Purine Auxotrophic Starvation Evokes Phenotype Similar to Stationary Phase Cells in Budding Yeast
Abstract
1. Introduction
2. Materials and Methods
2.1. Strains and Cultivation Conditions
2.2. NMR Analysis of Extracellular Amino Acids and Purines
2.3. Cell Morphology Measurements
2.4. Flow Cytometry
2.5. Fermentation and Metabolite Flux Measurements
2.6. FTIR Analysis
2.7. Cell Carbohydrate Extraction and Quantification
2.8. Transcriptomics
2.9. Sublethal Stresses
3. Results
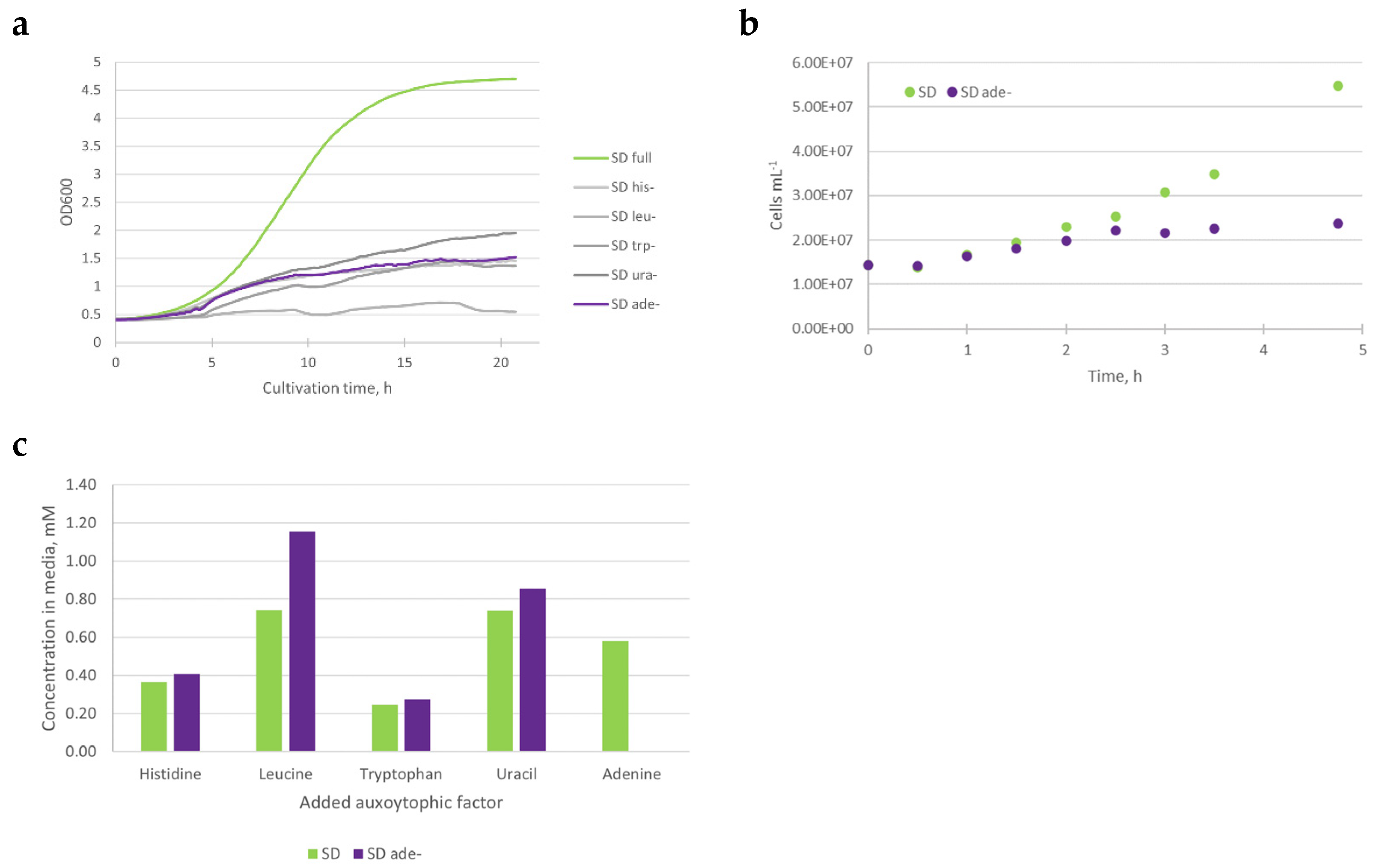
3.1. Growth of CEN.PK2-1D ade8 Ceases in the Absence of Purine
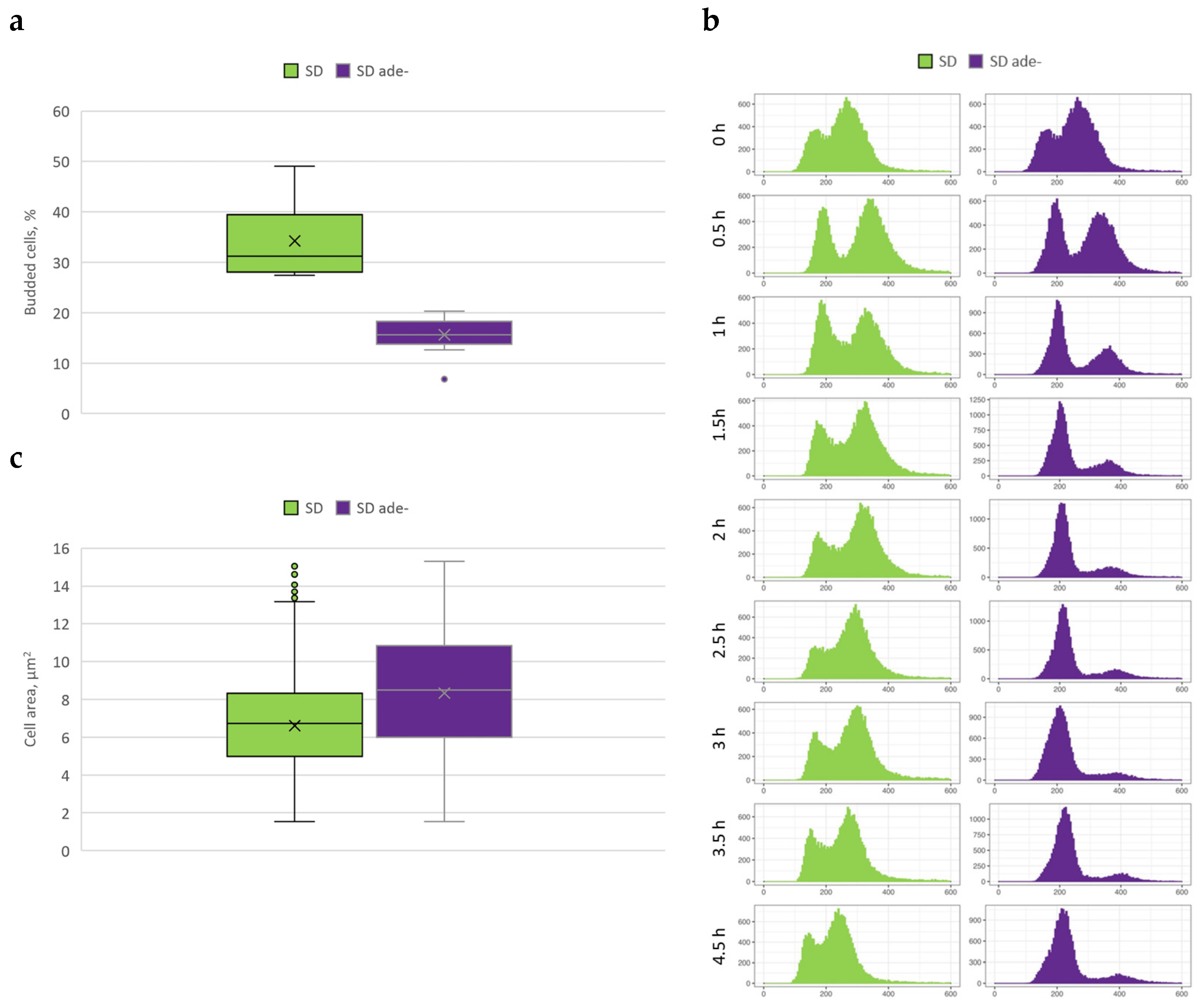
3.2. Cell Cycle Arrests in G1/0 during Purine Starvation
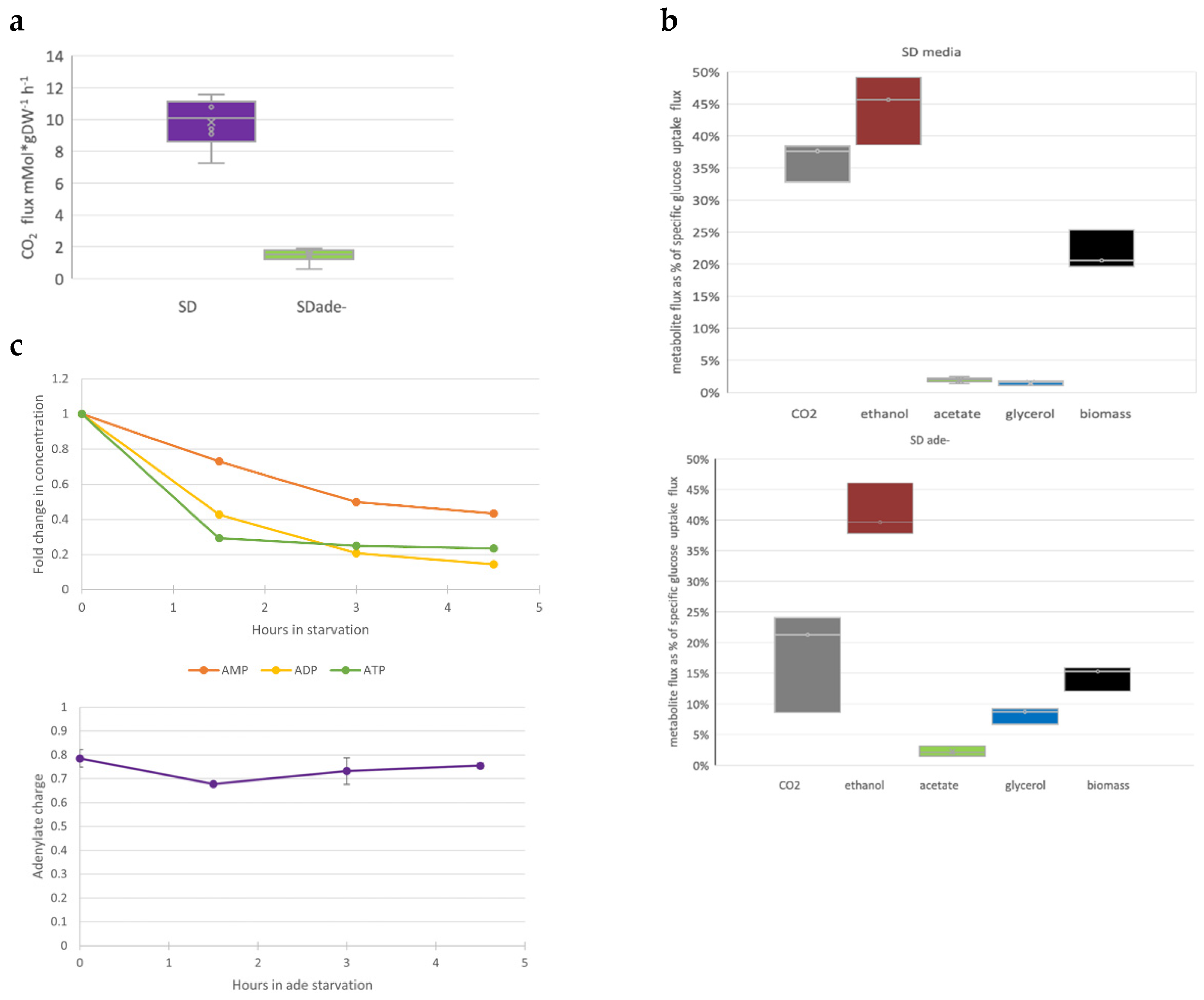
3.3. Purine Starvation Slows Glycolysis
3.4. Purine Starvation Elicits Strong Stress Resilience
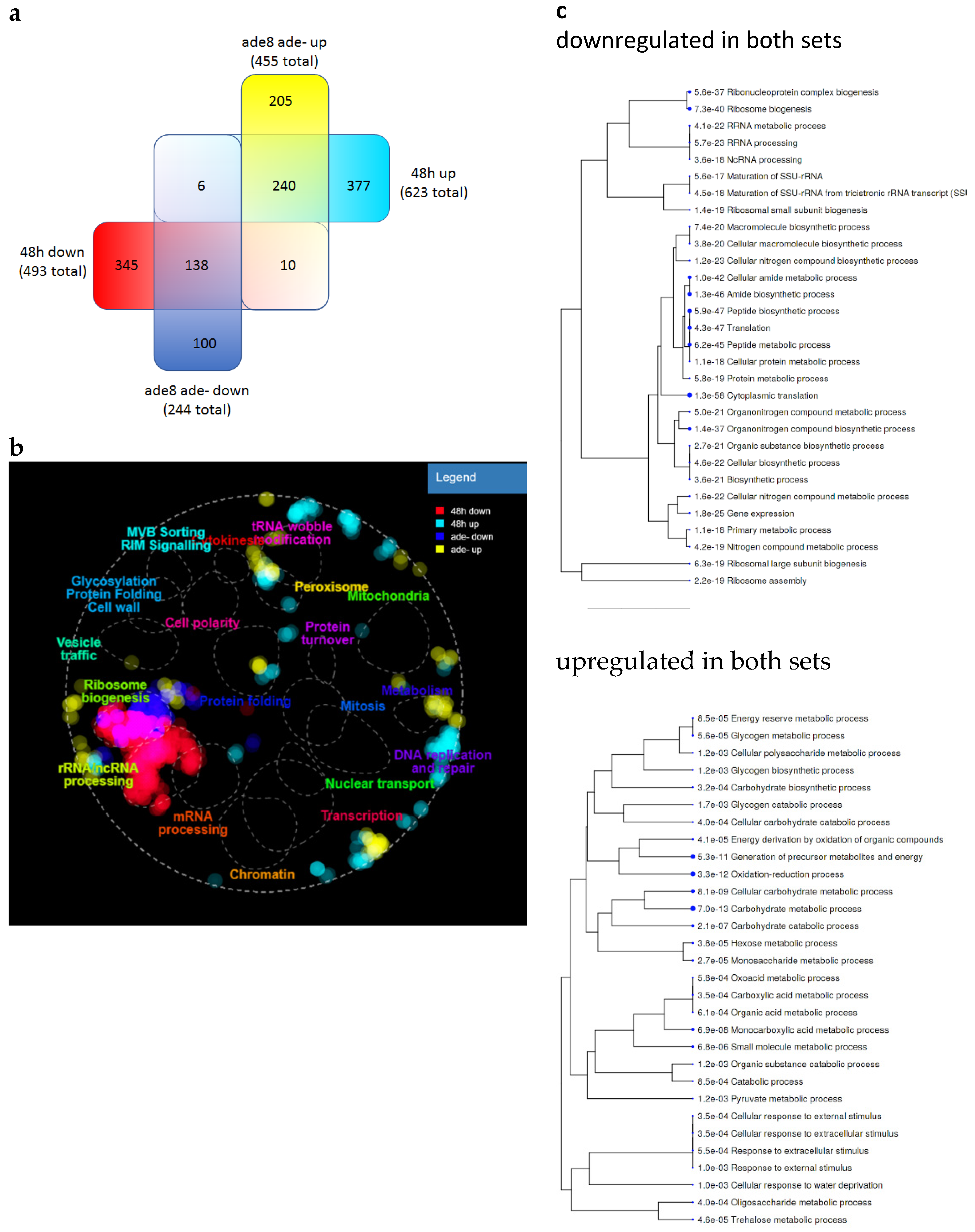
3.5. Purine-Starved Cells Exhibit a Distinct Transcriptome Resembling Stationary Phase Cells
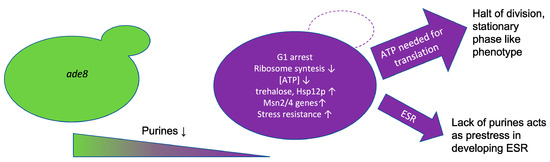
4. Discussion
4.1. Intracellular Adenylate Pool Is Not Sufficient to Sustain Cell Proliferation
4.2. Purine Starvation Induces Accumulation of Metabolites Capable of Increasing Stress Tolerance
4.3. Msn2/4p Are Master Regulators of Purine-Starved Cell Transcriptome
4.4. Intracellular Signalling of Purine Starvation
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Duan, S.F.; Han, P.J.; Wang, Q.M.; Liu, W.-Q.; Shi, J.-Y.; Li, K.; Zhang, X.-L.; Bai, F.-Y. The origin and adaptive evolution of domesticated populations of yeast from Far East Asia. Nat. Commun. 2018, 9, 2690. [Google Scholar] [CrossRef] [PubMed]
