A Combined Analysis of Transcriptome and Proteome Reveals the Inhibitory Mechanism of a Novel Oligosaccharide Ester against Penicillium italicum
Abstract
1. Introduction
2. Materials and Methods
2.1. MTE Preparation
2.2. Fungal Strain
2.3. Antifungal Activity of MTE-1 against P. italicum In Vitro
2.4. Effect of MTE-1 on Disease Development of Citrus Fruit Inoculated with P. italicum
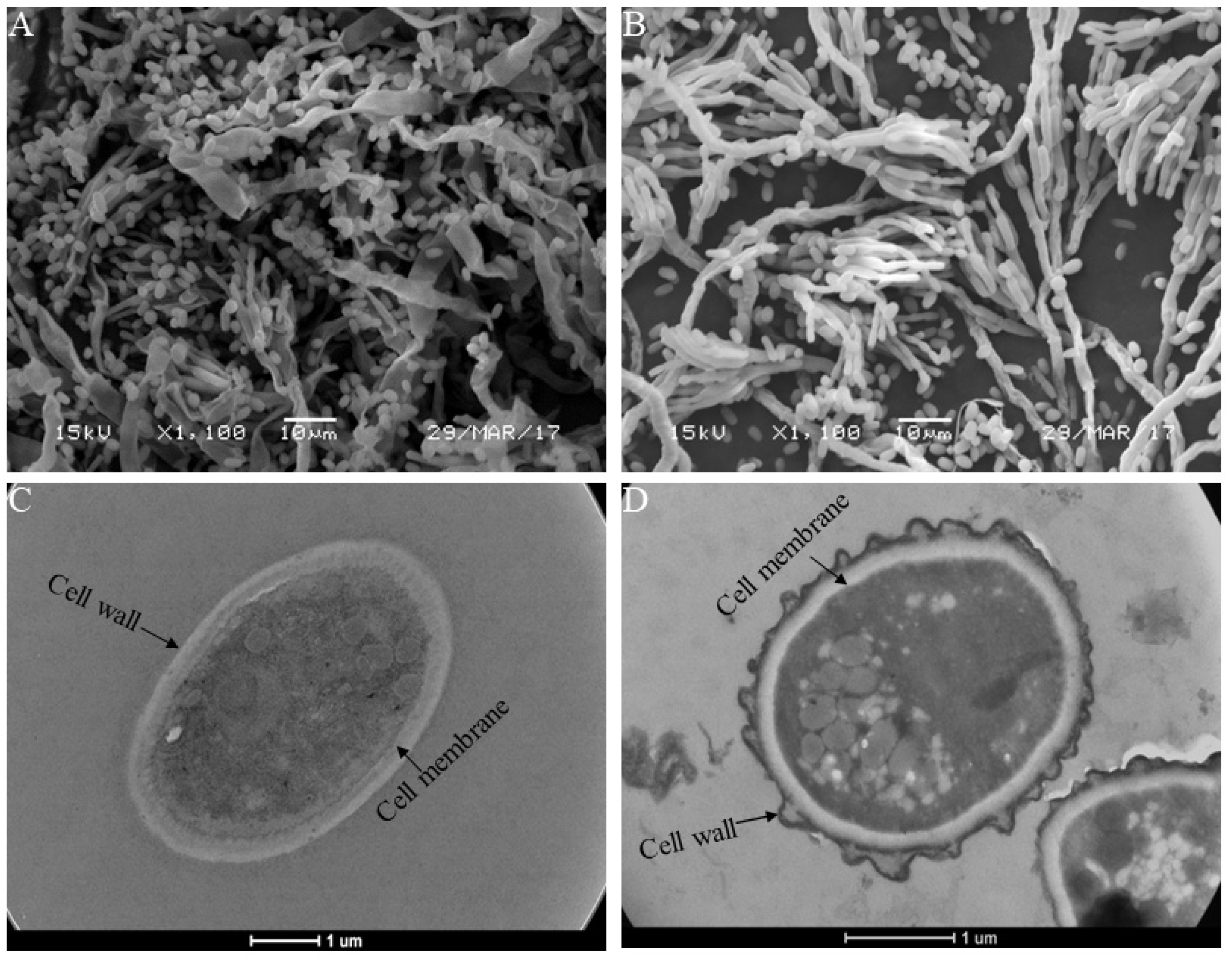
2.5. Scanning Electron Microscopy (SEM) and Transmission Electron Microscopy (TEM)
2.6. RNA Extraction and Transcriptome Analysis
2.7. Protein Extraction and Proteome Analysis
2.8. Data Analysis
3. Results
3.1. Effect of MTE-1 Treatment on the Growth of P. italicum
3.2. Effect of MTE-1 on Disease Development in Mandarin Fruit Inoculated with P. italicum
3.3. SEM and TEM Analysis
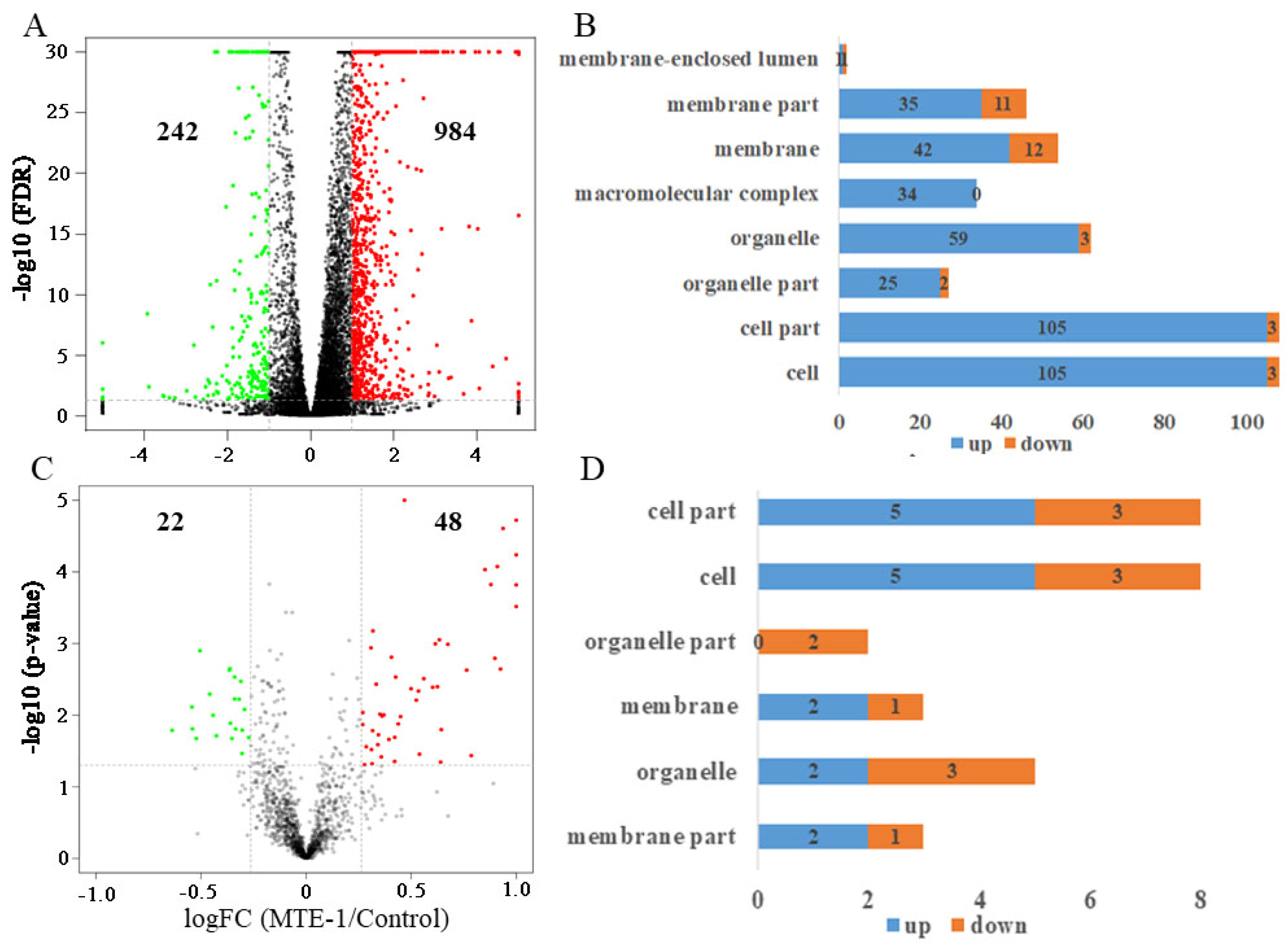
3.4. Transcriptome Analysis of P. italicum in Response to MTE-1
3.5. Proteome Analysis of P. italicum in Response to MTE-1
4. Discussion
4.1. MTE-1 Treatment Inhibited P. italicum Growth
4.2. MTE-1 Treatment Disrupted the Cell Structure of P. italicum
4.3. MTE-1 Treatment Regulated the Stress Response of P. italicum
4.4. MTE-1 Treatment Affected the Pathogenicity of P. italicum on Citrus Fruit
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ma, D.; Ji, D.; Liu, J.; Xu, Y.; Chen, T.; Tian, S. Efficacy of Methyl Thujate in Inhibiting Penicillium expansum Growth and Possible Mechanism Involved. Postharvest Biol. Technol. 2020, 161, 111070. [Google Scholar] [CrossRef]
- Xie, L.; Wu, Y.; Wang, Y.; Jiang, Y.; Yang, B.; Duan, X.; Li, T. Fumonisin B1 Induced Aggressiveness and Infection Mechanism of Fusarium proliferatum on Banana Fruit. Environ. Pollut. 2021, 288, 117793. [Google Scholar] [CrossRef]
- Pinto, L.; Cefola, M.; Bonifacio, M.A.; Cometa, S.; Bocchino, C.; Pace, B.; De Giglio, E.; Palumbo, M.; Sada, A.; Logrieco, A.F.; et al. Effect of Red Thyme Oil (Thymus vulgaris L.) Vapours on Fungal Decay, Quality Parameters and Shelf-Life of Oranges During Cold Storage. Food Chem. 2021, 336, 127590. [Google Scholar] [CrossRef] [PubMed]
