Discovery of Oleaginous Yeast from Mountain Forest Soil in Thailand
Abstract
1. Introduction
2. Materials and Methods
2.1. Yeast Isolation
2.2. Identification of Yeast
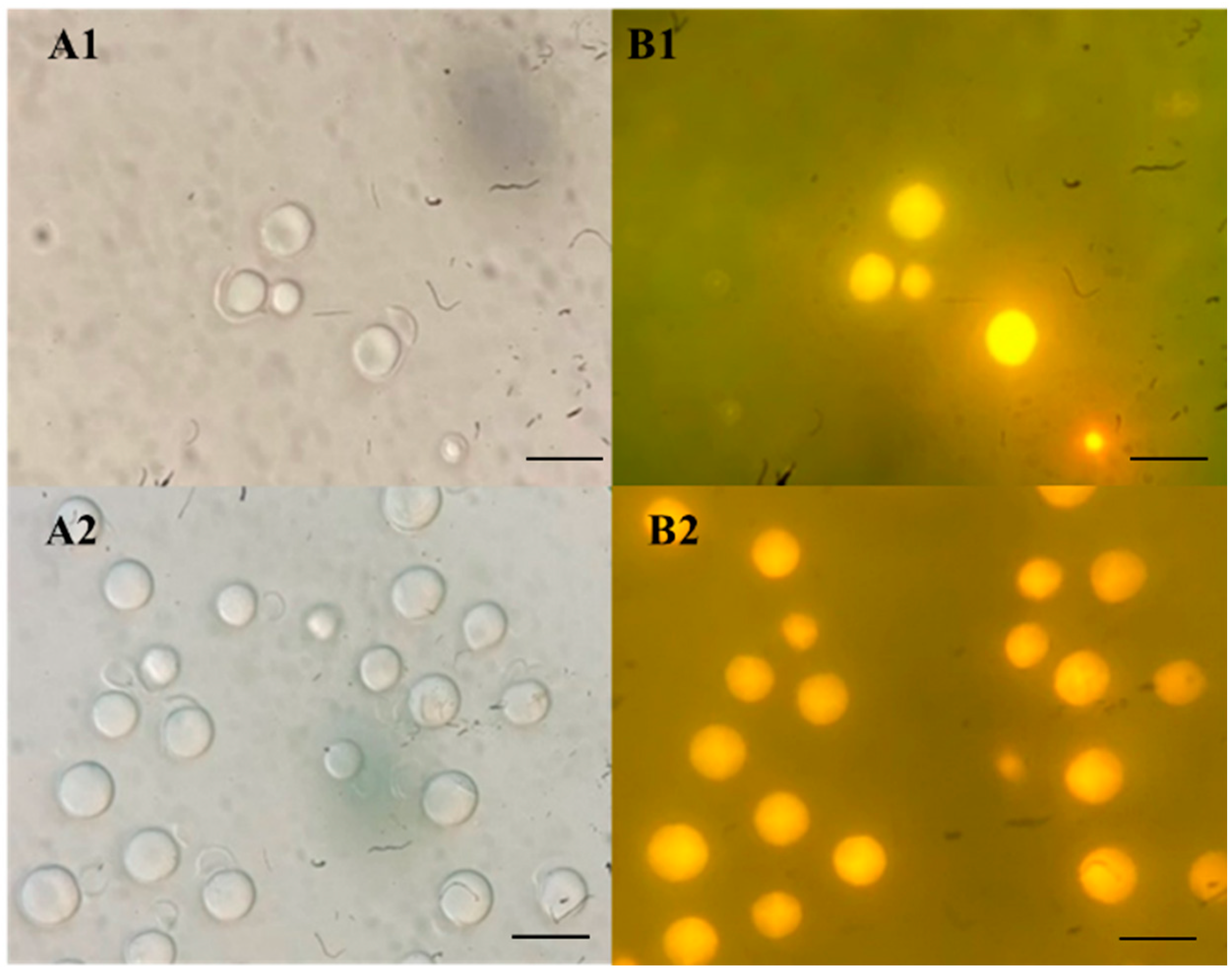
2.3. Screening for Oleaginous Yeast Strains
2.4. Analytical Methods
2.4.1. Biomass Analysis
2.4.2. Lipid Content and Fatty Acid Analyses
2.5. Statistical Analysis
3. Results and Discussion
3.1. Isolation and Identification of Yeasts
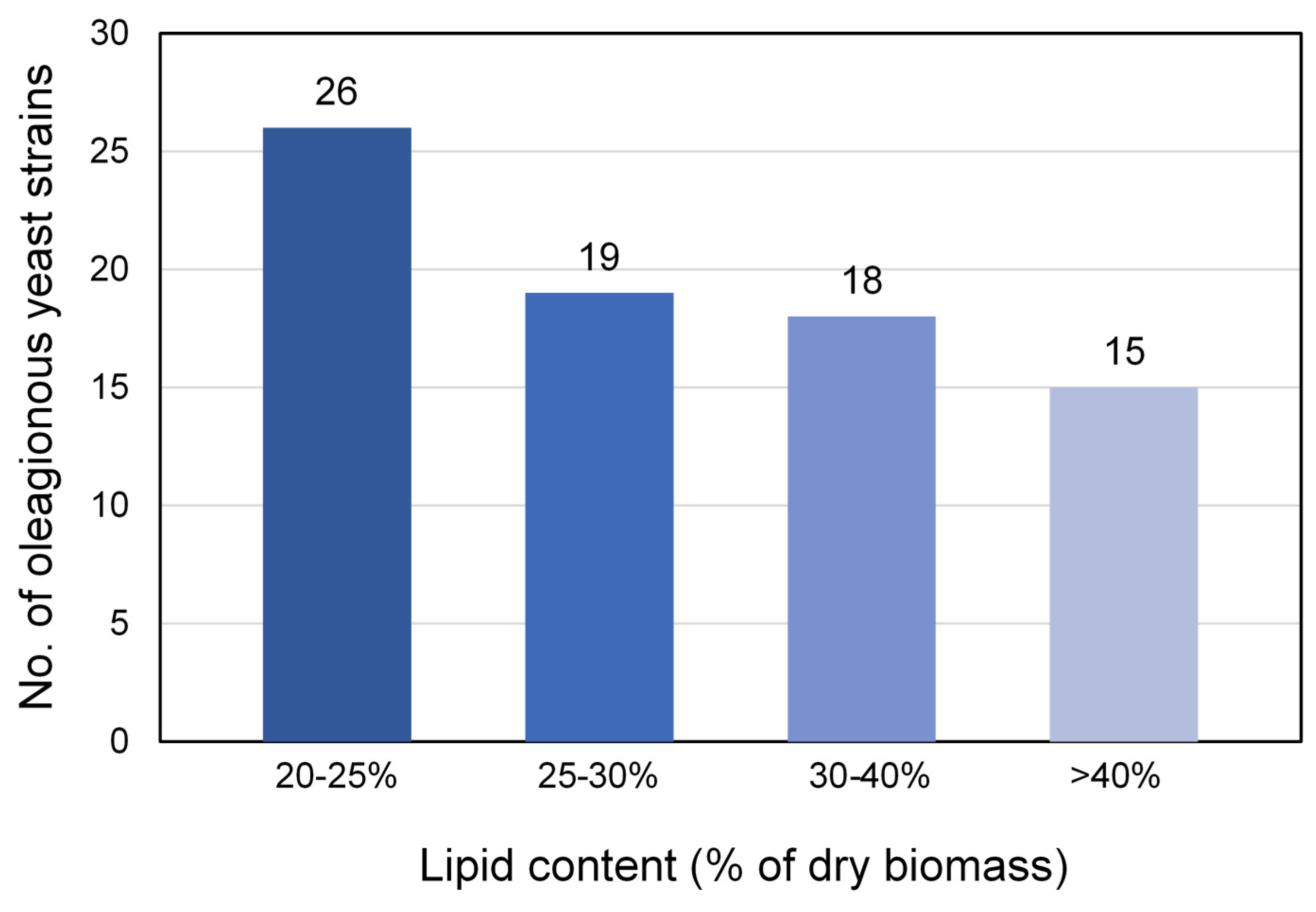
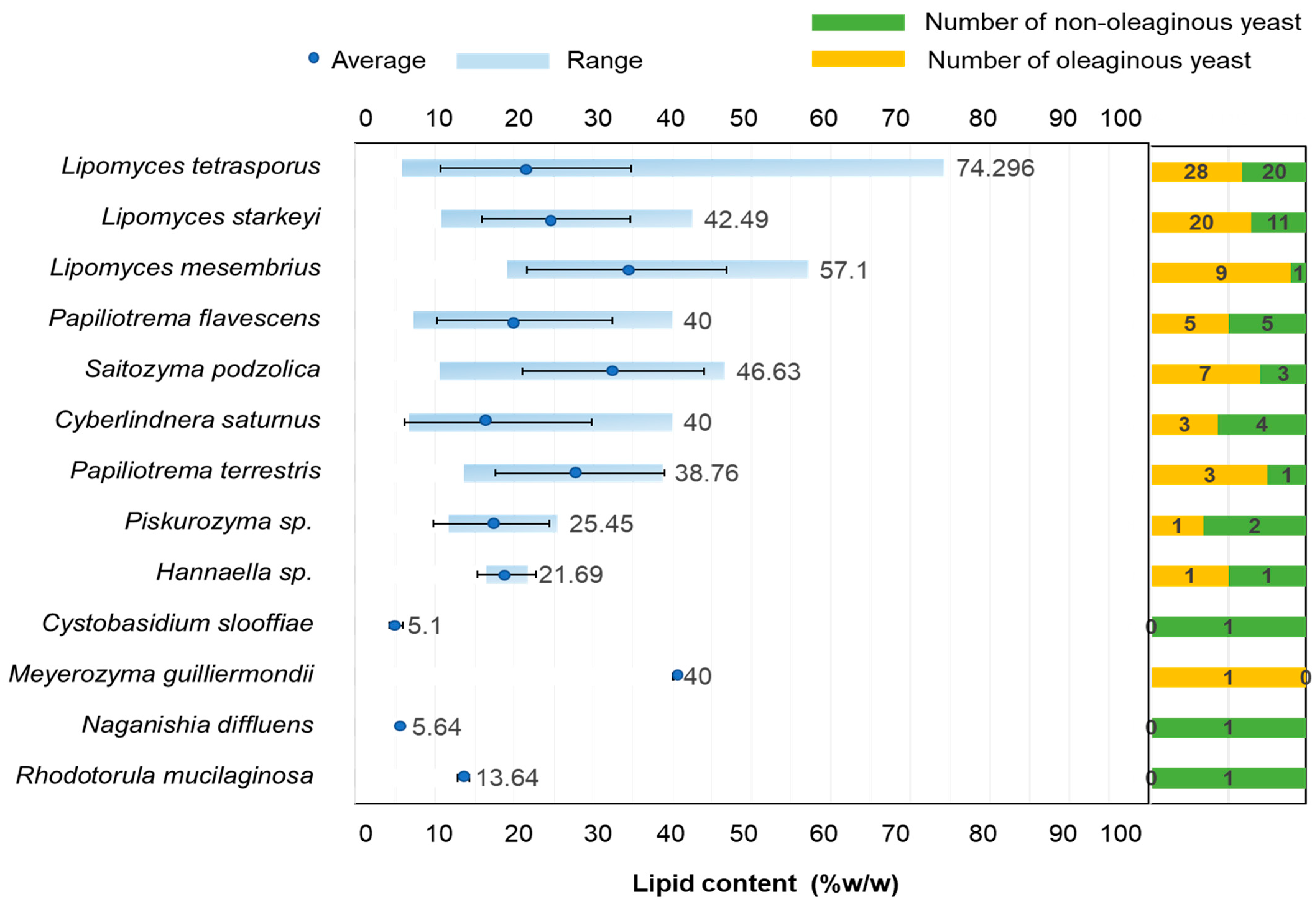
3.2. Screening for Oleaginous Yeast Strains
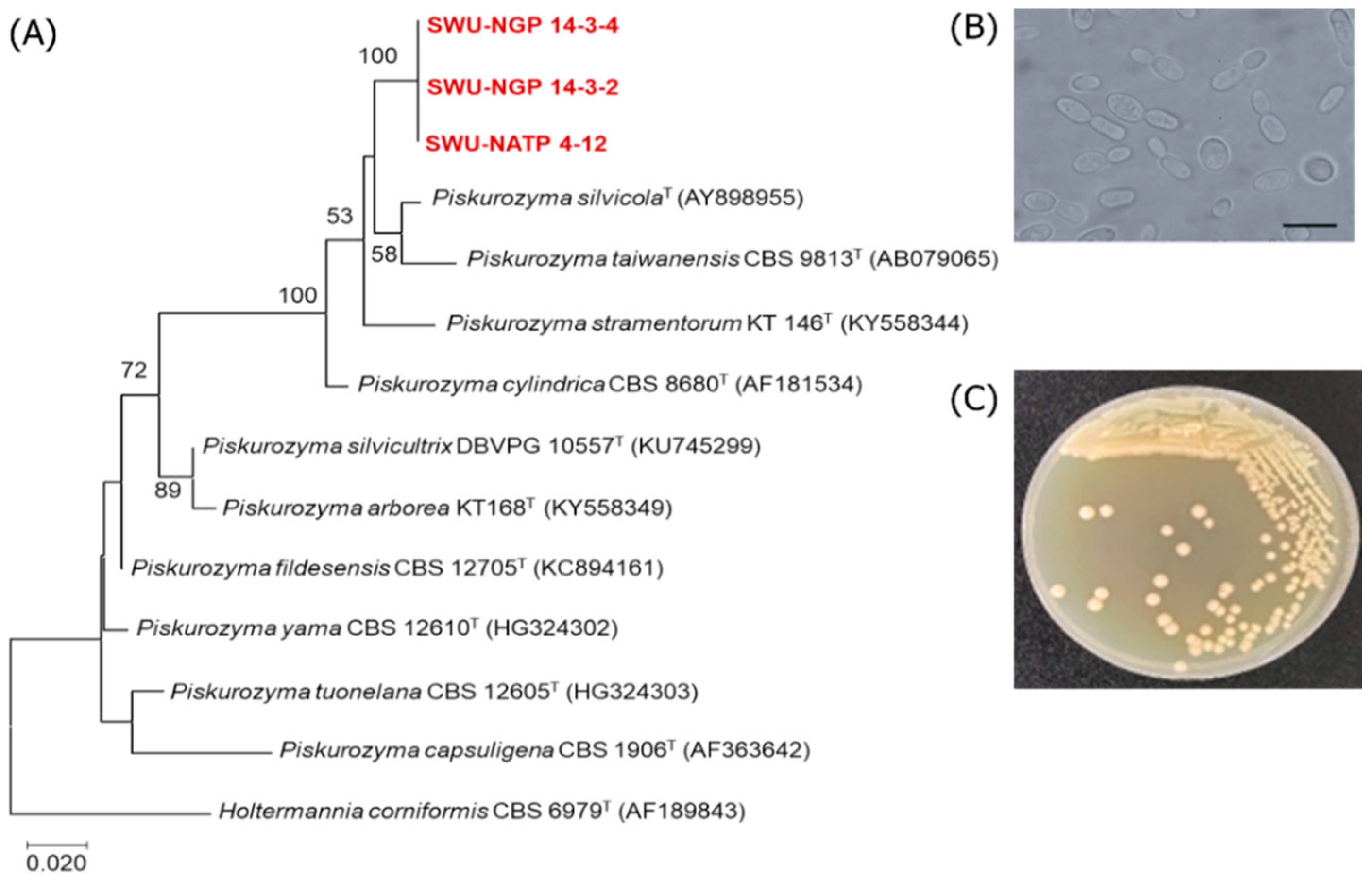
3.3. Discovery of Additional Oleaginous Yeast Species
3.4. Fatty Acid Composition Profiles
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Mohanty, A.K.; Misra, M.; Drzal, L. Sustainable bio-composites from renewable resources: Opportunities and challenges in the green materials world. J. Polym. Environ. 2002, 10, 19–26. [Google Scholar] [CrossRef]
- Singh, R.L.; Singh, P.K. Global environmental problems. In Principles and Applications of Environmental Biotechnology for a Sustainable Future; Springer: Singapore, 2017; pp. 13–41. [Google Scholar]
- Hoffmann, H.K. Bioenergy, development and food security in Sub-Saharan Africa. Qualifikationsarbeiten 2016, 67–73. [Google Scholar] [CrossRef]
- Anuar, M.R.; Abdullah, A.Z. Challenges in biodiesel industry with regards to feedstock, environmental, social and sustainability issues: A critical review. Renew. Sustain. Energy Rev. 2016, 58, 208–223. [Google Scholar] [CrossRef]
- Uthandi, S.; Kaliyaperumal, A.; Srinivasan, N.; Thangavelu, K.; Muniraj, I.K.; Zhan, X.; Gathergood, N.; Gupta, V.K. Microbial biodiesel production from lignocellulosic biomass: New insights and future challenges. Crit. Rev. Environ. Sci. Technol. 2022, 52, 2197–2225. [Google Scholar] [CrossRef]
- Szczepańska, P.; Hapeta, P.; Lazar, Z. Advances in production of high-value lipids by oleaginous yeasts. Crit. Rev. Biotechnol. 2022, 42, 1–22. [Google Scholar] [CrossRef] [PubMed]
- Shi, S.; Zhao, H. Metabolic engineering of oleaginous yeasts for production of fuels and chemicals. Front. Microbiol. 2017, 8, 1–16. [Google Scholar] [CrossRef]
- Ratledge, C. Microorganisms for lipids. Acta Biotechnol. 1991, 11, 429–438. [Google Scholar] [CrossRef]
- Sajish, S.; Singh, S.; Nain, L. Yeasts for Single Cell Oil Production from Non-conventional Bioresources. In Microbial Biotechnology for Renewable and Sustainable Energy; Springer: Singapore, 2022; pp. 337–364. [Google Scholar]
- Evans, C.T.; Ratledge, C. Influence of nitrogen metabolism on lipid accumulation by Rhodosporidium toruloides CBS 14. Microbiology 1984, 130, 1705–1710. [Google Scholar] [CrossRef][Green Version]
- Montet, D.; Ratomahenina, R.; Galzy, P.; Pina, M.; Graille, J. A study of the influence of the growth media on the fatty acid composition in Candida lipolytica diddens and lodder. Biotechnol. Lett. 1985, 7, 733–736. [Google Scholar] [CrossRef]
- Abeln, F.; Chuck, C.J. The history, state of the art and future prospects for oleaginous yeast research. Microb. Cell Factories 2021, 20, 1–31. [Google Scholar] [CrossRef]