- Gasch, A.P.; Spellman, P.T.; Kao, C.M.; Carmel-Harel, O.; Eisen, M.B.; Storz, G.; Botstein, D.; Brown, P.O. Genomic expression programs in the response of yeast cells to environmental changes. Mol. Biol. Cell 2000, 11, 4241–4257. [Google Scholar] [CrossRef]
- Brauer, M.J.; Huttenhower, C.; Airoldi, E.M.; Rosenstein, R.; Matese, J.C.; Gresham, D.; Boer, V.M.; Troyanskaya, O.G.; Botstein, D. Coordination of growth rate, cell cycle, stress response, and metabolic activity in yeast. Mol. Biol. Cell 2008, 19, 352–367. [Google Scholar] [CrossRef]
- Klosinska, M.M.; Crutchfield, C.A.; Bradley, P.H.; Rabinowitz, J.D.; Broach, J.R. Yeast cells can access distinct quiescent states. Genes Dev. 2011, 25, 336–349. [Google Scholar] [CrossRef]
- Galardini, M.; Busby, B.P.; Vieitez, C.; Dunham, A.S.; Typas, A.; Beltrao, P. The impact of the genetic background on gene deletion phenotypes in Saccharomyces cerevisiae. Mol. Syst. Biol. 2019, 15, e8831. [Google Scholar] [CrossRef] [PubMed]
- Pronk, J.T. Auxotrophic yeast strains in fundamental and applied research. Appl. Environ. Microbiol. 2002, 68, 2095–2100. [Google Scholar] [CrossRef]
- Puig-Castellví, F.; Alfonso, I.; Piña, B.; Tauler, R. 1H NMR metabolomic study of auxotrophic starvation in yeast using Multivariate Curve Resolution-Alternating Least Squares for Pathway Analysis. Sci. Rep. 2016, 6, 30982. [Google Scholar] [CrossRef] [PubMed]
- Petti, A.A.; Crutchfield, C.A.; Rabinowitz, J.D.; Botstein, D. Survival of starving yeast is correlated with oxidative stress response and nonrespiratory mitochondrial function. Proc. Natl. Acad. Sci. USA 2011, 108, E1089–E1098. [Google Scholar] [CrossRef]
- Boer, V.M.; Amini, S.; Botstein, D. Influence of genotype and nutrition on survival and metabolism of starving yeast. Proc. Natl. Acad. Sci. USA 2008, 105, 6930–6935. [Google Scholar] [CrossRef]