- Tian, S.; Torres, R.; Ballester, A.R.; Li, B.; Vilanova, L.; González-Candelas, L. Molecular Aspects in Pathogen-Fruit Interactions: Virulence and Resistance. Postharvest Biol. Technol. 2016, 122, 11–21. [Google Scholar] [CrossRef]
- Wu, Y.; Lin, H.; Lin, Y.; Shi, J.; Xue, S.; Hung, Y.-C.; Chen, Y.; Wang, H. Effects of Biocontrol Bacteria Bacillus amyloliquefaciens Ly-1 Culture Broth on Quality Attributes and Storability of Harvested Litchi Fruit. Postharvest Biol. Technol. 2017, 132, 81–87. [Google Scholar] [CrossRef]
- Bose, S.K.; Howlader, P.; Jia, X.; Wang, W.; Yin, H. Alginate Oligosaccharide Postharvest Treatment Preserve Fruit Quality and Increase Storage Life Via Abscisic Acid Signaling in Strawberry. Food Chem. 2019, 283, 665–674. [Google Scholar] [CrossRef] [PubMed]
- Bose, S.K.; Howlader, P.; Wang, W.; Yin, H. Oligosaccharide is a Promising Natural Preservative for Improving Postharvest Preservation of Fruit: A Review. Food Chem. 2021, 341, 128178. [Google Scholar] [CrossRef] [PubMed]
- Ibrahim, O.O. Functional Oligosaccharide: Chemicals Structure, Manufacturing, Health Benefits, Applications and Regulations. Food Chem. Nanotechnol. 2018, 4, 65–76. [Google Scholar] [CrossRef]
- Cano, A.; Moschou, E.A.; Daunert, S.; Coello, J.; Palet, C. Optimization of the Xylan Degradation Activity of Monolithic Enzymatic Membranes as a Function of Their Composition Using Design of Experiments. Bioprocess Biosyst. Eng. 2006, 29, 261–268. [Google Scholar] [CrossRef] [PubMed]
- Ke, Y.; Ding, B.; Zhang, M.; Dong, T.; Fu, Y.; Lv, Q.; Ding, W.; Wang, X. Study on Inhibitory Activity and Mechanism of Chitosan Oligosaccharides on Aspergillus flavus and Aspergillus fumigatus. Carbohydr. Polym. 2022, 275, 118673. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Wei, X.; Wu, P.; Xu, L.; Xue, J. New Pezicula Species SC1337 Strain in Preparing Trisaccharide Ester Derivatives. Patent CN106867917-A; Patent CN106867917-B, 15 March 2017. [Google Scholar]
- Yin, C.; Zhu, H.; Jiang, Y.; Shan, Y.; Gong, L. Silencing Dicer-Like Genes Reduces Virulence and Srna Generation in Penicillium italicum, the Cause of Citrus Blue Mold. Cells 2020, 9, 363. [Google Scholar] [CrossRef] [PubMed]
- Puskarova, A.; Buckova, M.; Krakova, L.; Pangallo, D.; Kozics, K. The antibacterial and antifungal activity of six essential oils and their cyto/genotoxicity to human HEL 12469 cells. Sci. Rep. 2017, 7, 8211. [Google Scholar] [CrossRef] [PubMed]