- Castanha, R.F.; Mariano, A.P.; Morais, L.A.S.d.; Scramin, S.; Monteiro, R.T.R. Optimization of lipids production by Cryptococcus laurentii 11 using cheese whey with molasses. Braz. J. Microbiol. 2014, 45, 379–387. [Google Scholar] [CrossRef] [PubMed]
- Kurtzman, C.P.; Fell, J.W.; Boekhout, T. The Yeasts: A Taxonomic Study; Elsevier: London, UK, 2011. [Google Scholar]
- Ratledge, C.; Cohen, Z. Microbial and algal oils: Do they have a future for biodiesel or as commodity oils? Lipid Technol. 2008, 20, 155–160. [Google Scholar] [CrossRef]
- Schulze, I.; Hansen, S.; Großhans, S.; Rudszuck, T.; Ochsenreither, K.; Syldatk, C.; Neumann, A. Characterization of newly isolated oleaginous yeasts-Cryptococcus podzolicus, Trichosporon porosum and Pichia segobiensis. AMB Express 2014, 4, 1–11. [Google Scholar] [CrossRef]
- Kitcha, S.; Cheirsilp, B. Screening of oleaginous yeasts and optimization for lipid production using crude glycerol as a carbon source. Energy Procedia 2011, 9, 274–282. [Google Scholar] [CrossRef]
- Kanti, A.; Sukara, E.; Latifah, K.; Sukarno, N.; Boundy-Mills, K. Indonesian oleaginous yeasts isolated from Piper betle and P. nigrum. Mycosphere 2013, 4, 363–454. [Google Scholar] [CrossRef]
- Pan, L.-X.; Yang, D.-F.; Shao, L.; Li, W.; Chen, G.-G.; Liang, Z.-Q. Isolation of the oleaginous yeasts from the soil and studies of their lipid-producing capacities. Food Technol. Biotechnol. 2009, 47, 215–220. [Google Scholar]
- Papanikolaou, S.; Aggelis, G. Lipids of oleaginous yeasts. Part I: Biochemistry of single cell oil production. Eur. J. Lipid Sci. Technol. 2011, 113, 1031–1051. [Google Scholar] [CrossRef]
- Ageitos, J.M.; Vallejo, J.A.; Veiga-Crespo, P.; Villa, T.G. Oily yeasts as oleaginous cell factories. Appl. Microbiol. Biotechnol. 2011, 90, 1219–1227. [Google Scholar] [CrossRef]
- Miranda, C.; Bettencourt, S.; Pozdniakova, T.; Pereira, J.; Sampaio, P.; Franco-Duarte, R.; Pais, C. Modified high-throughput Nile red fluorescence assay for the rapid screening of oleaginous yeasts using acetic acid as carbon source. BMC Microbiol. 2020, 20, 1–11. [Google Scholar] [CrossRef]
- Bettencourt, S.; Miranda, C.; Pozdniakova, T.A.; Sampaio, P.; Franco-Duarte, R.; Pais, C. Single cell oil production by oleaginous yeasts grown in synthetic and waste-derived volatile fatty acids. Microorganisms 2020, 8, 1809. [Google Scholar] [CrossRef]
- Wang, R.; Wang, J.; Xu, R.; Fang, Z.; Liu, A. Oil production by the oleaginous yeast Lipomyces starkeyi using diverse carbon sources. BioResources 2014, 9, 7027–7040. [Google Scholar] [CrossRef]
- Lin, J.; Shen, H.; Tan, H.; Zhao, X.; Wu, S.; Hu, C.; Zhao, Z.K. Lipid production by Lipomyces starkeyi cells in glucose solution without auxiliary nutrients. J. Biotechnol. 2011, 152, 184–188. [Google Scholar] [CrossRef] [PubMed]
- Lin, X.; Liu, S.; Bao, R.; Gao, N.; Zhang, S.; Zhu, R.; Zhao, Z.K. Development of an Agrobacterium-mediated transformation method and evaluation of two exogenous constitutive promoters in oleaginous yeast Lipomyces starkeyi. Appl. Biochem. Biotechnol. 2017, 183, 867–875. [Google Scholar] [CrossRef] [PubMed]
- Sutanto, S.; Zullaikah, S.; Tran-Nguyen, P.L.; Ismadji, S.; Ju, Y.-H. Lipomyces starkeyi: Its current status as a potential oil producer. Fuel Process. Technol. 2018, 177, 39–55. [Google Scholar] [CrossRef]