- Boer, V.M.; Crutchfield, C.A.; Bradley, P.H.; Botstein, D.; Rabinowitz, J.D. Growth-limiting intracellular metabolites in yeast growing under diverse nutrient limitations. Mol. Biol. Cell 2010, 21, 198–211. [Google Scholar] [CrossRef] [PubMed]
- Gomes, P.; Sampaio-Marques, B.; Ludovico, P.; Rodrigues, F.; Leão, C. Low auxotrophy-complementing amino acid concentrations reduce yeast chronological life span. Mech. Ageing Dev. 2007, 128, 383–391. [Google Scholar] [CrossRef] [PubMed]
- Sutter, B.M.; Wu, X.; Laxman, S.; Tu, B.P. Methionine inhibits autophagy and promotes growth by inducing the SAM-responsive methylation of PP2A. Cell 2013, 154, 403–415. [Google Scholar] [CrossRef]
- González, A.; Larroy, C.; Biosca, J.A.; Ariño, J. Use of the TRP1 auxotrophic marker for gene disruption and phenotypic analysis in yeast: A note of warning. FEMS Yeast Res. 2008, 8, 2–5. [Google Scholar] [CrossRef][Green Version]
- Young, M.J.; Court, D.A. Effects of the S288c genetic background and common auxotrophic markers on mitochondrial DNA function in Saccharomyces cerevisiae. Yeast 2008, 25, 903–912. [Google Scholar] [CrossRef] [PubMed]
- Alam, M.T.; Zelezniak, A.; Mülleder, M.; Shliaha, P.; Schwarz, R.; Capuano, F.; Vowinckel, J.; Radmanesfahar, E.; Krüger, A.; Calvani, E.; et al. The metabolic background is a global player in Saccharomyces gene expression epistasis. Nat. Microbiol. 2016, 1, 15030. [Google Scholar] [CrossRef] [PubMed]
- Rébora, K.; Laloo, B.; Daignan-Fornier, B. Revisiting purine-histidine cross-pathway regulation in Saccharomyces cerevisiae: A central role for a small molecule. Genetics 2005, 170, 61–70. [Google Scholar] [CrossRef]