- Li, T.; Jian, Q.; Chen, F.; Wang, Y.; Gong, L.; Duan, X.; Yang, B.; Jiang, Y. Influence of Butylated Hydroxyanisole on the Growth, Hyphal Morphology, and the Biosynthesis of Fumonisins in Fusarium proliferatum. Front. Microbiol. 2016, 7, 1038–1048. [Google Scholar] [CrossRef] [PubMed]
- Li, T.; Jiang, G.; Qu, H.; Wang, Y.; Xiong, Y.; Jian, Q.; Wu, Y.; Duan, X.; Zhu, X.; Hu, W.; et al. Comparative Transcriptome Analysis of Penicillium Citrinum Cultured with Different Carbon Sources Identifies Genes Involved in Citrinin Biosynthesis. Toxin 2017, 9, 69. [Google Scholar] [CrossRef] [PubMed]
- Yang, Q.; Qian, X.; Dhanasekaran, S.; Boateng, N.A.S.; Yan, X.; Zhu, H.; He, F.; Zhang, H. Study on the Infection Mechanism of Penicillium digitatum on Postharvest Citrus (Citrus reticulata Blanco) Based on Transcriptomics. Microorganisms 2019, 7, 672. [Google Scholar] [CrossRef] [PubMed]
- Xu, L.X.; Duan, X.W.; Wei, X.Y.; Xue, J.H.; Feng, L.Y.; Wu, P. Fungal Trisaccharide Ester Compound and Application Thereof in Preparation of Medicines for Preventing and Controlling Plant Fungus Diseases. Patent CN106946955-A; Patent CN106946955-B, 15 March 2017. [Google Scholar]
- Wang, Z.; Li, J.; Liu, J.; Tian, X.; Zhang, D.; Wang, Q. Management of Blue Mold (Penicillium italicum) on Mandarin Fruit with a Combination of the Yeast, Meyerozyma guilliermondii and an Alginate Oligosaccharide. Biol. Control 2021, 152, 104451. [Google Scholar] [CrossRef]
- Lu, H.; Chen, H.; Tang, X.; Yang, Q.; Zhang, H.; Chen, Y.Q.; Chen, W. Time-Resolved Multi-Omics Analysis Reveals the Role of Nutrient Stress-Induced Resource Reallocation for Tag Accumulation in Oleaginous Fungus Mortierella alpina. Biotechnol. Biofuels 2020, 13, 116. [Google Scholar] [CrossRef] [PubMed]
- Hoehamer, C.F.; Cummings, E.D.; Hilliard, G.M.; Rogers, P.D. Changes in the Proteome of Candida albicans in Response to Azole, Polyene, and Echinocandin Antifungal Agents. Antimicrob. Agents Chemother. 2010, 54, 1655–1664. [Google Scholar] [CrossRef] [PubMed]
- Palmieri, F. Encyclopedia of Biological Chemistry; Elsevier Inc.: Amsterdam, The Netherland, 2013; pp. 149–158. [Google Scholar]