- Yurkov, A.M. Yeasts of the soil–obscure but precious. Yeast 2018, 35, 369–378. [Google Scholar] [CrossRef] [PubMed]
- Smith, M.T.; Kurtzman, C.P. Chapter 43-Lipomyces Lodder & Kreger-van Rij (1952). In The Yeasts, 5th ed.; Kurtzman, C.P., Fell, J.W., Boekhout, T., Eds.; Elsevier: London, UK, 2011; pp. 545–560. [Google Scholar]
- Yamazaki, A.; Lorliam, W.; Kawasaki, H.; Uchino, M.; Suzuki, K.-i. Fourteen novel Lipomycetaceous yeast species isolated from soil in Japan and transfer of Dipodascopsis anomala to the genus Babjevia based on ascospore production phenotype. Int. J. Syst. Evol. Microbiol. 2020, 70, 1372–1397. [Google Scholar] [CrossRef]
- Sawangkeaw, R.; Ngamprasertsith, S. A review of lipid-based biomasses as feedstocks for biofuels production. Renew. Sustain. Energy Rev. 2013, 25, 97–108. [Google Scholar] [CrossRef]
- Botha, A. Yeasts in soil. In Biodiversity and Ecophysiology of Yeasts; Péter, G., Rosa, C., Eds.; Springer: Berlin/Heidelberg, Germany, 2006; pp. 221–240. [Google Scholar]
- Polburee, P.; Yongmanitchai, W.; Lertwattanasakul, N.; Ohashi, T.; Fujiyama, K.; Limtong, S. Characterization of oleaginous yeasts accumulating high levels of lipid when cultivated in glycerol and their potential for lipid production from biodiesel-derived crude glycerol. Fungal Biol. 2015, 119, 1194–1204. [Google Scholar] [CrossRef]
- Hoondee, P.; Wattanagonniyom, T.; Weeraphan, T.; Tanasupawat, S.; Savarajara, A. Occurrence of oleaginous yeast from mangrove forest in Thailand. World J. Microbiol. Biotechnol. 2019, 35, 108. [Google Scholar] [CrossRef]
- Planonth, S.; Chantarasiri, A. The oleaginous yeast Pichia manshurica isolated from Lansium domesticum fruit in Thailand and its fatty acid composition of single cell oil. Biodiversitas J. Biol. Divers. 2022, 23, 801–809. [Google Scholar] [CrossRef]
- Poontawee, R.; Yongmanitchai, W.; Limtong, S. Efficient oleaginous yeasts for lipid production from lignocellulosic sugars and effects of lignocellulose degradation compounds on growth and lipid production. Process Biochem. 2017, 53, 44–60. [Google Scholar] [CrossRef]
- Ngamsirisomsakul, M.; Reungsang, A.; Kongkeitkajorn, M.B. Assessing oleaginous yeasts for their potentials on microbial lipid production from sugarcane bagasse and the effects of physical changes on lipid production. Bioresour. Technol. Rep. 2021, 14, 100650. [Google Scholar] [CrossRef]
- Kunthiphun, S.; Chokreansukchai, P.; Hondee, P.; Tanasupawat, S.; Savarajara, A. Diversity and characterization of cultivable oleaginous yeasts isolated from mangrove forests. World J. Microbiol. Biotechnol. 2018, 34, 125. [Google Scholar] [CrossRef] [PubMed]
- Kurtzman, C.P.; Robnett, C.J. Identification and phylogeny of ascomycetous yeasts from analysis of nuclear large subunit (26S) ribosomal DNA partial sequences. Antonie Van Leeuwenhoek 1998, 73, 331–371. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 2018, 35, 1547. [Google Scholar] [CrossRef]
- Fell, J.W.; Boekhout, T.; Fonseca, A.; Scorzetti, G.; Statzell-Tallman, A. Biodiversity and systematics of basidiomycetous yeasts as determined by large-subunit rDNA D1/D2 domain sequence analysis. Int. J. Syst. Evol. Microbiol. 2000, 50, 1351–1371. [Google Scholar] [CrossRef]