- Kokina, A.; Kibilds, J.; Liepins, J. Adenine auxotrophy—Be aware: Some effects of adenine auxotrophy in Saccharomyces cerevisiae strain W303-1A. FEMS Yeast Res. 2014, 14, 697–707. [Google Scholar] [CrossRef] [PubMed]
- Sadowski, I.; Lourenco, P.; Parent, J. Dominant marker vectors for selecting yeast mating products. Yeast 2008, 25, 595–599. [Google Scholar] [CrossRef]
- Saldanha, A.J.; Brauer, M.J.; Botstein, D. Nutritional homeostasis in batch and steady-state culture of yeast. Mol. Biol. Cell 2004, 15, 4089–4104. [Google Scholar] [CrossRef]
- Sein, H.; Reinmets, K.; Peil, K.; Kristjuhan, K.; Värv, S.; Kristjuhan, A. Rpb9-deficient cells are defective in DNA damage response and require histone H3 acetylation for survival. Sci. Rep. 2018, 8, 2949. [Google Scholar] [CrossRef] [PubMed]
- Valgepea, K.; Adamberg, K.; Nahku, R.; Lahtvee, P.J.; Arike, L.; Vilu, R. Systems biology approach reveals that overflow metabolism of acetate in Escherichia coli is triggered by carbon catabolite repression of acetyl-CoA synthetase. BMC Syst. Biol. 2010, 4, 166. [Google Scholar] [CrossRef] [PubMed]
- Grube, M.; Gapes, J.R.; Schuster, K.C. Application of quantitative IR spectral analysis of bacterial cells to acetone-butanol-ethanol fermentation monitoring. Anal. Chim. Acta 2002, 471, 127–133. [Google Scholar] [CrossRef]
- Stewart, P.R. Chapter 8 analytical methods for yeasts. In Methods in Cell Biology; Prescott, D.M., Ed.; Academic Press: New York, NY, USA, 1975; Volume 12, pp. 111–147. [Google Scholar]
- Dubois, M.; Gilles, K.A.; Hamilton, J.K.; Robers, P.A.; Smith, F. Colorimetric method for determination of sugars and related substances. Anal. Chem. 1956, 28, 350–356. [Google Scholar] [CrossRef]