- Takagi, H. Metabolic Regulatory Mechanisms and Physiological Roles of Functional Amino Acids and Their Applications in Yeast. Biosci. Biotechnol. Biochem. 2019, 83, 1449–1462. [Google Scholar] [CrossRef]
- Plascencia-Jatomea, M.; Viniegra, G.; Olayo, R.; Castillo-Ortega, M.M.; Shirai, K. Effect of Chitosan and Temperature on Spore Germination of Aspergillus niger. Macromol. Biosci. 2003, 3, 582–586. [Google Scholar] [CrossRef]
- Oliveira Junior, E.N.D. Fungal Pathogenicity; Intechopen: London, UK, 2016; pp. 61–81. [Google Scholar]
- Oliveira Junior, E.N.D.; De Melo, I.S.; Franco, T.T. Changes in Hyphal Morphology Due to Chitosan Treatment in Some Fungal Species. Braz. Arch. Biol. Technol. 2012, 55, 637–646. [Google Scholar] [CrossRef]
- Li, Y.; Guo, L.; Zhou, Z. Exploring the Antifungal Mechanism of Limonin-Loaded Eugenol Emulsion Against Penicillium italicum: From the Perspective of Microbial Metabolism. Postharvest Biol. Technol. 2021, 182, 111704. [Google Scholar] [CrossRef]
- Hopke, A.; Brown, A.J.P.; Hall, R.A.; Wheeler, R.T. Dynamic Fungal Cell Wall Architecture in Stress Adaptation and Immune Evasion. Trends Microbiol. 2018, 26, 284–295. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Long, Y.; Mo, F.; Shu, R.; Yin, X.; Wu, X.; Zhang, R.; Zhang, Z.; He, L.; Chen, T.; et al. Antifungal Activity and Biocontrol Mechanism of Fusicolla violacea J-1 Against Soft Rot in Kiwifruit Caused by Alternaria alternata. J. Fungi 2021, 7, 937. [Google Scholar] [CrossRef] [PubMed]
- Kang, L.; Zhu, Y.; Bai, Y.; Yuan, S. Characteristics, Transcriptional Patterns and Possible Physiological Significance of Glycoside Hydrolase Family 16 Members in Coprinopsis cinerea. FEMS Microbiol. Lett. 2019, 366, 937. [Google Scholar] [CrossRef] [PubMed]
- Da Rocha Neto, A.C.; Maraschin, M.; Di Piero, R.M. Antifungal Activity of Salicylic Acid Against Penicillium expansum and its Possible Mechanisms of Action. Int. J. Food Microbiol. 2015, 215, 64–70. [Google Scholar] [CrossRef] [PubMed]
- Lin, Y.; Lin, H.; Chen, Y.; Wang, H.; Ritenour, M.A.; Lin, Y. Hydrogen Peroxide-Induced Changes in Activities of Membrane Lipids-Degrading Enzymes and Contents of Membrane Lipids Composition in Relation to Pulp Breakdown of Longan Fruit During Storage. Food Chem. 2019, 297, 124955. [Google Scholar] [CrossRef]
