Natural Populations from the Phytophthora palustris Complex Show a High Diversity and Abundance of ssRNA and dsRNA Viruses
Abstract
1. Introduction
2. Materials and Methods
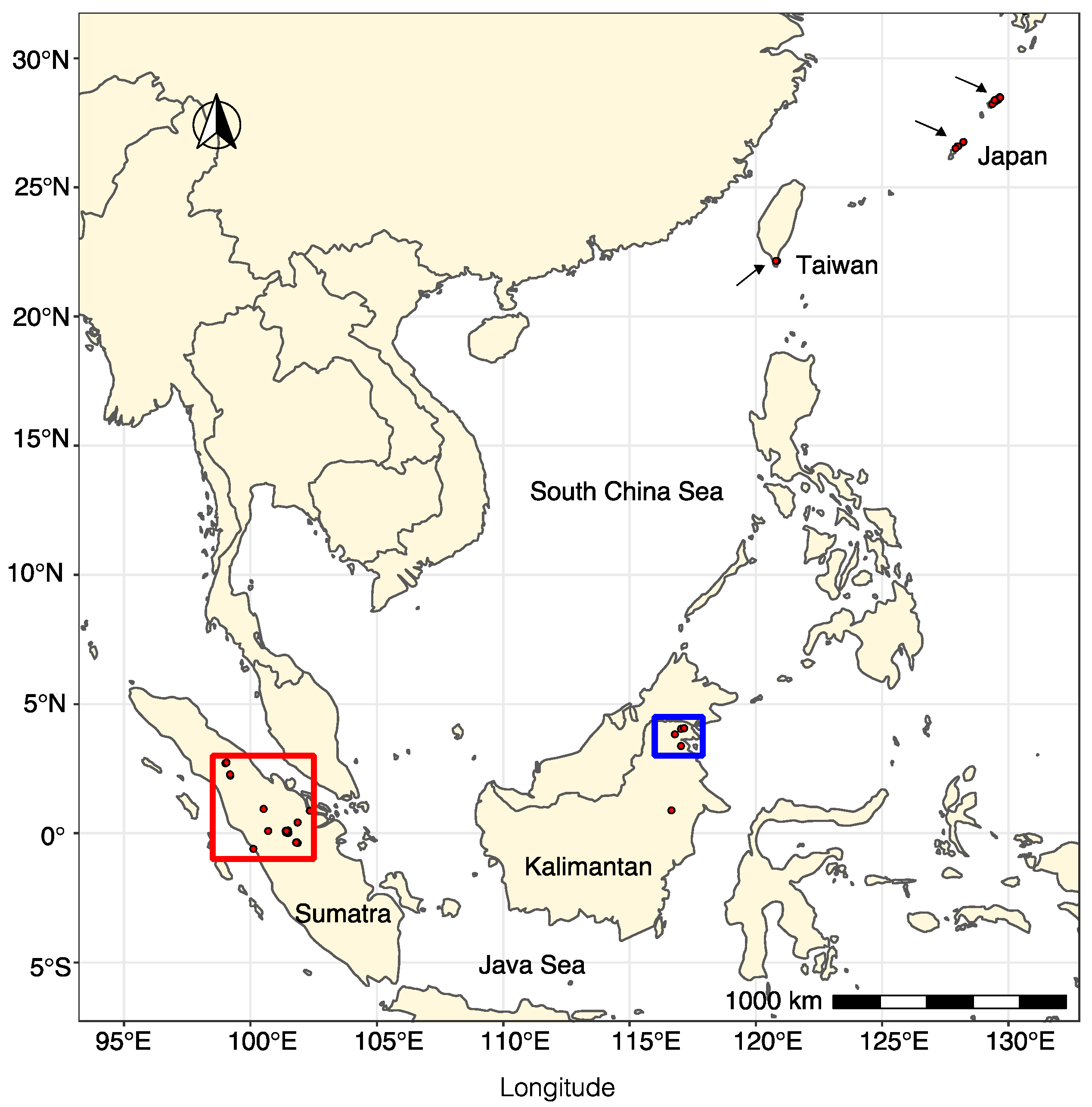
2.1. “Phytophthora palustris” Sampling and Isolation
2.2. RNA Extraction
2.3. RNA Library Preparation and Sequencing
2.4. De Novo Virus Assembly and Detection Workflow
2.5. Genetic Variability Analyses and Conserved Domains
2.6. Phylogenetic Trees
2.7. Rapid Amplification of cDNA Ends (RACE) and Confirmation of Viruses’ Occurrence by Direct Reverse-Transcription Polymerase Chain Reaction (RT-PCR)
2.8. Graphs and Visualizations
3. Results
3.1. Virus Identification
3.2. Virus Genomic Organization and Phylogenetic Relationships
3.2.1. (−)ssRNA Viruses
3.2.2. (+)ssRNA Viruses
3.2.3. dsRNA Viruses
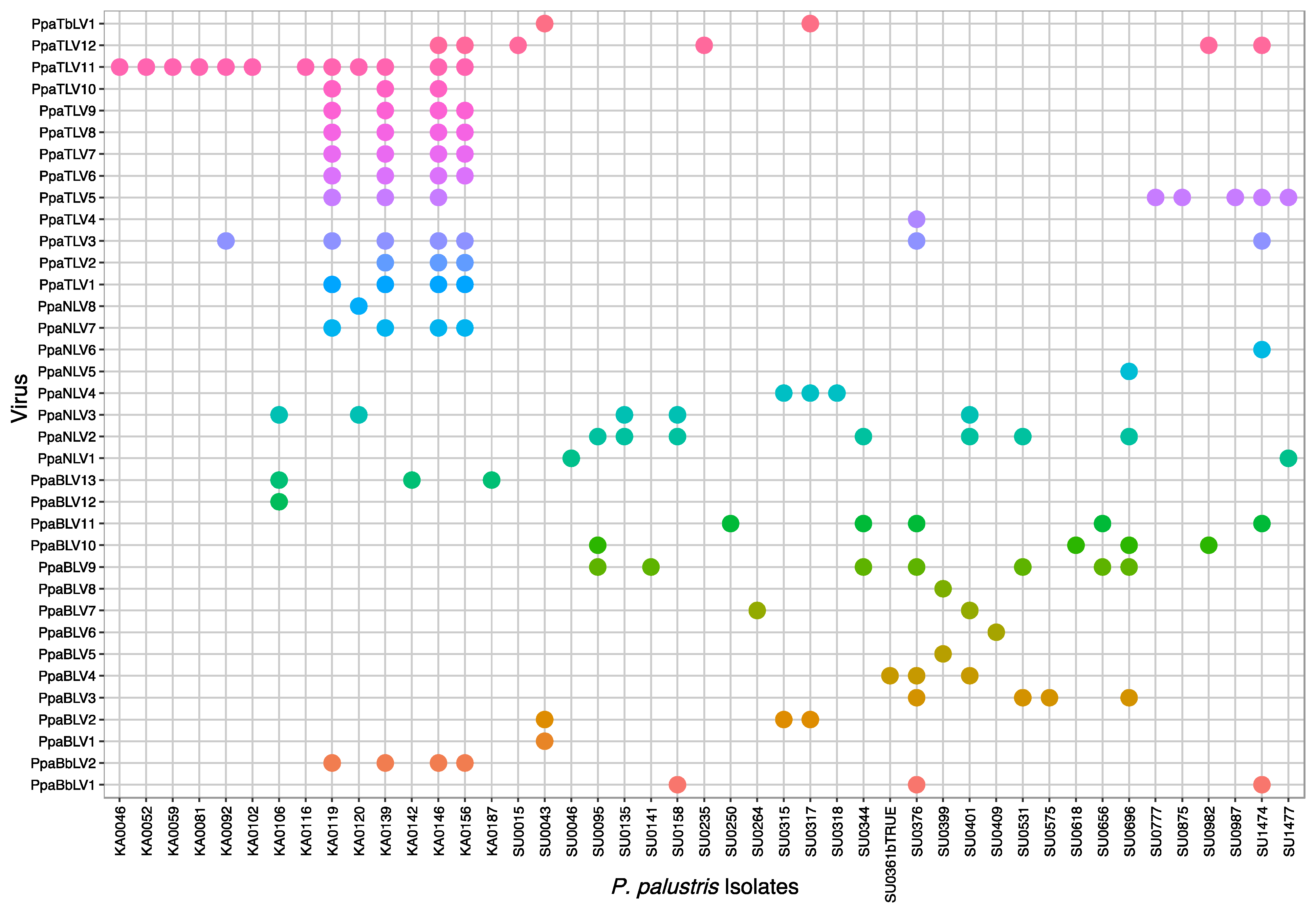
3.3. Virus Abundance, Diversity, and Geographical Distribution
3.3.1. Virus Occurrence Based on the RT-PCR Screening
3.3.2. Virus Read Abundance per Pool and Island

4. Discussion
4.1. Evolutionary Insights into Novel Taxa of (+)ssRNA Viruses
4.2. Discovery of New dsRNA Viruses and High Abundance of Toti-Like Viruses
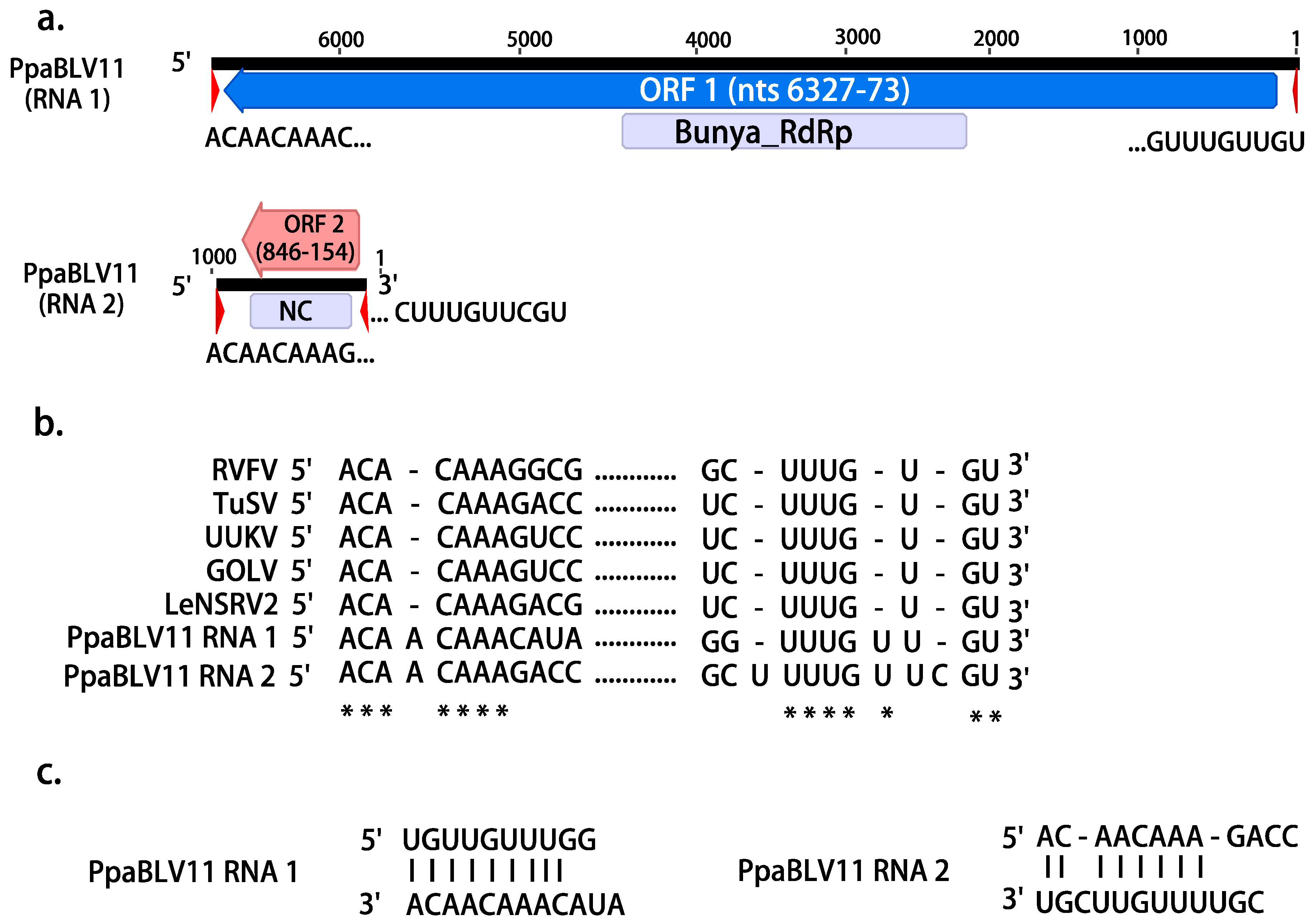
4.3. Persistence of Bunya-Like Viruses and the First Oomycete Bunyavirus with NC Protein
4.4. Virus Population Structure and Transmission Efficiency
4.5. Multiple Viral Infections at Isolate and Site Level
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Brasier, C.M. The biosecurity threat to the UK and global environment from international trade in plants. Plant Pathol. 2008, 57, 792–808. [Google Scholar] [CrossRef]
- Santini, A.; Ghelardini, L.; De Pace, C.; Desprez-Loustau, M.L.; Capretti, P.; Chandelier, A.; Cech, T.; Chira, D.; Diamandis, S.; Gaitniekis, T.; et al. Biogeographical patterns and determinants of invasion by forest pathogens in Europe. New Phytol. 2013, 197, 238–250. [Google Scholar] [CrossRef] [PubMed]
- Hantula, J.; Müller, M.M.; Uusivuori, J. International plant trade associated risks: Laissez-faire or novel solutions. Environ. Sci. Policy 2014, 37, 158–160. [Google Scholar] [CrossRef]
- Jung, T.; Orlikowski, L.; Henricot, B.; Abad-Campos, P.; Aday, A.G.; Aguín Casal, O.; Bakonyi, J.; Cacciola, S.O.; Cech, T.; Chavarriaga, D.; et al. Widespread Phytophthora infestations in European nurseries put forest, semi-natural and horticultural ecosystems at high risk of Phytophthora diseases. For. Pathol. 2016, 46, 134–163. [Google Scholar] [CrossRef]
- Boyd, I.L.; Freer-Smith, P.H.; Gilligan, C.A.; Godfray, H.C.J. The consequence of tree pests and diseases for ecosystem services. Science 2013, 342, 1235773. [Google Scholar] [CrossRef]
- Jung, T.; Pérez-Sierra, A.; Durán, A.; Jung, M.H.; Balci, Y.; Scanu, B. Canker and decline diseases caused by soil- and airborne Phytophthora species in forests and woodlands. Persoonia 2018, 40, 182. [Google Scholar] [CrossRef]
- Sakai, A.K.; Allendorf, F.W.; Holt, J.S.; Lodge, D.M.; Molofsky, J.; With, K.A.; Baughman, S.; Cabin, R.J.; Cohen, J.E.; Ellstrand, N.C.; et al. The population biology of invasive species. Annu. Rev. Ecol. Syst. 2001, 32, 305–332. [Google Scholar] [CrossRef]
- Roderick, G.K.; Navajas, M. Genes in new environments: Genetics and evolution in biological control. Nat. Rev. Genet. 2003, 4, 889–899. [Google Scholar] [CrossRef]
- Mitchell, C.E.; Power, A.O. Release of invasive plants from fungal and viral pathogens. Nature 2003, 421, 625–627. [Google Scholar] [CrossRef]
- Derelle, R.; López-García, P.; Timpano, H.; Moreira, D. A phylogenomic framework to study the diversity and evolution of Stramenopiles (=Heterokonts). Mol. Biol. Evol. 2016, 33, 2890–2898. [Google Scholar] [CrossRef]
- Richards, T.A.; Dacks, J.B.; Jenkinson, J.M.; Thornton, C.R.; Talbot, N.J. Evolution of filamentous plant pathogens: Gene exchange across eukaryotic kingdoms. Curr. Biol. 2006, 16, 1857–1864. [Google Scholar] [CrossRef]
- Brasier, C.M.; Vettraino, A.M.; Chang, T.T.; Vannini, A. Phytophthora lateralis discovered in an old growth Chamaecyparis forest in Taiwan. Plant Pathol. 2010, 59, 595–603. [Google Scholar] [CrossRef]