- Martynova, J.; Kokina, A.; Kibilds, J.; Liepins, J.; Scerbaka, R.; Vigants, A. Effects of acetate on Kluyveromyces marxianus DSM 5422 growth and metabolism. Appl. Microbiol. Biotechnol. 2016, 100, 4585–4594. [Google Scholar] [CrossRef] [PubMed]
- Larsson, C.; Påhlman, I.L.; Gustafsson, L. The importance of ATP as a regulator of glycolytic flux in Saccharomyces cerevisiae. Yeast 2000, 16, 797–809. [Google Scholar] [CrossRef]
- Gasch, A.P.; Werner-Washburne, M. The genomics of yeast responses to environmental stress and starvation. Funct. Integr. Genom. 2002, 2, 181–192. [Google Scholar] [CrossRef]
- Welch, A.Z.; Gibney, P.A.; Botstein, D.; Koshland, D.E. TOR and RAS pathways regulate desiccation tolerance in Saccharomyces cerevisiae. Mol. Biol. Cell 2013, 24, 115–128. [Google Scholar] [CrossRef]
- Ge, S.X.; Jung, D.; Yao, R. ShinyGO: A graphical gene-set enrichment tool for animals and plants. Bioinformatics 2020, 36, 2628–2629. [Google Scholar] [CrossRef]
- Parenteau, J.; Maignon, L.; Berthoumieux, M.; Catala, M.; Gagnon, V.; Abou Elela, S. Introns are mediators of cell response to starvation. Nature 2019, 565, 612–617. [Google Scholar] [CrossRef]
- Hong, K.-K.; Hou, J.; Shoaie, S.; Nielsen, J.; Bordel, S. Dynamic13C-labeling experiments prove important differences in protein turnover rate between two Saccharomyces cerevisiae strains. FEMS Yeast Res. 2012, 12, 741–747. [Google Scholar] [CrossRef]
- Vos, T.; Hakkaart, X.D.; de Hulster, E.A.; van Maris, A.J.; Pronk, J.T.; Daran-Lapujade, P. Maintenance-energy requirements and robustness of Saccharomyces cerevisiae at aerobic near-zero specific growth rates. Microb. Cell Fact. 2016, 15, 111. [Google Scholar] [CrossRef]