- Chen, A.; Zeng, G.; Chen, G.; Liu, L.; Shang, C.; Hu, X.; Lu, L.; Chen, M.; Zhou, Y.; Zhang, Q. Plasma Membrane Behavior, Oxidative Damage, and Defense Mechanism in Phanerochaete chrysosporium Under Cadmium Stress. Process Biochem. 2014, 49, 589–598. [Google Scholar] [CrossRef]
- Xu, D.; Deng, Y.; Xi, P.; Zhu, Z.; Kong, X.; Wan, L.; Situ, J.; Li, M.; Gao, L.; Jiang, Z. Biological Activity of Pterostilbene Against Peronophythora litchii, the Litchi Downy Blight Pathogen. Postharvest Biol. Technol. 2018, 144, 29–35. [Google Scholar] [CrossRef]
- Moore, C.B.; Sayers, N.; Mosquera, J.; Slaven, J.; Denning, D.W. Antifungal Drug Resistance in Aspergillus. J. Infect. 2000, 41, 203–220. [Google Scholar] [CrossRef] [PubMed]
- Szewczyk, R.; Soboń, A.; Sylwia, R.; Dzitko, K.; Waidelich, D.; Długoński, J. Intracellular Proteome Expression During 4-N-Nonylphenol Biodegradation by the Filamentous Fungus Metarhizium robertsii. Int. Biodeterior. Biodegrad. 2014, 93, 44–53. [Google Scholar] [CrossRef]
- Siozios, S.; Ioannidis, P.; Klasson, L.; Andersson, S.G.; Braig, H.R.; Bourtzis, K. The Diversity and Evolution of Wolbachia Ankyrin Repeat Domain Genes. PLoS ONE 2013, 8, e55390. [Google Scholar] [CrossRef]
- Lai, Y.L.; Cao, X.; Chen, J.J.; Wang, L.L.; Wei, G.; Wang, S.B. Coordinated Regulation of infection-Related Morphogenesis by Thekmt2-Cre1-Hyd4 Regulatory Pathway to Facilitate Fungal Infection. Sci. Adv. 2020, 6, 1659–1660. [Google Scholar] [CrossRef]
- Zhang, Z.; Chen, Y.; Li, B.; Chen, T.; Tian, S. Reactive Oxygen Species: A Generalist in Regulating Development and Pathogenicity of Phytopathogenic Fungi. Comput. Struct. Biotechnol. J. 2020, 18, 3344–3349. [Google Scholar] [CrossRef]
- Jacob, C.O.; Eisenstein, M.; Dinauer, M.C.; Ming, W.; Liu, Q.; John, S.; Quismorio, F.P., Jr.; Reiff, A.; Myones, B.L.; Kaufman, K.M.; et al. Lupus-Associated Causal Mutation in Neutrophil Cytosolic Factor 2 (Ncf2) Brings Unique Insights to the Structure and Function of Nadph Oxidase. Proc. Natl. Acad. Sci. USA 2012, 109, 59–67. [Google Scholar] [CrossRef] [PubMed]