- Brasier, C.M.; Franceschini, S.; Vettraino, A.M.; Hansen, E.M.; Green, S.; Robin, C.; Webber, J.F.; Vannini, A. Four phenotypically and phylogenetically distinct lineages in Phytophthora lateralis. Fungal Biol. 2012, 116, 1232–1249. [Google Scholar] [CrossRef]
- Jung, T.; Chang, T.T.; Bakonyi, J.; Seress, D.; Pérez-Sierra, A.; Yang, X.; Hong, C.; Scanu, B.; Fu, C.H.; Hsueh, K.L.; et al. Diversity of Phytophthora species in natural ecosystems of Taiwan and association with disease symptoms. Plant Pathol. 2017, 66, 194–211. [Google Scholar] [CrossRef]
- Jung, T.; Scanu, B.; Brasier, C.M.; Webber, J.; Milenković, I.; Corcobado, T.; Tomšovský, M.; Pánek, M.; Bakonyi, J.; Maia, C.; et al. A survey in natural forest ecosystems of Vietnam reveals high diversity of both new and described Phytophthora taxa including P. ramorum. Forests 2020, 11, 93. [Google Scholar] [CrossRef]
- Jung, T.; Jung, M.H.; Webber, J.F.; Kageyama, K.; Hieno, A.; Masuya, H.; Uematsu, S.; Pérez-Sierra, A.; Harris, A.R.; Forster, J.; et al. The destructive tree pathogen Phytophthora ramorum originates from the laurosilva forests of East Asia. J. Fungi. 2021, 7, 226. [Google Scholar] [CrossRef]
- Jung, T.; Milenković, I.; Corcobado, T.; Májek, T.; Janoušek, J.; Kudláček, T.; Tomšovský, M.; Nagy, Z.; Durán, A.; Tarigan, M.; et al. Extensive morphological and behavioural diversity among fourteen new and seven described species in Phytophthora Clade 10 and its evolutionary implications. Persoonia 2022, 49, 1–57. [Google Scholar] [CrossRef]
- Gower, D.J.; Johnson, K.G.; Richardson, J.E.; Rosen, B.R.; Ruber, L.W.S. Biotic Evolution and Environmental Change in Southeast Asia; Cambridge University Press: Cambrigde, UK, 2012; ISBN 9780511735882. [Google Scholar]
- Prospero, S.; Botella, L.; Santini, A.; Robin, C. Biological control of emerging forest diseases: How can we move from dreams to reality? For. Ecol. Manag. 2021, 496, 119377. [Google Scholar] [CrossRef]
- Rigling, D.; Prospero, S. Cryphonectria parasitica, the causal agent of chestnut blight: Invasion history, population biology and disease control. Mol. Plant Pathol. 2018, 19, 7–20. [Google Scholar] [CrossRef]
- Ghabrial, S.A.; Suzuki, N. Viruses of plant pathogenic fungi. Annu. Rev. Phytopathol. 2009, 47, 353–384. [Google Scholar] [CrossRef]
- Bryner, S.F.; Rigling, D.; Brunner, P.C. Invasion history and demographic pattern of Cryphonectria hypovirus 1 across European populations of the chestnut blight fungus. Ecol. Evol. 2012, 2, 3227–3241. [Google Scholar] [CrossRef] [PubMed]
- Lefeuvre, P.; Martin, D.P.; Elena, S.F.; Shepherd, D.N.; Roumagnac, P.; Varsani, A. Evolution and ecology of plant viruses. Nat. Rev. Microbiol. 2019, 17, 632–644. [Google Scholar] [CrossRef] [PubMed]
- Poisot, T.; Stouffer, D.B.; Gravel, D. Beyond species: Why ecological interaction networks vary through space and time. Oikos 2015, 124, 243–251. [Google Scholar] [CrossRef]
- Schoelz, J.E.; Stewart, L.R. The Role of Viruses in the Phytobiome. Annu. Rev. Virol. 2018, 5, 93–111. [Google Scholar] [CrossRef] [PubMed]
- French, R.K.; Holmes, E.C. An ecosystems perspective on virus evolution and emergence. Trends Microbiol. 2020, 28, 165–175. [Google Scholar] [CrossRef]
- Ihrmark, K.; Johannesson, H.; Stenström, E.; Stenlid, J. Transmission of double-stranded RNA in Heterobasidion annosum. Fungal Genet. Biol. 2022, 36, 147–154. [Google Scholar] [CrossRef]
- Ihrmark, K.; Stenström, E.; Stenlid, J. Double-stranded RNA transmission through basidiospores of Heterobasidion annosum. Mycol. Res. 2004, 108, 149–153. [Google Scholar] [CrossRef]
- Göker, M.; Scheuner, C.; Klenk, H.-P.; Stielow, J.B.; Menzel, W. Codivergence of mycoviruses with their hosts. PLoS ONE 2011, 6, e22252. [Google Scholar] [CrossRef]
- Voth, P.D.; Mairura, L.; Lockhart, B.E.; May, G. Phylogeography of Ustilago maydis virus H1 in the USA and Mexico. J. Gen. Virol. 2006, 87, 3433–3441. [Google Scholar] [CrossRef]
- Botella, L.; Tuomivirta, T.T.; Hantula, J.; Diez, J.J.; Jankovsky, L. The European race of Gremmeniella abietina hosts a single species of Gammapartitivirus showing a global distribution and possible recombinant events in its history. Fungal Biol. 2015, 119, 125–135. [Google Scholar] [CrossRef]
