.; Barral, Y. Compartmentalization of ER-bound chaperone confines protein deposit formation to the aging yeast cell. Curr. Biol. 2017, 27, 773–783. [Google Scholar] [CrossRef] [PubMed]
- Aguilaniu, H.; Gustafsson, L.; Rigoulet, M.; Nyström, T. Asymmetric inheritance of oxidatively damaged proteins during cytokinesis. Science 2003, 299, 1751–1753. [Google Scholar] [CrossRef]
- Tessarz, P.; Schwarz, M.; Mogk, A.; Bukau, B. The yeast AAA+ chaperone Hsp104 is part of a network that links the actin cytoskeleton with the inheritance of damaged proteins. Mol. Cell. Biol. 2009, 29, 3738–3745. [Google Scholar] [CrossRef] [PubMed]
- Liu, B.; Larsson, L.; Caballero, A.; Hao, X.; Öling, D.; Grantham, J.; Nyström, T. The polarisome is required for segregation and retrograde transport of protein aggregates. Cell 2010, 140, 257–267. [Google Scholar] [CrossRef]
- Newnam, G.P.; Birchmore, J.L.; Chernoff, Y.O. Destabilization and recovery of a yeast prion after mild heat shock. J. Mol. Biol. 2011, 408, 432–448. [Google Scholar] [CrossRef]
| Protein | Perturbation | Variant | [PSI+] | [URE3] | [PIN+] | Comment | References |
|---|---|---|---|---|---|---|---|
| Hsp104 | |||||||
| Hsp104 | OE | FL | ↓/none | none | [49,62,63] | ||
| KT * | [49,50] | ||||||
| N | ↑ | n.a. | n.a. | Increases [PSI+] induction rates | [42,65] | ||
| [49,51,103] | |||||||
| Hsp70 | |||||||
| Ssa1 | OE | FL | ↑/none | ↓/none | none | Prevents HSP104 OE curing of [PSI+]. Effects on [URE3-1] curing are not reproducible, the effect might be strain-specific | [62,73,78,104,105] |
| SSA1-21 | /none | none | none | Dominant [PSI+] curing is strain-specific. The effect is more visible in the absence of either SSA1 or SSA2 | [63,74,75,105],† | ||
| Ssa2 | OE | FL | ↑/none | ↓/none | n.a. | Prevents HSP104 OE curing | [62,79,104] |
| Substitution | SSA2-21 | n.a. | [63] | ||||
| Ssa3 | OE | FL | ↑/none | none | n.a. | Prevents HSP104 OE curing | [62,79] |
| Ssa4 | OE | FL | ↑/↓ | none | n.a. | Prevents HSP104 OE curing | [62,79] |
| Ssb1/2 | OE | FL | ↓ | none | n.a. | Enhances HSP104 OE curing | [62,77,78] |
| ↑ | n.a. | n.a. | Increases [PSI+] induction rates | [77,80] | |||
| Hsp40 | |||||||
| Sis1 | OE | FL | ↓/none | none | none | Enhances HSP104 OE curing | [100,106,107] |
| Depletion | FL | ↑/↓ | Effect on [PSI+] depends on depletion method (Tet-Off or relocalization) | [86,87,100,108] | |||
| Substitution | DD | ↓/none | ↓ | none | Prevents HSP104 OE curing | [44,89],† | |
| CTD | ↓ | Prevents HSP104 OE curing | [44,89] | ||||
| JGF | ↓ | Prevents HSP104 OE curing | [44,89] | ||||
| K199A | ↓ | ↓ | n.a. | Prevents HSP104 OE curing | [44,89] | ||
| Ydj1 | OE | FL | ↑/↓/none | none | Strain-dependent effect on [PSI+] | [51,78,100,109] | |
| none | none | none | [75,86,110] | ||||
| Apj1 | OE | FL | none | none | n.a. | Enhances HSP104 OE curing | [94,106,111] |
| none | none | none | [86,110] | ||||
| Zuo1 | ↑ | none | none | Increases [PSI+] induction rates and rates of [PSI+] curing by mild heat shock | [40,80,86,110,112] | ||
| Swa2 | none | ↓ | none | [86,110] | |||
| Protein-sorting factors | |||||||
| Cur1 | OE | FL | ↑/↓ | ↓ | none | Strain-dependent effect on [PSI+] | [98,100,109] |
| OE | 3-22 | n.a. | [62,100] | ||||
| ↓/none | ↑ | n.a. | Increases [URE3] induction rates; slightly weakens strong [PSI+] phenotype and enhances weak [PSI+] curing by mild heat shock | [98,100,113] | |||
| Btn2 | OE | FL | none | none | [98,100,113] | ||
| ↓ | ↑ | n.a. | Increases [URE3] induction rates; enhances weak [PSI+] curing by mild heat shock | [98,100,113] | |||
| Hsp42 | OE | FL | ↑/↓/none | n.a. | [100,113,114] | ||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Barbitoff, Y.A.; Matveenko, A.G.; Zhouravleva, G.A. Differential Interactions of Molecular Chaperones and Yeast Prions. J. Fungi 2022, 8, 122. https://doi.org/10.3390/jof8020122
Barbitoff YA, Matveenko AG, Zhouravleva GA. Differential Interactions of Molecular Chaperones and Yeast Prions. Journal of Fungi. 2022; 8(2):122. https://doi.org/10.3390/jof8020122
Chicago/Turabian StyleBarbitoff, Yury A., Andrew G. Matveenko, and Galina A. Zhouravleva. 2022. "Differential Interactions of Molecular Chaperones and Yeast Prions" Journal of Fungi 8, no. 2: 122. https://doi.org/10.3390/jof8020122
APA StyleBarbitoff, Y. A., Matveenko, A. G., & Zhouravleva, G. A. (2022). Differential Interactions of Molecular Chaperones and Yeast Prions. Journal of Fungi, 8(2), 122. https://doi.org/10.3390/jof8020122