- Meena, M.; Prasad, V.; Zehra, A.; Gupta, V.K.; Upadhyay, R.S. Mannitol Metabolism During Pathogenic Fungal-Host Interactions Under Stressed Conditions. Front. Microbiol. 2015, 6, 1019–1031. [Google Scholar] [CrossRef] [PubMed]
- Voegele, R.T.; Hahn, M.; Lohaus, G.; Link, T.; Heiser, I.; Mendgen, K. Possible Roles for Mannitol And Mannitol Dehydrogenase In The Biotrophic Plant Pathogen Uromyces fabae. Plant Physiol. 2005, 137, 190–198. [Google Scholar] [CrossRef] [PubMed][Green Version]




| GO ID | Description | Gene Number | p-Value |
|---|---|---|---|
| GO:0018208 | peptidyl-proline modification | 6 | 0.000379783 |
| GO:0006520 | cellular amino-acid metabolic process | 29 | 0.000873782 |
| GO:0044710 | single-organism metabolic process | 145 | 0.001147218 |
| GO:0019752 | carboxylic-acid metabolic process | 37 | 0.002046061 |
| GO:0006082 | organic acid metabolic process | 37 | 0.003157122 |
| GO:0043436 | oxoacid metabolic process | 37 | 0.003157122 |
| GO:0006790 | sulfur-compound metabolic process | 10 | 0.003760795 |
| GO:0009081 | branched-chain amino-acid metabolic process | 5 | 0.003761958 |
| GO:0044699 | single-organism process | 201 | 0.012854284 |
| GO:1901605 | alpha-amino-acid metabolic process | 15 | 0.013183352 |
| GO:0018193 | peptidyl-amino-acid modification | 6 | 0.015477395 |
| GO:0006549 | isoleucine metabolic process | 3 | 0.020871202 |
| GO:0072525 | pyridine-containing compound biosynthetic process | 3 | 0.020871202 |
| GO:0000096 | sulfur amino-acid metabolic process | 6 | 0.02121723 |
| GO:0006733 | oxidoreduction coenzyme metabolic process | 4 | 0.025168398 |
| GO:0044281 | small-molecule metabolic process | 66 | 0.030835746 |
| GO:0009066 | aspartate-family amino-acid metabolic process | 5 | 0.033230704 |
| GO:0044272 | sulfur-compound biosynthetic process | 5 | 0.033230704 |
| GO:1901564 | organonitrogen-compound metabolic process | 56 | 0.037409962 |
| GO:0006007 | glucose catabolic process | 4 | 0.037491393 |
| GO:0009069 | serine-family amino-acid metabolic process | 4 | 0.037491393 |
| GO:0019320 | hexose catabolic process | 4 | 0.037491393 |
| GO:0006875 | cellular-metal-ion homeostasis | 3 | 0.037529777 |
