The Microbiome Structure of the Symbiosis between the Desert Truffle Terfezia boudieri and Its Host Plant Helianthemum sessiliflorum
Abstract
1. Introduction
2. Materials and Methods
2.1. Subjects and Sampling
2.2. Total Genomic DNA Extraction
2.3. Next-Generation Amplicon Sequencing
2.4. Basic Processing of Amplicon Sequence Data
2.5. Alpha Diversity Analyses
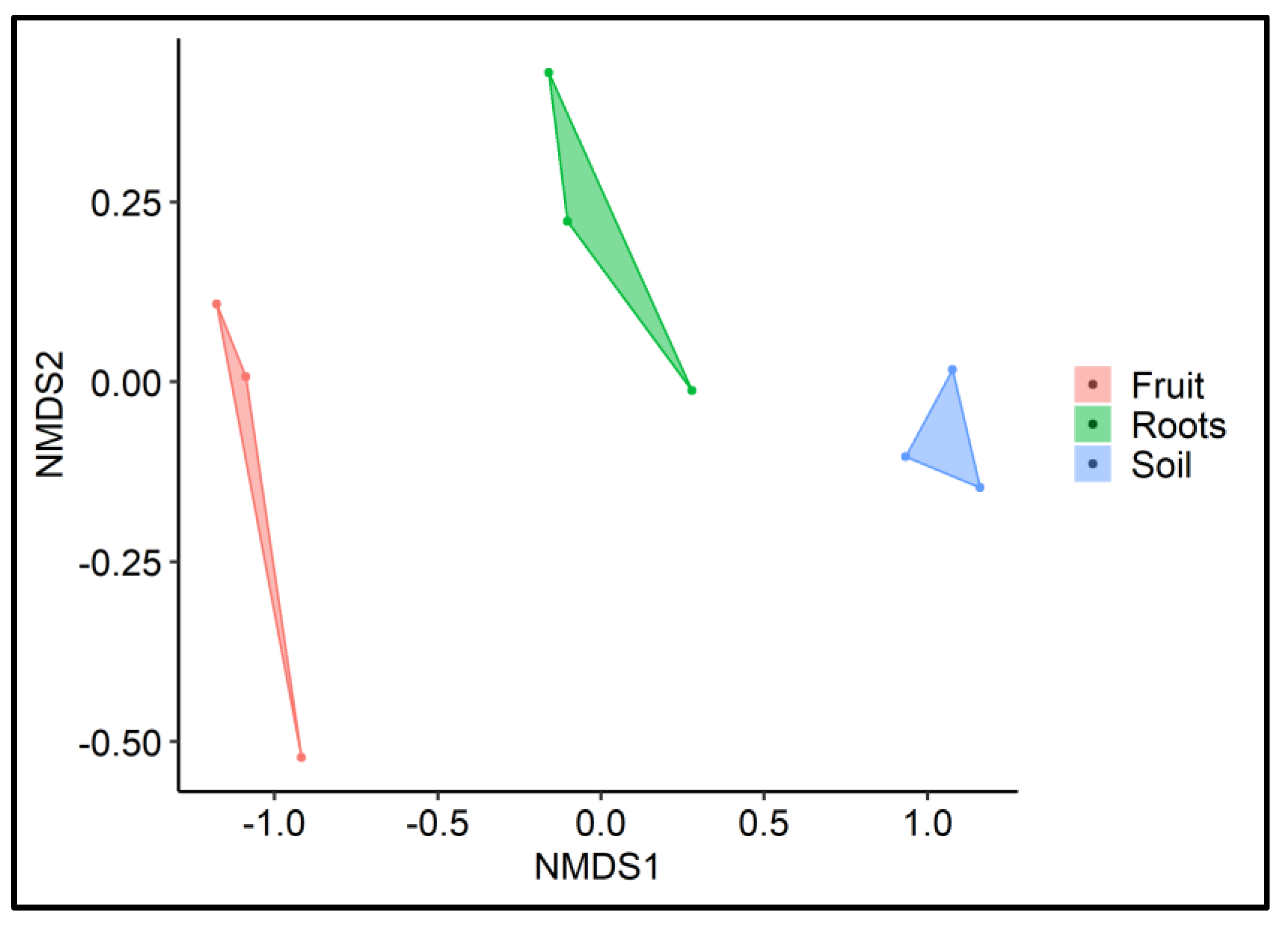
2.6. Beta Diversity/Dissimilarity Analyses
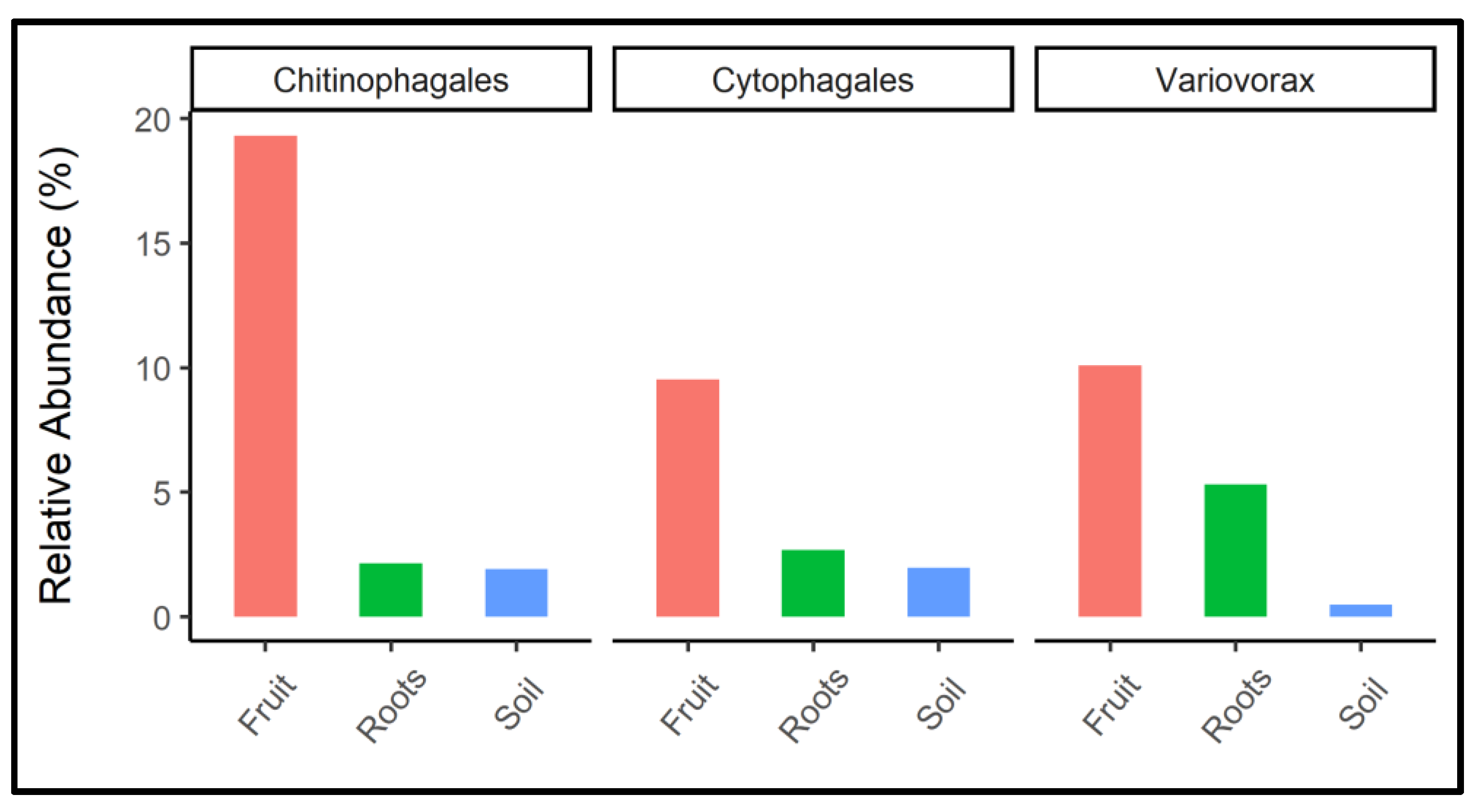
2.7. Differential Analysis of Microbial Taxa
2.8. Determination of Soil N, P, and K Levels
3. Results
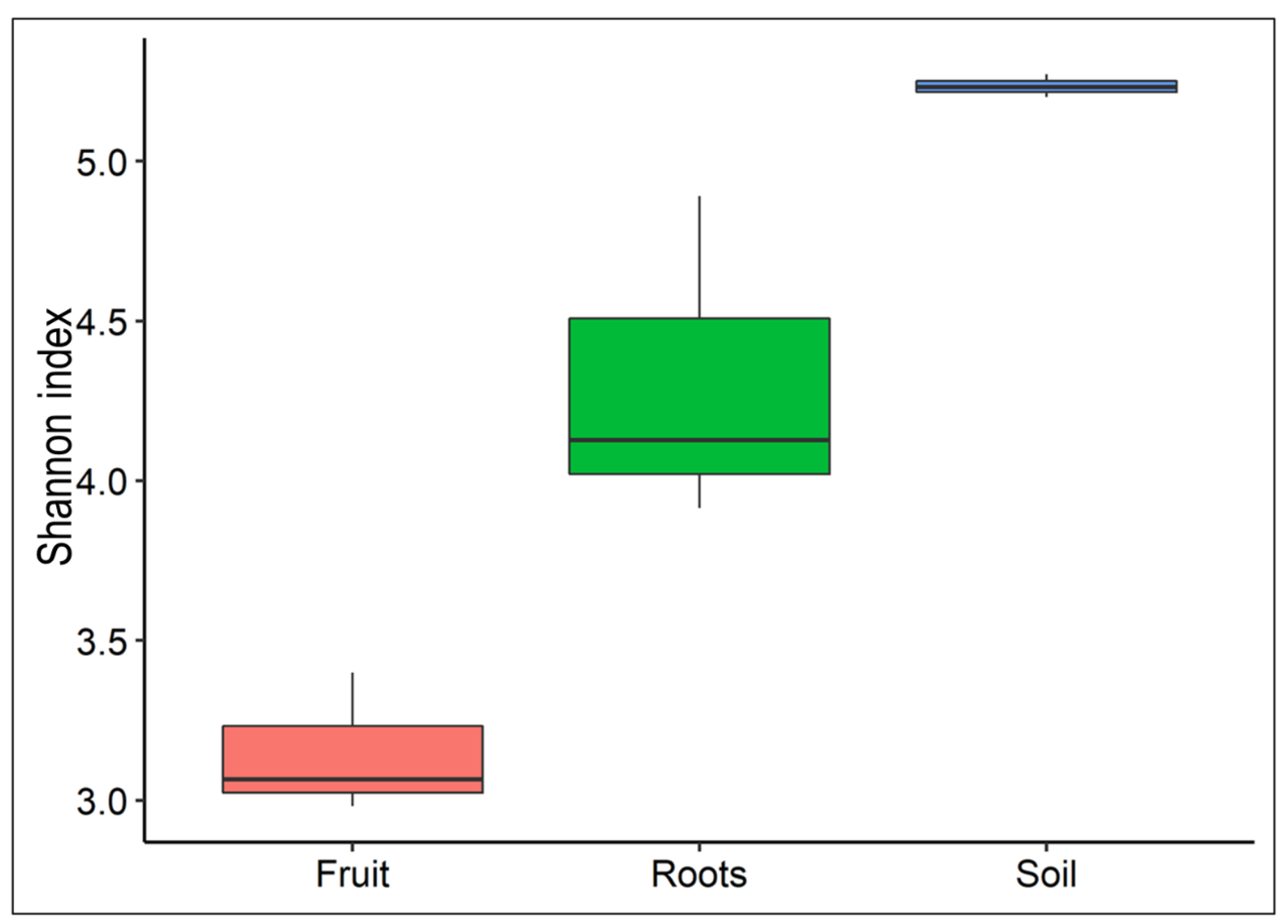
3.1. Bacterial Diversity Varied among T. boudieri Fruit Bodies, Ectomycorrhizal Roots, and Rhizosphere Soil Samples
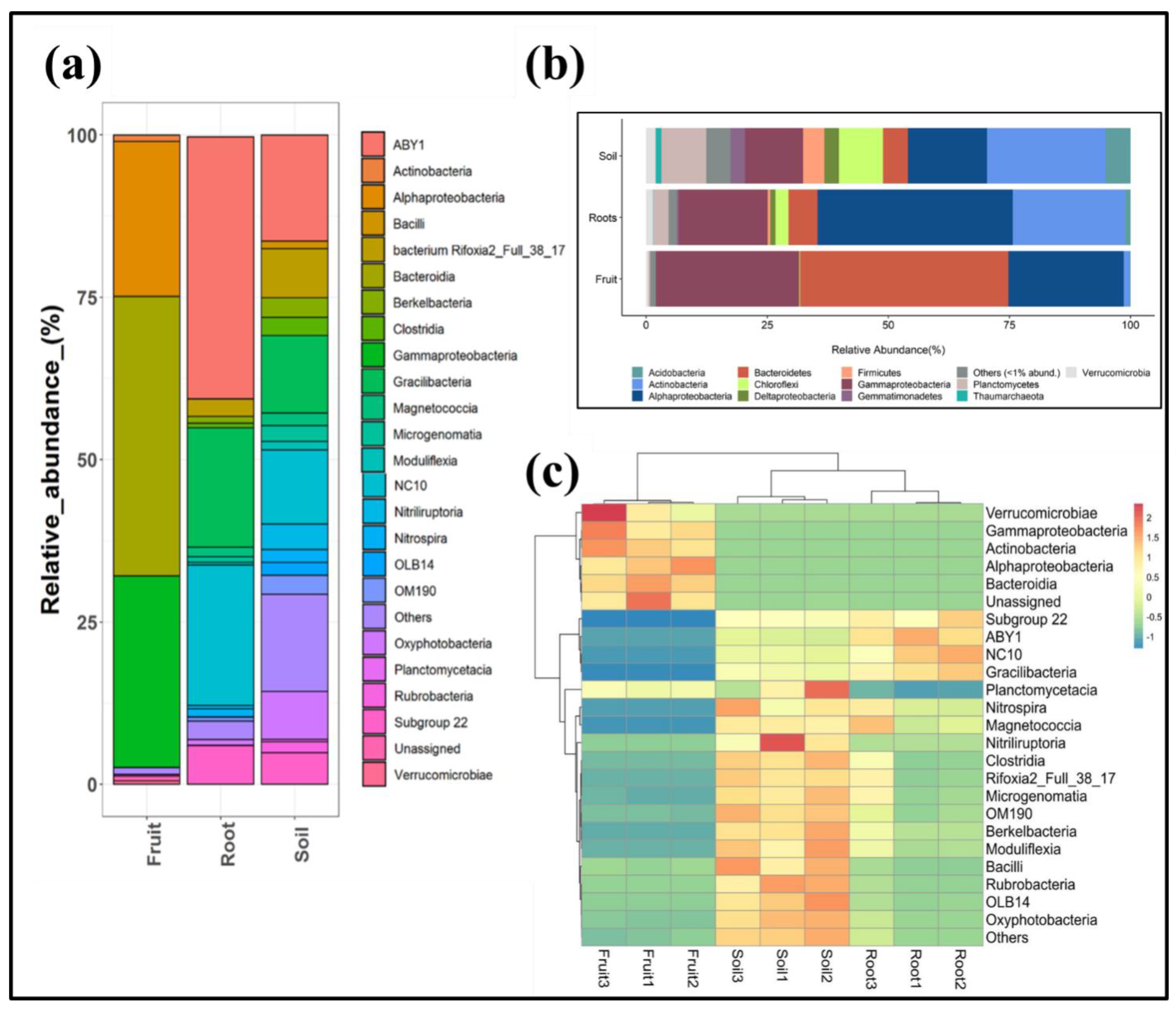
3.2. Bacterial Community Composition of T. boudieri Fruit Bodies, Ectomycorrhizal Roots, and Rhizosphere Soil Samples
3.3. Bacterial Community Composition of T. boudieri Fruit Bodies, Ectomycorrhizal Roots, and Rhizosphere Soil Samples
3.4. Soil N and K Level Changes after Mycorrhiza Colonization
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Moreno, G.; Alvarado, P.; Manjón, J.L. Hypogeous desert fungi. In Desert Truffles; Kagan-Zur, V., Roth-Bejerano, N., Sitrit, Y., Morte, A., Eds.; Springer: Berlin/Heidelberg, Germany, 2014; Volume 38, pp. 3–20. [Google Scholar]