- Franco-Duarte, R.; Mendes, I.; Gomes, A.C.; Santos, M.A.; de Sousa, B.; Schuller, D. Genotyping of Saccharomyces cerevisiae strains by interdelta sequence typing using automated microfluidics. Electrophoresis 2011, 32, 1447–1455. [Google Scholar] [CrossRef]
- Yamazaki, A.; Kanti, A.; Kawasaki, H. Three novel lipomycetaceous yeasts, Lipomyces maratuensis sp. nov., Lipomyces tropicalis sp. nov., and Lipomyces kalimantanensis f.a., sp. nov. isolated from soil from the Maratua and Kalimantan Islands, Indonesia. Mycoscience 2017, 58, 413–423. [Google Scholar] [CrossRef]
- Wierzchowska, K.; Zieniuk, B.; Nowak, D.; Fabiszewska, A. Phosphorus and Nitrogen Limitation as a Part of the Strategy to Stimulate Microbial Lipid Biosynthesis. Appl. Sci. 2021, 11, 11819. [Google Scholar] [CrossRef]
- Bligh, E.G.; Dyer, W.J. A rapid method of total lipid extraction and purification. Can. J. Biochem. Physiol. 1959, 37, 911–917. [Google Scholar] [CrossRef]
- Holub, B.; Bakker, D.; Skeaff, C. Alterations in molecular species of cholesterol esters formed via plasma lecithin-cholesterol acyltransferase in human subjects consuming fish oil. Atherosclerosis 1987, 66, 11–18. [Google Scholar] [CrossRef]
- Starmer, W.T.; Lachance, M.-A. Chapter 6-Yeast Ecology. In The Yeasts, 5th ed.; Kurtzman, C.P., Fell, J.W., Boekhout, T., Eds.; Elsevier: London, UK, 2011; pp. 65–83. [Google Scholar]
- Li, A.H.; Yuan, F.X.; Groenewald, M.; Bensch, K.; Yurkov, A.M.; Li, K.; Han, P.J.; Guo, L.D.; Aime, M.C.; Sampaio, J.P.; et al. Diversity and phylogeny of basidiomycetous yeasts from plant leaves and soil: Proposal of two new orders, three new families, eight new genera and one hundred and seven new species. Stud. Mycol. 2020, 96, 17–140. [Google Scholar] [CrossRef]
- Glushakova, A.; Kachalkin, A.; Tiunov, A.; Chernov, I.Y. Distribution of yeast complexes in the profiles of different soil types. Eurasian Soil Sci. 2017, 50, 820–825. [Google Scholar] [CrossRef]
- Boekhout, T.; Amend, A.S.; El Baidouri, F.; Gabaldón, T.; Geml, J.; Mittelbach, M.; Robert, V.; Tan, C.S.; Turchetti, B.; Vu, D. Trends in yeast diversity discovery. Fungal Divers. 2022, 114, 491–537. [Google Scholar] [CrossRef]
- Liu, Z.; Li, X.; Sun, Z.; Wang, Z.; Li, G. Papiliotrema flavescens colonized in biochars inhibits wheat crown rot and Fusarium head blight. Biochar 2021, 3, 625–639. [Google Scholar] [CrossRef]
- Crestani, J.; Fontes Landell, M.; Faganello, J.; Henning Vainstein, M.; Simpson Vishniac, H.; Valente, P. Cryptococcus terrestris sp. nov., a tremellaceous, anamorphic yeast phylogenetically related to Cryptococcus flavescens. Int. J. Syst. Evol. Microbiol. 2009, 59, 631–636. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Junyapate, K.; Jindamorakot, S.; Limtong, S. Yamadazyma ubonensis fa, sp. nov., a novel xylitol-producing yeast species isolated in Thailand. Antonie Van Leeuwenhoek 2014, 105, 471–480. [Google Scholar] [CrossRef] [PubMed]
- Moradian, J.M.; Xu, Z.-A.; Shi, Y.-T.; Fang, Z.; Yong, Y.-C. Efficient biohydrogen and bioelectricity production from xylose by microbial fuel cell with newly isolated yeast of Cystobasidium slooffiae. Int. J. Energy Res. 2020, 44, 325–333. [Google Scholar] [CrossRef]