- Schoebel, C.N.; Botella, L.; Lygis, V.; Rigling, D. Population genetic analysis of a parasitic mycovirus to infer the invasion history of its fungal host. Mol. Ecol. 2017, 26, 2482–2497. [Google Scholar] [CrossRef]
- Botella, L.; Jung, T. Multiple viral infections detected in Phytophthora condilina by total and small RNA sequencing. Viruses 2021, 13, 620. [Google Scholar] [CrossRef]
- Cai, G.; Hillman, B.I. Phytophthora Viruses, 1st ed.; Elsevier Inc: Amsterdam, The Netherlands, 2013; Volume 86, ISBN 9780123943156. [Google Scholar]
- Hacker, C.V.; Brasier, C.M.; Buck, K.W. A double-stranded RNA from a Phytophthora species is related to the plant endornaviruses and contains a putative UDP glycosyltransferase gene. J. Gen. Virol. 2005, 86, 1561–1570. [Google Scholar] [CrossRef]
- Kozlakidis, Z.; Brown, N.A.; Jamal, A.; Phoon, X.; Coutts, R.H.A. Incidence of endornaviruses in Phytophthora taxon douglasfir and Phytophthora ramorum. Virus Genes 2010, 40, 130–134. [Google Scholar] [CrossRef]
- Poimala, A.; Parikka, P.; Hantula, J.; Vainio, E.J. Viral diversity in Phytophthora cactorum population infecting strawberry. Environ. Microbiol. 2021, 23, 5200–5221. [Google Scholar] [CrossRef]
- Uchida, K.; Sakuta, K.; Ito, A.; Takahashi, Y.; Katayama, Y.; Omatsu, T.; Mizutani, T.; Arie, T.; Komatsu, K.; Fukuhara, T.; et al. Two novel endornaviruses co-infecting a phytophthora pathogen of Asparagus officinalis modulate the developmental stages and fungicide sensitivities of the host oomycete. Front. Microbiol. 2021, 12, 633502. [Google Scholar] [CrossRef]
- Xu, Z.; Khalifa, M.E.; Frampton, R.A.; Smith, G.R.; McDougal, R.L.; Macdiarmid, R.M.; Kalamorz, F. Characterization of a novel double-stranded RNA virus from Phytophthora pluvialis in New Zealand. Viruses 2022, 14, 247. [Google Scholar] [CrossRef]
- Raco, M.; Vainio, E.; Sutela, S.; Eichmeier, A.; Hakalová, E.; Jung, T.; Botella, L. High diversity of novel viruses in the tree pathogen Phytophthora castaneae revealed by high-throughput sequencing of total and small RNA. Front. Microbiol. 2022, 13, 911474. [Google Scholar] [CrossRef]
- Hannat, S.; Pontarotti, P.; Colson, P.; Kuhn, M.L.; Galiana, E.; La Scola, B.; Aherfi, S.; Panabières, F. Diverse trajectories drive the expression of a giant virus in the oomycete plant pathogen Phytophthora parasitica. Front. Microbiol. 2021, 12, 1–13. [Google Scholar] [CrossRef]
- Grasse, W.; Spring, O. Occurrence and genetic diversity of the Plasmopara halstedii virus in sunflower downy mildew populations of the world. Fungal Biol. 2015, 119, 170–178. [Google Scholar] [CrossRef]
- Chiapello, M.; Rodríguez-Romero, J.; Nerva, L.; Forgia, M.; Chitarra, W.; Ayllón, M.A.; Turina, M. Putative new plant viruses associated with Plasmopara viticola -infected grapevine samples. Ann. Appl. Biol. 2020, 176, 180–191. [Google Scholar] [CrossRef]
- Shiba, K.; Hatta, C.; Sasai, S.; Tojo, M.; Ohki, T.S.; Mochizuki, T. Genome sequence of a novel partitivirus identified from the oomycete Pythium nunn. Arch. Virol. 2018, 163, 2561–2563. [Google Scholar] [CrossRef] [PubMed]
- Sasai, S.; Tamura, K.; Tojo, M.; Herrero, M.L.; Hoshino, T.; Ohki, S.T.; Mochizuki, T. A novel non-segmented double-stranded RNA virus from an Arctic isolate of Pythium polare. Virology 2018, 522, 234–243. [Google Scholar] [CrossRef] [PubMed]
- Botella, L.; Janoušek, J.; Maia, C.; Jung, M.H.; Raco, M.; Jung, T. Marine oomycetes of the genus Halophytophthora harbor viruses related to bunyaviruses. Front. Microbiol. 2020, 11, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Fukunishi, M.; Sasai, S.; Tojo, M.; Mochizuki, T. Novel fusari- and toti-like viruses, with probable different origins, in the plant pathogenic oomycete Globisporangium ultimum. Viruses 2021, 13, 1931. [Google Scholar] [CrossRef]
- Chomczynski, P.; Wilfinger, W.; Kennedy, A.; Rymaszewski, M.; Mackey, K. RNAzol® RT: A new single-step method for isolation of RNA. Nat. Methods 2010, 7, 4–5. [Google Scholar] [CrossRef]
- Brister, J.R.; Ako-Adjei, D.; Bao, Y.; Blinkova, O. NCBI viral genomes resource. Nucleic Acids Res. 2015, 43, D571–D577. [Google Scholar] [CrossRef]
- Stamatakis, A. RAxML-VI-HPC: Maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 2006, 22, 2688–2690. [Google Scholar] [CrossRef]