- Ljungdahl, P.O.; Daignan-Fornier, B. Regulation of Amino Acid, Nucleotide, and Phosphate Metabolism in Saccharomyces cerevisiae. Genetics 2012, 190, 885–929. [Google Scholar] [CrossRef] [PubMed]
- Von der Haar, T. A quantitative estimation of the global translational activity in logarithmically growing yeast cells. BMC Syst. Biol. 2008, 16, 87. [Google Scholar] [CrossRef]
- Reichert, U.; Winter, M. Uptake and accumulation of purine bases by stationary yeast cells pretreated with glucose. Biochim. Biophys. Acta 1974, 356, 108–116. [Google Scholar] [CrossRef]
- Berry, D.B.; Gasch, A.P. Stress-activated genomic expression changes serve a preparative role for impending stress in yeast. Mol. Biol. Cell 2008, 19, 4580–4587. [Google Scholar] [CrossRef] [PubMed]
- Kitichantaropas, Y.; Boonchird, C.; Sugiyama, M.; Kaneko, Y.; Harashima, S.; Auesukaree, C. Cellular mechanisms contributing to multiple stress tolerance in Saccharomyces cerevisiae strains with potential use in high-temperature ethanol fermentation. AMB Expr. 2016, 6, 107. [Google Scholar] [CrossRef]
- Tapia, H.; Young, L.; Fox, D.; Bertozzi, C.R.; Koshland, D. Increasing intracellular trehalose is sufficient to confer desiccation tolerance to Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 2015, 112, 6122–6127. [Google Scholar] [CrossRef]
- Kim, S.X.; Çamdere, G.; Hu, X.; Koshland, D.; Tapia, H. Synergy between the small intrinsically disordered protein Hsp12 and trehalose sustain viability after severe desiccation. eLife 2018, 7, 38337. [Google Scholar] [CrossRef]
- Lu, C.; Brauer, M.J.; Botstein, D. Slow growth induces heat-shock resistance in normal and respiratory-deficient yeast. Mol. Biol. Cell 2009, 20, 891–903. [Google Scholar] [CrossRef]