- Prajapati, V.D.; Jani, G.K.; Zala, B.S.; Khutliwala, T.A. An insight into the emerging exopolysaccharide gellan gum as a novel polymer. Carbohydr. Polym. 2013, 93, 670–678. [Google Scholar] [CrossRef]
- Zhang, L.; Du, J.; Jiang, Y. Low acyl gellan gum as a gelling agent in medium of Saccharomyces yeasts. Int. J. Food Eng. 2018, 14, 20170292. [Google Scholar] [CrossRef]
- Tamaki, H.; Hanada, S.; Sekiguchi, Y.; Tanaka, Y.; Kamagata, Y. Effect of gelling agent on colony formation in solid cultivation of microbial community in lake sediment. Environ. Microbiol. 2009, 11, 1827–1834. [Google Scholar] [CrossRef] [PubMed]
- Das, N.; Tripathi, N.; Basu, S.; Bose, C.; Maitra, S.; Khurana, S. Progress in the development of gelling agents for improved culturability of microorganisms. Front. Microbiol. 2015, 6, 698. [Google Scholar] [CrossRef] [PubMed]
- Fujitani, H.; Kumagai, A.; Ushiki, N.; Momiuchi, K.; Tsuneda, S. Selective isolation of ammonia-oxidizing bacteria from autotrophic nitrifying granules by applying cell-sorting and sub-culturing of microcolonies. Front. Microbiol. 2015, 6, 1159. [Google Scholar] [CrossRef]
- Klein, T.; Poghosyan, L.; Barclay, J.E.; Murrell, J.C.; Hutchings, M.I.; Lehtovirta-Morley, L.E. Cultivation of ammonia-oxidising archaea on solid medium. FEMS Microbiol. Lett. 2022, 369, fnac029. [Google Scholar] [CrossRef] [PubMed]
- Sitepu, I.R.; Garay, L.A.; Sestric, R.; Levin, D.; Block, D.E.; German, J.B.; Boundy-Mills, K.L. Oleaginous yeasts for biodiesel: Current and future trends in biology and production. Biotechnol. Adv. 2014, 32, 1336–1360. [Google Scholar] [CrossRef]
- Xue, Y.-P.; Jin, M.; Orjuela, A.; Slininger, P.J.; Dien, B.S.; Dale, B.E.; Balan, V. Microbial lipid production from AFEX™ pretreated corn stover. RSC Adv. 2015, 5, 28725–28734. [Google Scholar] [CrossRef]
- Slininger, P.J.; Dien, B.S.; Kurtzman, C.P.; Moser, B.R.; Bakota, E.L.; Thompson, S.R.; O’Bryan, P.J.; Cotta, M.A.; Balan, V.; Jin, M. Comparative lipid production by oleaginous yeasts in hydrolyzates of lignocellulosic biomass and process strategy for high titers. Biotechnol. Bioeng. 2016, 113, 1676–1690. [Google Scholar] [CrossRef]
- Caporusso, A.; De Bari, I.; Valerio, V.; Albergo, R.; Liuzzi, F. Conversion of cardoon crop residues into single cell oils by Lipomyces tetrasporus and Cutaneotrichosporon curvatus: Process optimizations to overcome the microbial inhibition of lignocellulosic hydrolysates. Ind. Crops Prod. 2021, 159, 113030. [Google Scholar] [CrossRef]
- Juanssilfero, A.B.; Kahar, P.; Amza, R.L.; Miyamoto, N.; Otsuka, H.; Matsumoto, H.; Kihira, C.; Thontowi, A.; Ogino, C.; Prasetya, B. Selection of oleaginous yeasts capable of high lipid accumulation during challenges from inhibitory chemical compounds. Biochem. Eng. J. 2018, 137, 182–191. [Google Scholar] [CrossRef]
- Huang, C.; Yang, Y.; Qin, H.; Feng, S.; Guo, M.; Xu, Q.; Qiao, D.; Cao, Y. An oleaginous yeast strain: Screening, identification and optimization of fermentation conditions. Chin. J. Appl. Environ. Biol. 2014, 20, 609–614. [Google Scholar]