- Kamle, M.; Bar, E.; Lewinsohn, D.; Shavit, E.; Roth-Bejerano, N.; Kagan-Zur, V.; Barak Ze Guy, O.; Zaady, E.; Lewinsohn, E. Characterization of morphology, volatile profiles, and molecular markers in edible desert truffles from the negev desert. J. Agric. Food Chem. 2017, 65, 2977–2983. [Google Scholar] [CrossRef]
- Morte, A.; Zamora, M.; Gutiérrez, A.; Honrubia, M. Desert truffle cultivation in semiarid Mediterranean areas. In Mycorrhizas-Functional Processes and Ecological Impact; Springer: Berlin/Heidelberg, Germany, 2009; pp. 221–233. [Google Scholar]
- Shavit, E.; Shavit, E. The medicinal value of desert truffles. In Desert Truffles; Springer: Berlin/Heidelberg, Germany, 2014; pp. 323–340. [Google Scholar]
- Al Obaydi, M.F.; Hamed, W.M.; Al Kury, L.T.; Talib, W.H. Terfezia boudieri: A desert truffle with anticancer and immunomodulatory activities. Front. Nutr. 2020, 7, 38. [Google Scholar] [CrossRef] [PubMed]
- Tagnamas, Z.; Bahammou, Y.; Kouhila, M.; Hilali, S.; Idlimam, A.; Lamharrar, A. Conservation of Moroccan truffle (Terfezia boudieri) using solar drying method. Renew. Energy 2020, 146, 16–24. [Google Scholar] [CrossRef]
- Murcia, M.A.; Martinez-Tome, M.; Jimenez, A.M.; Vera, A.M.; Honrubia, M.; Parras, P. Antioxidant activity of edible fungi (truffles and mushrooms): Losses during industrial processing. J. Food Protect. 2002, 65, 1614–1622. [Google Scholar] [CrossRef]
- Hamza, A.; Zouari, N.; Zouari, S.; Jdir, H.; Zaidi, S.; Gtari, M.; Neffati, M. Nutraceutical potential, antioxidant and antibacterial activities of Terfezia boudieri Chatin, a wild edible desert truffle from Tunisia arid zone. Arab. J. Chem. 2016, 9, 383–389. [Google Scholar] [CrossRef]