- Abdulrehman, D.; Monteiro, P.T.; Teixeira, M.C.; Mira, N.P.; Lourenço, A.B.; dos Santos, S.C.; Cabrito, T.R.; Francisco, A.P.; Madeira, S.C.; Aires, R.S.; et al. YEASTRACT: Providing a programmatic access to curated transcriptional regulatory associations in Saccharomyces cerevisiae through a web services interface. Nucleic Acids Res. 2011, 39, 136–140. [Google Scholar] [CrossRef] [PubMed]
- Ozolina, Z.; Kokina, A.; Liepins, J. Adenine starvation is signalled through environmental stress response system in budding yeast Saccharomyces cerevisiae. Environ. Experim. Biol. 2017, 15, 283–286. [Google Scholar] [CrossRef]
- Pelletier, J.; Riaño-Canalias, F.; Almacellas, E.; Mauvezin, C.; Samino, S.; Feu, S.; Menoyo, S.; Domostegui, A.; Garcia-Cajide, M.; Salazar, R.; et al. Nucleotide depletion reveals the impaired ribosome biogenesis checkpoint as a barrier against DNA damage. EMBO J. 2020, 39, 103838. [Google Scholar] [CrossRef]
- Li, P.; Liu, X.; Hao, Z.; Jia, Y.; Zhao, X.; Xie, D.; Dong, J.; Zeng, F. Dual Repressive Function by Cip1, a Budding Yeast Analog of p21, in Cell-Cycle START Regulation. Front. Microbiol. 2020, 11, 1623. [Google Scholar] [CrossRef]
- Chang, Y.L.; Tseng, S.F.; Huang, Y.C.; Shen, Z.J.; Hsu, P.H.; Hsieh, M.H.; Yang, C.W.; Tognetti, S.; Canal, B.; Subirana, L.; et al. Yeast Cip1 is activated by environmental stress to inhibit Cdk1-G1 cyclins via Mcm1 and Msn2/4. Nat. Commun. 2017, 8, 56. [Google Scholar] [CrossRef]
- Hoxhaj, G.; Hughes-Hallett, J.; Timson, R.C.; Ilagan, E.; Yuan, M.; Asara, J.M.; Ben-Sahra, I.; Manning, B.D. The mTORC1 Signaling Network Senses Changes in Cellular Purine Nucleotide Levels. Cell Rep. 2017, 21, 1331–1346. [Google Scholar] [CrossRef] [PubMed]