- Sitepu, I.; Selby, T.; Lin, T.; Zhu, S.; Boundy-Mills, K. Carbon source utilization and inhibitor tolerance of 45 oleaginous yeast species. J. Ind. Microbiol. Biotechnol. 2014, 41, 1061–1070. [Google Scholar] [CrossRef] [PubMed]
- Dien, B.S.; Slininger, P.J.; Kurtzman, C.P.; Moser, B.R.; O’Bryan, P.J. Identification of superior lipid producing Lipomyces and Myxozyma yeasts. AIMS Environ. Sci 2016, 3, 1–20. [Google Scholar] [CrossRef]
- Qian, X.; Gorte, O.; Chen, L.; Zhang, W.; Dong, W.; Ma, J.; Xin, F.; Jiang, M.; Ochsenreither, K. Continuous self-provided fermentation for microbial lipids production from acetate by using oleaginous yeasts Cryptococcus podzolicus and Trichosporon porosum. Renew. Energy 2020, 146, 737–743. [Google Scholar] [CrossRef]
- Qian, X.; Gorte, O.; Chen, L.; Zhang, W.; Dong, W.; Ma, J.; Jiang, M.; Xin, F.; Ochsenreither, K. Co-production of single cell oil and gluconic acid using oleaginous Cryptococcus podzolicus DSM 27192. Biotechnol. Biofuels 2019, 12, 1–9. [Google Scholar] [CrossRef]
- Ramírez-Castrillón, M.; Jaramillo-Garcia, V.P.; Rosa, P.D.; Landell, M.F.; Vu, D.; Fabricio, M.F.; Ayub, M.A.; Robert, V.; Henriques, J.A.; Valente, P. The oleaginous yeast Meyerozyma guilliermondii BI281A as a new potential biodiesel feedstock: Selection and lipid production optimization. Front. Microbiol. 2017, 8, 1776. [Google Scholar] [CrossRef]
- Ganapathy, B.; Yahya, A.; Ibrahim, N. Bioremediation of palm oil mill effluent (POME) using indigenous Meyerozyma guilliermondii. Environ. Sci. Pollut. Res. 2019, 26, 11113–11125. [Google Scholar] [CrossRef]
- Yan, W.; Gao, H.; Qian, X.; Jiang, Y.; Zhou, J.; Dong, W.; Xin, F.; Zhang, W.; Jiang, M. Biotechnological applications of the non-conventional yeast Meyerozyma guilliermondii. Biotechnol. Adv. 2021, 46, 107674. [Google Scholar] [CrossRef]
- Llamas, M.; Dourou, M.; González-Fernández, C.; Aggelis, G.; Tomás-Pejó, E. Screening of oleaginous yeasts for lipid production using volatile fatty acids as substrate. Biomass Bioenergy 2020, 138, 105553. [Google Scholar] [CrossRef]
- Vieira, N.M.; Dos Santos, R.C.V.; Germano, V.K.d.C.; Ventorim, R.Z.; de Almeida, E.L.M.; da Silveira, F.A.; Ribeiro Júnior, J.I.; da Silveira, W.B. Isolation of a new Papiliotrema laurentii strain that displays capacity to achieve high lipid content from xylose. 3 Biotech. 2020, 10, 1–14. [Google Scholar] [CrossRef]
- Park, Y.; Maeng, S.; Srinivasan, S. Isolation and characterization of unrecorded yeasts species in the family Metschnikowiaceae and Bulleribasidiaceae in Korea. J. Species Res. 2020, 9, 198–203. [Google Scholar]
- Beopoulos, A.; Cescut, J.; Haddouche, R.; Uribelarrea, J.-L.; Molina-Jouve, C.; Nicaud, J.-M. Yarrowia lipolytica as a model for bio-oil production. Prog. Lipid Res. 2009, 48, 375–387. [Google Scholar] [CrossRef] [PubMed]
- Taskin, M.; Ortucu, S.; Aydogan, M.N.; Arslan, N.P. Lipid production from sugar beet molasses under non-aseptic culture conditions using the oleaginous yeast Rhodotorula glutinis TR29. Renew. Energy 2016, 99, 198–204. [Google Scholar] [CrossRef]
- Li, Y.; Zhao, Z.K.; Bai, F. High-density cultivation of oleaginous yeast Rhodosporidium toruloides Y4 in fed-batch culture. Enzym. Microb. Technol. 2007, 41, 312–317. [Google Scholar] [CrossRef]