- Miller, M.A.; Pfeiffer, W.; Schwartz, T. Creating the CIPRES Science Gateway for inference of large phylogenetic trees. In Proceedings of the Gateway Computing Environments Workshop (GCE), New Orleans, LA, USA, 14 November 2010; pp. 1–8. [Google Scholar]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2021; Available online: https://www.R-project.org/ (accessed on 28 August 2022).
- Muller, R.; Poch, O.; Delarue, M.; Bishop, D.H.L.; Bouloy, M. Rift valley fever virus L segment: Correction of the sequence and possible functional role of newly identified regions conserved in RNA-dependent polymerases. J. Gen. Virol. 1994, 75, 1345–1352. [Google Scholar] [CrossRef]
- Wille, M.; Eden, J.; Shi, M.; Klaassen, M.; Hurt, A.C.; Holmes, E.C. Virus–virus interactions and host ecology are associated with RNA virome structure in wild birds. Mol. Ecol. 2018, 27, 5263–5278. [Google Scholar] [CrossRef]
- Yokoi, T.; Yamashita, S.; Hibi, T. The nucleotide sequence and genome organization of Sclerophthora macrospora virus A. Virology 2003, 311, 394–399. [Google Scholar] [CrossRef]
- Starr, E.P.; Nuccio, E.E.; Pett-Ridge, J.; Banfield, J.F.; Firestone, M.K. Metatranscriptomic reconstruction reveals RNA viruses with the potential to shape carbon cycling in soil. Proc. Natl. Acad. Sci. USA 2019, 116, 25900–25908. [Google Scholar] [CrossRef]
- Preisig, O.; Moleleki, N.; Smit, W.A.; Wingfield, B.D.; Wingfield, M.J. A noval RNA mycovirus in a hypovirulent isolate of the plant pathogen Diaporthe ambigua. J. Gen. Virol. 2000, 81, 3107–3114. [Google Scholar] [CrossRef]
- Kondo, H.; Botella, L.; Suzuki, N. Mycovirus Diversity and Evolution Revealed/Inferred from Recent Studies. Annu. Rev. Phytopathol. 2022, 60, 1–30. [Google Scholar] [CrossRef]
- Zhou, L.; Li, X.; Kotta-Loizou, I.; Dong, K.; Li, S.; Ni, D.; Hong, N.; Wang, G.; Xu, W. A mycovirus modulates the endophytic and pathogenic traits of a plant associated fungus. ISME J. 2021, 15, 1893–1906. [Google Scholar] [CrossRef]
- Forgia, M.; Isgandarli, E.; Aghayeva, D.N.; Huseynova, I.; Turina, M. Virome characterization of Cryphonectria parasitica isolates from Azerbaijan unveiled a new mymonavirus and a putative new RNA virus unrelated to described viral sequences. Virology 2021, 553, 51–61. [Google Scholar] [CrossRef]
- Chiapello, M.; Rodríguez-Romero, J.; Ayllón, M.A.; Turina, M. Analysis of the virome associated to grapevine downy mildew lesions reveals new mycovirus lineages. Virus Evol. 2020, 6, 1–18. [Google Scholar] [CrossRef]
- Jones, R.A.C. Plant and insect viruses in managed and natural environments: Novel and neglected transmission pathways. Adv. Virus Res. 2018, 101, 149–187. [Google Scholar]
- Issaka, S.; Traoré, O.; Longué, R.D.S.; Pinel-Galzi, A.; Gill, M.S.; Dellicour, S.; Bastide, P.; Guindon, S.; Hébrard, E.; Dugué, M.J.; et al. Rivers and landscape ecology of a plant virus, Rice yellow mottle virus along the Niger Valley. Virus Evol. 2021, 7, veab072. [Google Scholar] [CrossRef]
- Büttner, C.; Jacobi, V.; Koenig, R. Isolation of Carnation Italian Ringspot Virus from a creek in a forested area south west of Bonn. J. Phytopathol. 1987, 118, 131–134. [Google Scholar] [CrossRef]
- Culley, A.I.; Lang, A.S.; Suttla, C.A. Metagenomic analysis of coastal RNA virus communities. Science 2006, 312, 1795–1798. [Google Scholar] [CrossRef]
- Shi, M.; Lin, X.D.; Tian, J.H.; Chen, L.J.; Chen, X.; Li, C.X.; Qin, X.C.; Li, J.; Cao, J.P.; Eden, J.S.; et al. Redefining the invertebrate RNA virosphere. Nature 2016, 540, 539–543. [Google Scholar] [CrossRef]
- Zhang, Y.Y.; Chen, Y.; Wei, X.; Cui, J. Viromes in marine ecosystems reveal remarkable invertebrate RNA virus diversity. Sci. China Life Sci. 2021, 2021, 1–12. [Google Scholar] [CrossRef]
- Li, H.; Roossinck, M.J. Genetic bottlenecks reduce population variation in an experimental RNA virus population. Society 2004, 78, 10582–10587. [Google Scholar] [CrossRef]
- Vives, M.C.; Rubio, L.; Galipienso, L.; Navarro, L.; Moreno, P.; Guerri, J. Low genetic variation between isolates of Citrus leaf blotch virus from different host species and of different geographical origins. J. Gen. Virol. 2002, 83, 2587–2591. [Google Scholar] [CrossRef]