- Patel, S.; Rauf, A.; Khan, H.; Khalid, S.; Mubarak, M.S. Potential health benefits of natural products derived from truffles: A review. Trends Food Sci. Technol. 2017, 70, 1–8. [Google Scholar] [CrossRef]
- Navarro-Ródenas, A.; Pérez-Gilabert, M.; Torrente, P.; Morte, A. The role of phosphorus in the ectendomycorrhiza continuum of desert truffle mycorrhizal plants. Mycorrhiza 2012, 22, 565–575. [Google Scholar] [CrossRef]
- Wang, Y.; Chen, Y.L. Recent advances in cultivation of edible mycorrhizal mushrooms. In Mycorrhizal Fungi: Use in Sustainable Agriculture and Land Restoration; Springer: Berlin/Heidelberg, Germany, 2014; pp. 375–397. [Google Scholar]
- Morte, A.; Kagan-Zur, V.; Navarro-Ródenas, A.; Sitrit, Y. Cultivation of desert truffles—A crop suitable for arid and semi-arid zones. Agronomy 2021, 11, 1462. [Google Scholar] [CrossRef]
- Smith, S.E.; Read, D.J. Mycorrhizal Symbiosis; Academic Press: Cambridge, UK, 2008. [Google Scholar]
- Zaretsky, M.; Kagan-Zur, V.; Mills, D.; Roth-bejerano, N. Analysis of mycorrhizal associations formed by Cistus incanus transformed root clones with Terfezia boudieri isolates. Plant Cell Rep. 2006, 25, 62–70. [Google Scholar] [CrossRef]
- Kovács, G.M.; Balázs, T.K.; Calonge, F.D.; Martin, M.P. The diversity of Terfezia desert truffles: New species and a highly variable species complex with intrasporocarpic nrDNA ITS heterogeneity. Mycologia 2011, 103, 841–853. [Google Scholar] [CrossRef]