- Calvey, C.H.; Su, Y.K.; Willis, L.B.; McGee, M.; Jeffries, T.W. Nitrogen limitation, oxygen limitation, and lipid accumulation in Lipomyces Starkeyi. Bioresour. Technol. 2016, 200, 780–788. [Google Scholar] [CrossRef] [PubMed]
- Deeba, F.; Pruthi, V.; Negi, Y.S. Fostering triacylglycerol accumulation in novel oleaginous yeast Cryptococcus psychrotolerans IITRFD utilizing groundnut shell for improved biodiesel production. Bioresour. Technol. 2017, 242, 113–120. [Google Scholar] [CrossRef] [PubMed]
- Arous, F.; Azabou, S.; Triantaphyllidou, I.E.; Aggelis, G.; Jaouani, A.; Nasri, M.; Mechichi, T. Newly isolated yeasts from Tunisian microhabitats: Lipid accumulation and fatty acid composition. Eng. Life Sci. 2017, 17, 226–236. [Google Scholar] [CrossRef]
- Palazzolo, M.A.; Garcia-Perez, M. Microbial lipid biosynthesis from lignocellulosic biomass pyrolysis products. Biotechnol. Adv. 2022, 54, 107791. [Google Scholar] [CrossRef]
- Poontawee, R.; Limtong, S. Feeding strategies of two-stage fed-batch cultivation processes for microbial lipid production from sugarcane top hydrolysate and crude glycerol by the oleaginous red yeast Rhodosporidiobolus fluvialis. Microorganisms 2020, 8, 151. [Google Scholar] [CrossRef]
- Huang, X.-f.; Shen, Y.; Luo, H.-j.; Liu, J.-n.; Liu, J. Enhancement of extracellular lipid production by oleaginous yeast through preculture and sequencing batch culture strategy with acetic acid. Bioresour. Technol. 2018, 247, 395–401. [Google Scholar] [CrossRef]
- Wu, H.; Li, Y.; Chen, L.; Zong, M. Production of microbial oil with high oleic acid content by Trichosporon capitatum. Appl. Energy 2011, 88, 138–142. [Google Scholar] [CrossRef]

| Phylum and Subphylum | Species | Number of Strains | FO (%) a |
|---|---|---|---|
| Ascomycota (97 strains) | |||
| Saccharomycotina | Cyberlindnera saturnus | 7 | 5.5 |
| Lipomyces mesembrius | 10 | 7.9 | |
| Lipomyces starkeyi | 31 | 24.4 | |
| Lipomyces tetrasporus | 48 | 37.8 | |
| Meyerozyma guilliermondii | 1 | 0.8 | |
| Basidiomycota (31 strains) | |||
| Pucciniomycotina | Rhodotorula mucilaginosa | 1 | 0.8 |
| Cystobasidium slooffiae | 1 | 0.8 | |
| Agaricomycotina | Naganishia diffluens | 1 | 0.8 |
| Papiliotrema flavescens | 10 | 7.9 | |
| Papiliotrema terrestris | 4 | 3.2 | |
| Saitozyma podzolica | 8 | 6.3 | |
| Piskurozyma sp. | 3 | 2.4 | |
| Hannaella sp. | 2 | 1.6 | |
| Total number of strains | 127 | 100.0 |
| Yeast Strain | Biomass (g L−1) | Lipid (g L−1) | Lipid Content (%) |
|---|---|---|---|
| L. tetrasporus SWU-NGP 2-5 | 1.49 ± 0.05 g | 1.10 ± 0.06 gh | 74.26 ± 6.30 a |
| L. tetrasporus SWU-NAP 4-1 | 4.50 ± 0.08 ef | 2.57 ± 0.04 d | 57.10 ± 1.82 b |
| L. mesembrius SWU-NGP 14-6 | 9.13 ± 0.36 a | 5.20 ± 0.03 a | 57.10 ± 1.89 b |
| L. mesembrius SWU-NGP 6-4 | 7.83 ± 0.17 b | 3.98 ± 0.02 b | 50.80 ± 0.80 b |
| L. tetrasporus SWU-NGP 5-8-1 | 9.07 ± 0.16 a | 4.27 ± 0.01 b | 47.06 ± 0.66 bcd |
| S. podzolica SWU-NAP 5-4-2 | 3.88 ± 0.19 ef | 1.81 ± 0.19 ef | 46.63 ± 2.51 cde |
| L. starkeyi SWU-NGP 14-3-1 | 3.53 ± 0.56 f | 1.47 ± 0.07 fg | 42.49 ± 4.65 cde |
| L. starkeyi SWU-NGTP 4-5 | 5.92 ± 0.14 cd | 2.45 ± 0.22 d | 41.56 ± 4.67 de |