- Barr, J.N.; Fearns, R. How RNA viruses maintain their genome integrity. J. Gen. Virol. 2010, 91, 1373–1387. [Google Scholar] [CrossRef]
- Botella, L.; Hantula, J. Description, Distribution, and Relevance of Viruses of the Forest Pathogen Gremmeniella abietina. Viruses 2018, 10, 654. [Google Scholar] [CrossRef]
- Wu, M.; Zhang, J.; Yang, L.; Li, G. RNA mycoviruses and their role in Botrytis biology. In Botrytis—The Fungus, the Pathogen and Its Management in Agricultural Systems; Fillinger, S., Elad, Y., Eds.; Springer: Cham, Switzerland, 2015; pp. 71–90. [Google Scholar]
- Marzano, S.-Y.L.; Nelson, B.D.; Ajayi-Oyetunde, O.; Bradley, C.A.; Hughes, T.J.; Hartman, G.L.; Eastburn, D.M.; Domier, L.L. Identification of diverse mycoviruses through metatranscriptomics characterization of the viromes of five major fungal plant pathogens. J. Virol. 2016, 90, 6846–6863. [Google Scholar] [CrossRef]
- Briese, T.; Calisher, C.H.; Higgs, S. Viruses of the family Bunyaviridae: Are all available isolates reassortants? Virology 2013, 446, 207–216. [Google Scholar] [CrossRef] [PubMed]
- Neriya, Y.; Morikawa, T.; Hamamoto, K.; Noguchi, K.; Kobayashi, T.; Suzuki, T.; Nishigawa, H.; Natsuaki, T. Characterization of tulip streak virus, a novel virus associated with the family Phenuiviridae. J. Gen. Virol. 2021, 102, 001525. [Google Scholar] [CrossRef] [PubMed]
- Linn, Y.H.; Fujita, M.; Sotaro, C.; Hyodo, K.; Andika, I.B.; Suzuki, N.; Kondo, H. Two novel fungal negative-strand RNA viruses related to mymonaviruses and phenuiviruses in the shiitake mushroom (Lentinula edodes). Virology 2019, 533, 125–136. [Google Scholar] [CrossRef] [PubMed]
- Feau, N.; Dutech, C.; Brusini, J.; Rigling, D.; Robin, C. Multiple introductions and recombination in Cryphonectria hypovirus 1: Perspective for a sustainable biological control of chestnut blight. Evol. Appl. 2014, 7, 580–596. [Google Scholar] [CrossRef] [PubMed]
- Jung, T.; Stukely, M.J.C.; Hardy, G.E.S.J.; White, D.; Paap, T.; Dunstan, W.A.; Burgess, T.I. Multiple new Phytophthora species from ITS Clade 6 associated with natural ecosystems in Australia: Evolutionary and ecological implications. Pers. Mol. Phylogeny Evol. Fungi 2011, 26, 13–39. [Google Scholar] [CrossRef]
- Thines, M.; Choi, Y.J. Evolution, diversity, and taxonomy of the Peronosporaceae, with focus on the genus Peronospora. Phytopathology 2016, 106, 6–18. [Google Scholar] [CrossRef]
- Cai, G.; Fry, W.E.; Hillman, B.I. PiRV-2 stimulates sporulation in Phytophthora infestans. Virus Res. 2019, 271, 197674. [Google Scholar] [CrossRef]
- Deakin, G.; Dobbs, E.; Bennett, J.M.; Jones, I.M.; Grogan, H.M.; Burton, K.S. Multiple viral infections in Agaricus bisporus-Characterisation of 18 unique RNA viruses and 8 ORFans identified by deep sequencing. Sci. Rep. 2017, 7, 2469. [Google Scholar] [CrossRef]
- Schardl, C.L.; Craven, K.D. Interspecific hybridization in plant-associated fungi and oomycetes: A review. Mol. Ecol. 2003, 12, 2861–2873. [Google Scholar] [CrossRef]
- Burgess, T.I. Molecular characterization of natural hybrids formed between five related indigenous clade 6 Phytophthora species. PLoS ONE 2015, 10, e0134225. [Google Scholar] [CrossRef]
- Jung, T.; Jung, M.H.; Scanu, B.; Seress, D.; Kovács, G.M.; Maia, C.; Pérez-Sierra, A.; Chang, T.T.; Chandelier, A.; Heungens, K.; et al. Six new Phytophthora species from ITS clade 7a including two sexually functional heterothallic hybrid species detected in natural ecosystems in Taiwan. Pers. Mol. Phylogeny Evol. Fungi 2017, 38, 100–135. [Google Scholar] [CrossRef]
 Fungi,
Fungi,  Nematoda,
Nematoda,  Oomycota,
Oomycota,  Arthropoda,
Arthropoda,  Plants,
Plants,  Mammalia,
Mammalia,  (Fishes) Chordata,
(Fishes) Chordata,  Ochrophyta (Heterokonta),
Ochrophyta (Heterokonta),  Excavata. Scale bars represent expected changes per site per branch.
Excavata. Scale bars represent expected changes per site per branch.
 Fungi,
Fungi,  Nematoda,
Nematoda,  Oomycota,
Oomycota,  Arthropoda,
Arthropoda,  Plants,
Plants,  Mammalia,
Mammalia,  (Fishes) Chordata,
(Fishes) Chordata,  Ochrophyta (Heterokonta),
Ochrophyta (Heterokonta),  Excavata. Scale bars represent expected changes per site per branch.