- Murat, C.; Payen, T.; Noel, B.; Kuo, A.; Morin, E.; Chen, J.; Kohler, A.; Krizsán, K.; Balestrini, R.; Da Silva, C.; et al. Pezizomycetes genomes reveal the molecular basis of ectomycorrhizal truffle lifestyle. Nat. Ecol. Evol. 2018, 2, 1956–1965. [Google Scholar] [CrossRef]
- Turgeman, T.; Asher, J.B.; Roth-Bejerano, N.; Kagan-Zur, V.; Kapulnik, Y.; Sitrit, Y. Mycorrhizal association between the desert truffle Terfezia boudieri and Helianthemum sessiliflorum alters plant physiology and fitness to arid conditions. Mycorrhiza 2011, 21, 623–630. [Google Scholar] [CrossRef]
- Morte, A.; Lovisolo, C.; Schubert, A. Effect of drought stress on growth and water relations of the mycorrhizal association Helianthemum almeriense-Terfezia claveryi. Mycorrhiza 2000, 10, 115–119. [Google Scholar] [CrossRef]
- Perotto, S.; Bonfante, P. Bacterial associations with mycorrhizal fungi: Close and distant friends in the rhizosphere. Trends Microbiol. 1997, 5, 496–501. [Google Scholar] [CrossRef]
- Verma, K.K.; Wu, K.C.; Singh, P.; Malviya, M.K.; Singh, R.K.; Song, X.P.; Li, Y.R. The protective role of silicon in sugarcane under water stress: Photosynthesis and antioxidant enzymes. Biomed. J. Sci. Tech. Res. 2019, 15, 002685. [Google Scholar]
- Shobana, N.; Thangappan, S.; Uthandi, S. Plant growth-promoting Bacillus sp. cahoots moisture stress alleviation in rice genotypes by triggering antioxidant defense system. Microbiol. Res. 2020, 239, 126518. [Google Scholar]