- Dibble, C.C.; Manning, B.D. The TSC1-TSC2 complex: A key signal-integrating node upstream of TOR. In The Enzymes; Tamanoi, F., Hall, M.N., Eds.; Academic Press: Cambridge, MA, USA, 2010; Volume 28, pp. 21–48. [Google Scholar]
- Koltin, Y.; Faucette, L.; Bergsma, D.J.; Levy, M.A.; Cafferkey, R.; Koser, P.L.; Johnson, R.K.; Livi, G.P. Rapamycin sensitivity in Saccharomyces cerevisiae is mediated by a peptidyl-prolyl cis-trans isomerase related to human FK506-binding protein. Mol. Cell. Biol. 1991, 11, 1718–1723. [Google Scholar] [CrossRef] [PubMed]



Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kokina, A.; Tanilas, K.; Ozolina, Z.; Pleiko, K.; Shvirksts, K.; Vamza, I.; Liepins, J. Purine Auxotrophic Starvation Evokes Phenotype Similar to Stationary Phase Cells in Budding Yeast. J. Fungi 2022, 8, 29. https://doi.org/10.3390/jof8010029
Kokina A, Tanilas K, Ozolina Z, Pleiko K, Shvirksts K, Vamza I, Liepins J. Purine Auxotrophic Starvation Evokes Phenotype Similar to Stationary Phase Cells in Budding Yeast. Journal of Fungi. 2022; 8(1):29. https://doi.org/10.3390/jof8010029
Chicago/Turabian StyleKokina, Agnese, Kristel Tanilas, Zane Ozolina, Karlis Pleiko, Karlis Shvirksts, Ilze Vamza, and Janis Liepins. 2022. "Purine Auxotrophic Starvation Evokes Phenotype Similar to Stationary Phase Cells in Budding Yeast" Journal of Fungi 8, no. 1: 29. https://doi.org/10.3390/jof8010029
APA StyleKokina, A., Tanilas, K., Ozolina, Z., Pleiko, K., Shvirksts, K., Vamza, I., & Liepins, J. (2022). Purine Auxotrophic Starvation Evokes Phenotype Similar to Stationary Phase Cells in Budding Yeast. Journal of Fungi, 8(1), 29. https://doi.org/10.3390/jof8010029