Excavata. Scale bars represent expected changes per site per branch.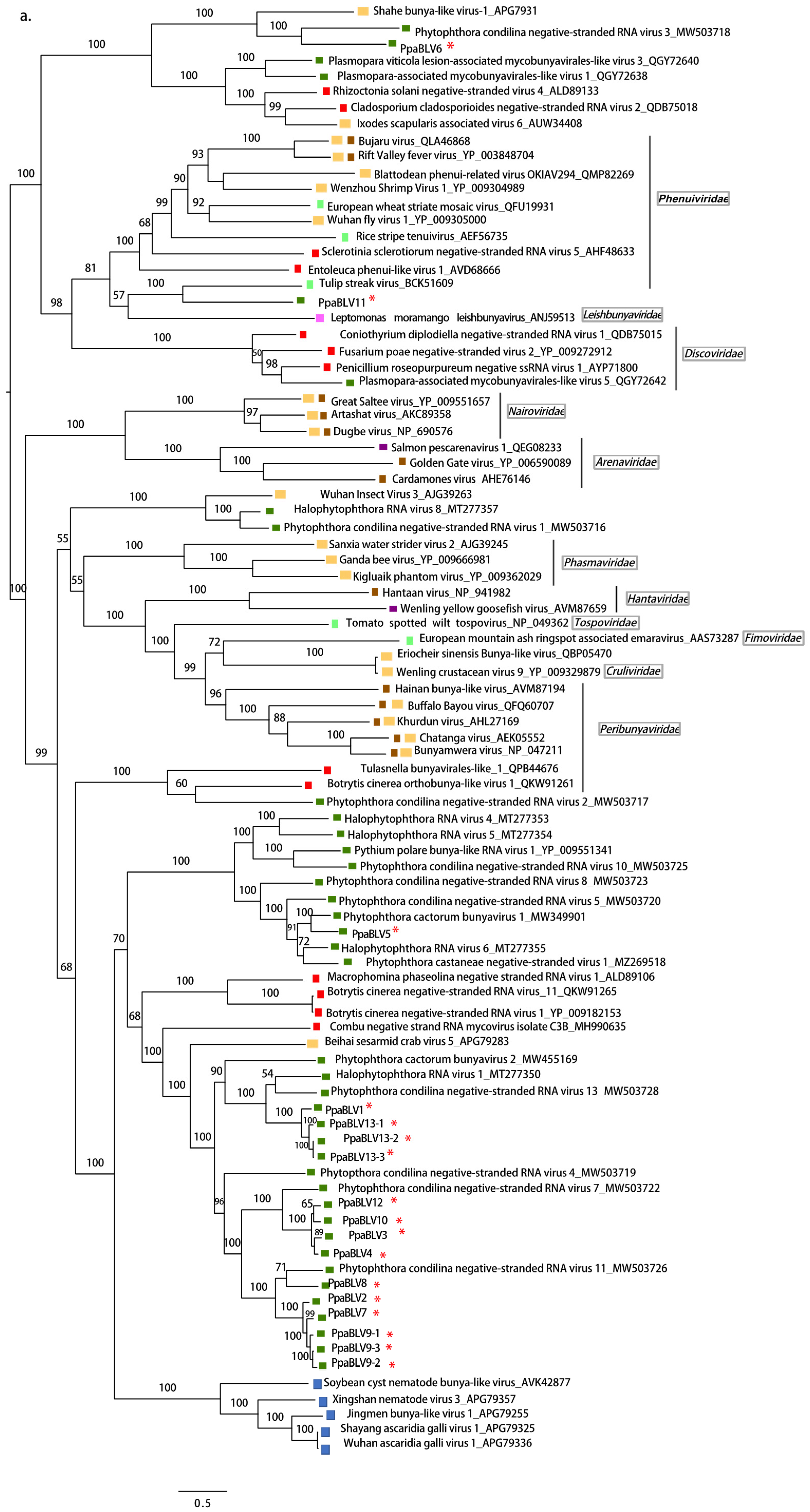

 Fungi,
Fungi,  Oomycota,
Oomycota,  Arthropoda,
Arthropoda,  Animalia,
Animalia,  Plants. Scale bar = 0.4 expected changes per site per branch.
Plants. Scale bar = 0.4 expected changes per site per branch.
 Fungi,
Fungi,  Oomycota,
Oomycota,  Arthropoda,
Arthropoda,  Animalia,
Animalia,  Plants. Scale bar = 0.4 expected changes per site per branch.
Plants. Scale bar = 0.4 expected changes per site per branch.
 Fungi,
Fungi,  Oomycota,
Oomycota,  Arthropoda,
Arthropoda,  Animalia,
Animalia,  Plants,
Plants,  Ochrophyta (Heterokonta),
Ochrophyta (Heterokonta),  Apicomplexa (Protists),
Apicomplexa (Protists),  Bacteria. Scale bar = 0.6 expected changes per site per branch.
Bacteria. Scale bar = 0.6 expected changes per site per branch.
 Fungi,
Fungi,  Oomycota,
Oomycota,  Arthropoda,
Arthropoda,  Animalia,
Animalia,  Plants,
Plants,  Ochrophyta (Heterokonta),
Ochrophyta (Heterokonta),  Apicomplexa (Protists),
Apicomplexa (Protists),  Bacteria. Scale bar = 0.6 expected changes per site per branch.
Bacteria. Scale bar = 0.6 expected changes per site per branch.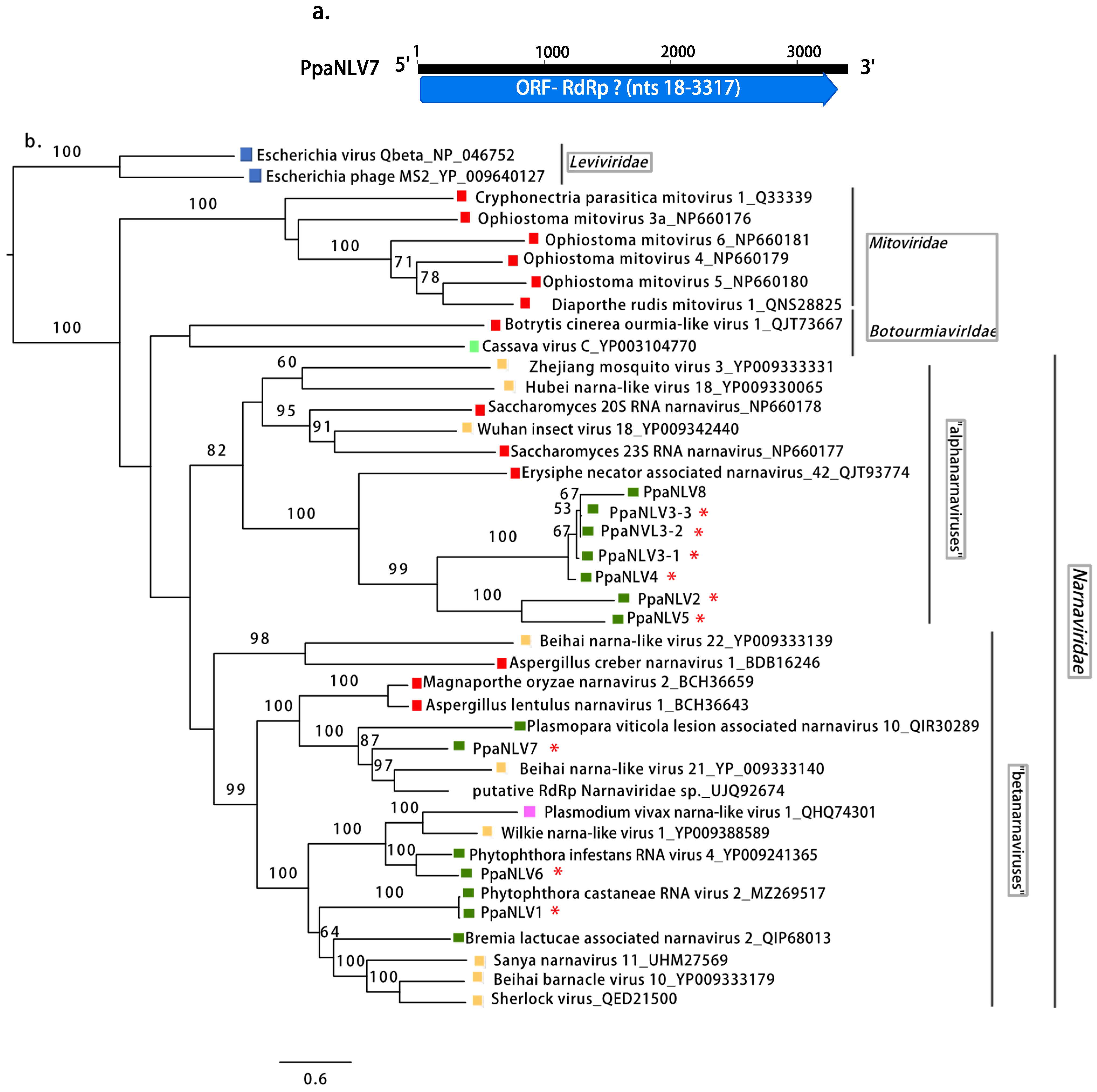
 Fungi,
Fungi,  Oomycota,
Oomycota,  Arthropoda,
Arthropoda,  Chordata,
Chordata,  Ochrophyta (Heterokonta),
Ochrophyta (Heterokonta),  Excavata. Scale bar = 0.6 expected changes per site per branch.
Excavata. Scale bar = 0.6 expected changes per site per branch.
 Fungi,
Fungi,  Oomycota,
Oomycota,  Arthropoda,
Arthropoda,  Chordata,
Chordata,  Ochrophyta (Heterokonta),
Ochrophyta (Heterokonta),  Excavata. Scale bar = 0.6 expected changes per site per branch.