- Schachtman, D.P.; Reid, R.J.; Ayling, S.M. Phosphorus uptake by plants: From soil to cell. Plant Physiol. 1998, 116, 447–453. [Google Scholar] [CrossRef]
- Yadav, R.; Tarafdar, J.J.S.B. Phytase and phosphatase producing fungi in arid and semi-arid soils and their efficiency in hydrolyzing different organic P compounds. Soil Biol. Biochem. 2003, 35, 745–751. [Google Scholar] [CrossRef]
- Piñuela, Y.; GAlday, J.; Oliach, D.; Bolaño, F.; Colinas, C.; Bonet, J.A. Use of inoculator bacteria to promote Tuber melanosporum root colonization and growth on Quercus faginea saplings. Forests 2020, 11, 792. [Google Scholar] [CrossRef]
- Navarro-Ródenas, A.; Berná, L.M.; Lozano-Carrillo, C.; Andrino, A.; Morte, A. Beneficial native bacteria improve survival and mycorrhization of desert truffle mycorrhizal plants in nursery conditions. Mycorrhiza 2016, 26, 769–779. [Google Scholar] [CrossRef]
- Akyol, T.Y.; Niwa, R.; Hirakawa, H.; Maruyama, H.; Sato, T.; Suzuki, T.; Fukunaga, A.; Sato, T.; Yoshida, S.; Tawaray, K.; et al. Impact of introduction of arbuscular Mycorrhizal fungi on the root microbial community in agricultural fields. Microbes Environ. 2018, 34, 23–32. [Google Scholar] [CrossRef] [PubMed]
- Mekonnen, H.; Kibret, M. The roles of plant growth promoting rhizobacteria in sustainable vegetable production in Ethiopia. Chem. Biol. Technol. Agric. 2021, 8, 15. [Google Scholar]