| Pathway | Gene Number | p-Value | Pathway ID |
|---|---|---|---|
| 2-Oxocarboxylic acid metabolism | 14 | 0.001152635 | ko01210 |
| Lysine biosynthesis | 6 | 0.001282578 | ko00300 |
| Valine, leucine, and isoleucine degradation | 13 | 0.001659644 | ko00280 |
| Biosynthesis of antibiotics | 57 | 0.001778891 | ko01130 |
| Biosynthesis of amino acids | 30 | 0.006832061 | ko01230 |
| Nicotinate and nicotinamide metabolism | 6 | 0.01082372 | ko00760 |
| Fructose and mannose metabolism | 14 | 0.01136807 | ko00051 |
| Biosynthesis of secondary metabolites | 69 | 0.01390241 | ko01110 |
| mRNA surveillance pathway | 13 | 0.01922519 | ko03015 |
| Ribosome | 21 | 0.02501491 | ko03010 |
| Metabolic pathways | 155 | 0.02545925 | ko01100 |
| Mismatch repair | 7 | 0.02694715 | ko03430 |
| Phenylalanine, tyrosine, and tryptophan biosynthesis | 8 | 0.02985515 | ko00400 |
| Tyrosine metabolism | 13 | 0.03127059 | ko00350 |
| Cysteine and methionine metabolism | 13 | 0.03629479 | ko00270 |
| Valine, leucine, and isoleucine biosynthesis | 6 | 0.03977772 | ko00290 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Feng, L.; Xu, L.; Li, X.; Xue, J.; Li, T.; Duan, X. A Combined Analysis of Transcriptome and Proteome Reveals the Inhibitory Mechanism of a Novel Oligosaccharide Ester against Penicillium italicum. J. Fungi 2022, 8, 111. https://doi.org/10.3390/jof8020111
Feng L, Xu L, Li X, Xue J, Li T, Duan X. A Combined Analysis of Transcriptome and Proteome Reveals the Inhibitory Mechanism of a Novel Oligosaccharide Ester against Penicillium italicum. Journal of Fungi. 2022; 8(2):111. https://doi.org/10.3390/jof8020111
Chicago/Turabian StyleFeng, Linyan, Liangxiong Xu, Xiaojie Li, Jinghua Xue, Taotao Li, and Xuewu Duan. 2022. "A Combined Analysis of Transcriptome and Proteome Reveals the Inhibitory Mechanism of a Novel Oligosaccharide Ester against Penicillium italicum" Journal of Fungi 8, no. 2: 111. https://doi.org/10.3390/jof8020111
APA StyleFeng, L., Xu, L., Li, X., Xue, J., Li, T., & Duan, X. (2022). A Combined Analysis of Transcriptome and Proteome Reveals the Inhibitory Mechanism of a Novel Oligosaccharide Ester against Penicillium italicum. Journal of Fungi, 8(2), 111. https://doi.org/10.3390/jof8020111