Excavata. Scale bar = 0.6 expected changes per site per branch.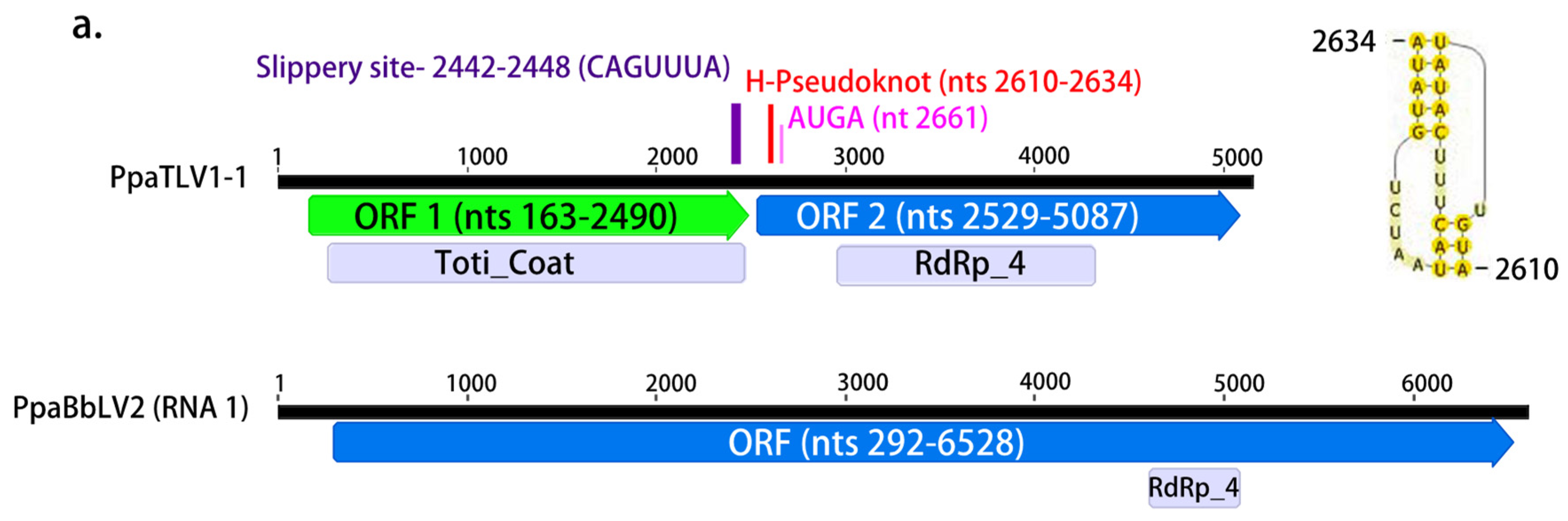

| Acronym * | N N | Genome | L L (bp) | Most Similar Virus in GenBank | E-Value | I (%) | QC (%) | CDD | CDD Position P | R (bp) R |
|---|---|---|---|---|---|---|---|---|---|---|
| PpaBbLV1-1 | OL795338 | dsRNA | 6680 | Plasmopara viticola lesion associated botybirnavirus 1 | 8.00 × 10−27 | 28.53 | 16 | RdRp_4 | 4815–5834 | 2729 |
| PpaBbLV1-2 | OL795339 | 6680 | Sclerotinia sclerotiorum botybirnavirus 3 | 3.00 × 10−31 | 27.94 | 18 | RdRp_4 | 4821–5834 | 1831 | |
| PpaBbLV1-3 | OL795340 | 6581 | Plasmopara viticola lesion associated botybirnavirus 1 | 3.00 × 10−32 | 29.59 | 17 | RdRp_4 | 4796–5809 | 6030 | |
| PpaBbLV2 | OL795341 | 6514 | Plasmopara viticola lesion associated botybirnavirus 1 | 4.00 × 10−32 | 29.13 | 18 | RdRp_4 | 4669–5808 | 2804 | |
| PpaBLV1 | OL795342 | (−)ssRNA | 9042 | Phytophthora condilina negative-stranded RNA virus 13 | 0.0 | 48.86 | 98 | Bunya_RdRp | 4123–5085 | 173,414 |
| N terminus | 7912–8166 | 7720 | ||||||||
| PpaBLV2 | OL795343 | 9024 | Phytophthora condilina negative-stranded RNA virus 11 | 0.0 | 56.29 | 97 | Bunya_RdRp | 4437–5114 | 114,896 | |
| PpaBLV3 | OL795344 | 9303 | Phytophthora condilina negative-stranded RNA virus 7 | 0.0 | 61.99 | 98 | Bunya_RdRp | 4338–5111 | 72,926 | |
| PpaBLV4 | OL795345 | 9302 | Phytophthora condilina negative-stranded RNA virus 7 | 0.0 | 56.27 | 98 | Bunya_RdRp | 4338–5828 | 43,584 | |
| PpaBLV5 | OL795346 | 8362 | Phytophthora cactorum bunyavirus 1 | 0.0 | 67.27 | 97 | Bunya_RdRp | 4082–5494 | 62,939 | |
| PpaBLV6 | OL795347 | 6396 | Phytophthora condilina negative-stranded RNA virus 3 | 0.0 | 33.97 | 84 | Bunya_RdRp | 2305–3342 | 10,639 | |
| PpaBLV7 | OL795348 | 9036 | Phytophthora condilina negative-stranded RNA virus 11 | 0.0 | 51.87 | 97 | Bunya_RdRp | n.d. | 47,152 | |
| PpaBLV8 | OL795349 | 9737 | Phytophthora condilina negative-stranded RNA virus 11 | 0.0 | 59.64 | 85 | Bunya_RdRp | 4091–5116 | 16,049 | |
| PpaBLV9-1 | OL795350 | 9016 | Phytophthora condilina negative-stranded RNA virus 11 | 0.0 | 52.06 | 98 | Bunya_RdRp | 4434–5045 | 59,989 | |
| PpaBLV9-2 | OL795351 | 9047 | Phytophthora condilina negative-stranded RNA virus 11 | 0.0 | 52.46 | 97 | Bunya_RdRp | n.d. | 271,404 | |
| PpaBLV9-3 | OL795352 | 9015 | Phytophthora condilina negative-stranded RNA virus 11 | 0.0 | 52.29 | 98 | Bunya_RdRp | n.d. | 93,848 | |
| PpaBLV10 | OL795353 | 9417 | Phytophthora condilina negative-stranded RNA virus 7 | 0.0 | 57.27 | 98 | Bunya_RdRp | 4451–5224 | 146,562 | |
| PpaBLV11 | OL795355 | 6448 | Tulip streak virus | 1.00 × 10−133 | 27.69 | 67 | Bunya_RdRp | 2443–4482 | 223,284 | |
| OL795354 | 878 | Wuhan Fly virus 1 | 1.00 × 10−06 | 28.29 | 54 | Tenui-Phenui NC | 210–806 | 154,294 | ||
| PpaBLV12 | OL795357 | 9386 | Phytophthora condilina negative-stranded RNA virus 7 | 0.0 | 55.64 | 99 | Bunya_RdRp | n.d. | 29,824 | |
| PpaBLV13-1 | OL795358 | 8997 | Phytophthora condilina negative-stranded RNA virus 13 | 0.0 | 50.63 | 95 | Bunya_RdRp | 3982–5055 | 14,451 | |
| PpaBLV13-2 | OL795359 | 8997 | Phytophthora condilina negative-stranded RNA virus 13 | 0.0 | 50.58 | 95 | Bunya_RdRp | 3982–5055 | 10,073 | |
| PpaBLV13-3 | OL795360 | 8995 | Phytophthora condilina negative-stranded RNA virus 13 | 0.0 | 50.54 | 95 | Bunya_RdRp | 3980–5053 | 9885 | |
| PpaTbLV1 | OL795371 | (+)ssRNA | 4324 | RdRp [Riboviria sp.] QDH88579 | 1.00 × 10−139 | 52.09 | 33 | RdRp_3 | 2042–2845 | 173,414 |
| RNA_dep_RNAP | 2060–2854 | |||||||||
| PpaNV1 | OL795361 | (+)ssRNA | 2818 | Phytophthora castaneae RNA virus 2 | 0.0 | 97.01 | 90 | RdRp | n.d. | 135 |
| PpaNV2 | OL795362 | 2545 | Erysiphe-necator-associated narnavirus 42 | 3.00 × 10−31 | 33.33 | 37 | RdRp | 243,122 | ||
| PpaNV3-1 | OL795363 | 2725 | Erysiphe-necator-associated narnavirus 42 | 3.00 × 10−21 | 30.48 | 38 | RdRp | 138404 | ||
| PpaNV3-2 | OL795364 | 2719 | Erysiphe-necator-associated narnavirus 42 | 3.00 × 10−22 | 31.02 | 39 | RdRp | 206,048 | ||
| PpaNV3-3 | OL795365 | 2736 | Erysiphe-necator-associated narnavirus 42 | 2.00 × 10−22 | 31.28 | 38 | RdRp | 266,624 | ||
| PpaNV4 | OL795366 | 2761 | Erysiphe-necator-associated narnavirus 42 | 1.00 × 10−19 | 33.33 | 27 | RdRp | 916,928 | ||
| PpaNV5 | OL795367 | 2576 | Erysiphe-necator-associated narnavirus 42 | 1.00 × 10−31 | 37.34 | 28 | RdRp | 104,172 | ||