| L. starkeyi SWU-NGP 5-7 | 8.39 ± 0.85 ab | 3.35 ± 0.00 c | 40.30 ± 4.11 e |
| Cyb. saturnus SWU-NGTP 5-3-3 | 1.00 ± 0.04 gh | 0.4 ± 0.014 ij | 40.00 ± 0.00 e |
| S. podzolica SWU-NGP 5-3-8 | 6.59 ± 0.89 c | 2.63 ± 0.35 d | 40.00 ± 0.00 e |
| P. flavescens SWU-NGTP 4-1 | 1.87 ± 0.24 g | 0.74 ± 0.10 hi | 40.00 ± 0.00 e |
| S. podzolica SWU-NGP 14-2-2 | 4.99 ± 0.19 de | 1.99 ± 0.07 e | 40.00 ± 0.00 e |
| M. guilliermondii SWU-NATP 2-4 | 0.25 ± 0.04 h | 0.10 ± 0.02 j | 40.00 ± 0.00 e |
| P. flavescens SWU-NATP 3-3 | 1.15 ± 0.06 gh | 0.46 ± 0.02 ij | 40.00 ± 0.00 e |
| Yeast Strain | Closely Related Species (Accession Number) | Similarity (%) | Gaps/Total Nucleotide | Nucleotide Substitution | Biomass (g L−1) | Lipid (g L−1) | Lipid Content (% of Dry Biomass) |
|---|---|---|---|---|---|---|---|
| Hannaella sp. SWU-YGP 11-1 | Hannaella oryzae CBS 7194T (AF075511) | 98.9 | 1/597 | 12 | 1.70 ± 0.15 | 0.36 ± 0.08 | 21.70 |
| Hannaella sp. SWU-NAPS 5-1 | Hannaella oryzae CBS 7194T (AF075511) | 98.0 | 1/501 | 12 | 6.41 ± 0.65 | 1.10 ± 0.15 | 16.46 |
| Piskurozyma sp. SWU-NATP 4-12 | Piskurozyma taiwanensis CBS 9813T (AF079035) | 96.5 | 2/596 | 19 | 2.92 ± 0.24 | 0.74 ± 0.04 | 25.45 |
| Piskurozyma sp. SWU-NGP 14-3-2 | Piskurozyma taiwanensis CBS 9813T (AF079035) | 96.8 | 2/517 | 18 | 0.53 ± 0.15 | 0.07 ± 0.01 | 14.12 |
| Piskurozyma sp. SWU-NGP 14-3-4 | Piskurozyma taiwanensis CBS 9813T (AF079035) | 96.8 | 2/583 | 19 | 0.67 ± 0.01 | 0.08 ± 0.01 | 11.72 |
| Strains | Relative Content of Fatty Acid (% w/w) | |||||||
|---|---|---|---|---|---|---|---|---|
| C14:0 | C16:0 | C16:1 | C18:0 | C18:1 | C18:2 | C18:3α | C18:3β | |
| L. mesembrius SWU-NAP 3-4 | 0.55 | 41.7 | 1.5 | 8 | 0.7 | 15.7 | - | 1.6 |
| L. mesembrius SWU-NAP 8-4 | 0.38 | 33.4 | 0.1 | 7.7 | 1 | 55.1 | 0.3 | 1.9 |
| L. tetrasporus SWU-NAP 5-3-1 | 0.32 | 41.1 | 0.6 | - | 6.8 | 48.6 | 0.9 | 1.6 |
| L. tetrasporus SWU-NAP 13-8 | 0.22 | 34.5 | 0.3 | 11.4 | 3.2 | 47.1 | 0.6 | 2.6 |
| P. terrestris SWU-NGPui 12-9 | 0.24 | 19.5 | - | 17.9 | 0.8 | 49.1 | - | 12.4 |
| P. terrestris SWU-NAPui 14-5 | - | 30.2 | - | 24.8 | 13.8 | 23.4 | 1.3 | 6.5 |
| S. podzolica SWU-YGP 8-1-2 | - | 20.3 | - | 11.6 | 0.3 | 63.1 | - | 4.6 |
| S. podzolica SWU-NGP 5-3-2 | 0.13 | 19.4 | 0.2 | 17.0 | 0.6 | 55.8 | - | 6.9 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sapsirisuk, S.; Polburee, P.; Lorliam, W.; Limtong, S. Discovery of Oleaginous Yeast from Mountain Forest Soil in Thailand. J. Fungi 2022, 8, 1100. https://doi.org/10.3390/jof8101100
Sapsirisuk S, Polburee P, Lorliam W, Limtong S. Discovery of Oleaginous Yeast from Mountain Forest Soil in Thailand. Journal of Fungi. 2022; 8(10):1100. https://doi.org/10.3390/jof8101100
Chicago/Turabian StyleSapsirisuk, Sirawich, Pirapan Polburee, Wanlapa Lorliam, and Savitree Limtong. 2022. "Discovery of Oleaginous Yeast from Mountain Forest Soil in Thailand" Journal of Fungi 8, no. 10: 1100. https://doi.org/10.3390/jof8101100
APA StyleSapsirisuk, S., Polburee, P., Lorliam, W., & Limtong, S. (2022). Discovery of Oleaginous Yeast from Mountain Forest Soil in Thailand. Journal of Fungi, 8(10), 1100. https://doi.org/10.3390/jof8101100