| PpaNV6 U | OL795370 | 1344 | Phytophthora infestans RNA virus 4 | 4.00 × 10−149 | 59.45 | 97 | RdRp | 162 | ||
| PpaNV7 | OL795368 | 3381 | Beihai narna-like virus 21 | 0.0 | 37.63 | 91 | RdRp | 125,312 | ||
| PpaNV8 U | OL795369 | 916 | Erysiphe-necator-associated narnavirus 42 | 1.00 × 10−11 | 51.19 | 27 | RdRp | 46,432 | ||
| PpaTLV1-1 | OL795372 | dsRNA | 5134 | Drosophila-associated totivirus 2 | 6.00 × 10−171 | 38.63 | 46 | RdRp_4 | 2958–4325 | 5030 |
| Totivirus_coat | 283–2487 | |||||||||
| PpaTLV1-2 | OL795373 | 5199 | Drosophila-associated totivirus 2 | 8.00 × 10−171 | 38.63 | 46 | RdRp_4 | 3023–4390 | 5464 | |
| Totivirus_coat | 348–2552 | |||||||||
| PpaTLV2-1 | OL795374 | 5183 | Red algae totivirus 1 | 2.00 × 10−162 | 37.56 | 47 | RdRp_4 | 3026–4381 | 4044 | |
| Totivirus_coat | 363–2624 | |||||||||
| PpaTLV2-2 | OL795375 | 5183 | Red algae totivirus 1 | 5.00 × 10−152 | 38.31 | 47 | RdRp_4 | 3026–4381 | 2962 | |
| Totivirus_coat | 363–2624 | |||||||||
| PpaTLV2-3 | OL795376 | 5183 | Red algae totivirus 1 | 1.00 × 10−151 | 38.23 | 47 | RdRp_4 | 3026–4381 | 2504 | |
| Totivirus_coat | 363–2624 | |||||||||
| PpaTLV3-1 | OL795377 | 4491 | Conidiobolus heterosporus totivirus 1 | 0.0 | 42.03 | 49 | RdRp_4 | 2548–3822 | 7076 | |
| PpaTLV3-2 | OL795378 | 4491 | Wuhan insect virus 26 | 0.0 | 41.34 | 49 | RdRp_4 | 2623–3822 | 2174 | |
| PpaTLV3-3 | OL795379 | 4495 | Wuhan insect virus 26 | 0.0 | 41.03 | 49 | RdRp_4 | 2627–3826 | 3750 | |
| PpaTLV4 U | OL795380 | 1553 | Diatom-colony-associated dsRNA virus 17 genome type B | 6.00 × 10−28 | 32.59 | 57 | RdRp_4 | 41–784 | 9026 | |
| PpaTLV5-1 | OL795381 | 5188 | Diatom-colony-associated dsRNA virus 11 | 8.00 × 10−149 | 38.41 | 47 | RdRp_4 | 2969–4375 | 7926 | |
| Totivirus_coat | 486–2672 | 1546 | ||||||||
| PpaTLV5-2 | OL795382 | 5199 | Diatom-colony-associated dsRNA virus 7 | 5.00 × 10−148 | 39.47 | 43 | RdRp_4 | 3019–4386 | ||
| Totivirus_coat | 497–2554 | 8402 | ||||||||
| PpaTLV6 | OL795383 | 5312 | Diatom-colony-associated dsRNA virus 11 | 1.00 × 10−94 | 35.22 | 37 | RdRp_4 | 3118–4497 | ||
| Totivirus_coat | 389–2566 | 17,680 | ||||||||
| PpaTLV7 | OL795384 | 5703 | Diatom-colony-associated dsRNA virus 17 genome type B | 2.00 × 10−36 | 27.67 | 27 | RdRp_4 | 3663–4625 | ||
| PpaTLV8 U | OL795385 | 2964 | Diatom-colony-associated dsRNA virus 17 genome type A | 4.00 × 10−35 | 27.29 | RdRp_4 | 1216–2178 | 4168 | ||
| PpaTLV9 | OL795386 | 5268 | Diatom-colony-associated dsRNA virus 11 | 0.0 | 48.39 | 52 | RdRp_4 | 3075–4457 | 12,384 | |
| Totivirus_coat | 439–2550 | |||||||||
| PpaTLV10 | OL795387 | 5205 | Red algae totivirus 1 | 3.00 × 10−132 | 36.31 | 41 | RdRp_4 | 2979–4376 | 13,180 | |
| Totivirus_coat | 313–2355 | |||||||||
| PpaTLV11 | OL795388 | 5408 | Red algae totivirus 1 | 6.00 × 10−165 | 40.69 | 39 | RdRp_4 | 3219–4583 | 18,930 | |
| Totivirus_coat | 403–2424 | |||||||||
| PpaTLV12-1 | OL795389 | 4825 | Plasmopara viticola lesion associated totivirus-like 1 | 0.0 | 46.10 | 44 | RdRp_4 | 3030–3737 | 1786 | |
| PpaTLV12-2 | OL795390 | 5833 | Plasmopara viticola lesion associated totivirus-like 1 | 6.00 × 10−179 | 45.96 | 36 | RdRp_4 | 4034–4741 | 13,412 | |
| PpaTLV12-3 | OL795391 | 5833 | Plasmopara viticola lesion associated totivirus-like 1 | 8.00 × 10−178 | 45.54 | 36 | RdRp_4 | 4046–4753 | 15,479 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Botella, L.; Jung, M.H.; Rost, M.; Jung, T. Natural Populations from the Phytophthora palustris Complex Show a High Diversity and Abundance of ssRNA and dsRNA Viruses. J. Fungi 2022, 8, 1118. https://doi.org/10.3390/jof8111118
Botella L, Jung MH, Rost M, Jung T. Natural Populations from the Phytophthora palustris Complex Show a High Diversity and Abundance of ssRNA and dsRNA Viruses. Journal of Fungi. 2022; 8(11):1118. https://doi.org/10.3390/jof8111118
Chicago/Turabian StyleBotella, Leticia, Marília Horta Jung, Michael Rost, and Thomas Jung. 2022. "Natural Populations from the Phytophthora palustris Complex Show a High Diversity and Abundance of ssRNA and dsRNA Viruses" Journal of Fungi 8, no. 11: 1118. https://doi.org/10.3390/jof8111118
APA StyleBotella, L., Jung, M. H., Rost, M., & Jung, T. (2022). Natural Populations from the Phytophthora palustris Complex Show a High Diversity and Abundance of ssRNA and dsRNA Viruses. Journal of Fungi, 8(11), 1118. https://doi.org/10.3390/jof8111118