- Jacoby, R.; Peukert, M.; Succurro, A.; Koprivova, A.; Kopriva, S. The role of soil microorganisms in plant mineral nutrition—Current knowledge and future directions. Front. Plant Sci. 2017, 8, 1617. [Google Scholar] [CrossRef] [PubMed]
- Barbieri, E.; Bertini, L.; Rossi, I.; Ceccaroli, P.; Saltarelli, R.; Guidi, C.; Zambonelli, A.; Stocchi, V. New evidence for bacterial diversity in the ascoma of the ectomycorrhizal fungus Tuber borchii Vittad. FEMS Microbiol. Lett. 2005, 247, 23–35. [Google Scholar] [CrossRef] [PubMed]
- Splivallo, R.; Deveau, A.; Valdez, N.; Kirchhoff, N.; Frey-Klett, P.; Karlovsky, P. Bacteria associated with truffle-fruiting bodies contribute to truffle aroma. Environ. Microbiol. 2015, 8, 2647–2660. [Google Scholar] [CrossRef] [PubMed]
- Barbieri, E.; Guidi, C.; Bertaux, J.; Frey-Klett, P.; Garbaye, J.; Ceccaroli, P.; Saltarelli, R.; Zambonelli, A.; Stocchi, V. Occurrence and diversity of bacterial communities in Tuber magnatum during truffle maturation. Environ. Microbiol. 2007, 9, 2234–2246. [Google Scholar]
- Gryndler, M.; Soukupová, L.; Hršelová, H.; Gryndlerová, H.; Borovička, J.; Streiblová, E.; Jansa, J. A quest for indigenous truffle helper prokaryotes. Environ. Microbiol. Rep. 2013, 5, 346–352. [Google Scholar] [CrossRef] [PubMed]
- Splivallo, R.; Vahdatzadeh, M.; Maciá-Vicente, J.G.; Molinier, V.; Peter, M.; Egli, S.; Uroz, S.; Paolocci, F.; Deveau, A. Orchard conditions and fruiting body characteristics drive the microbiome of the black truffle Tuber aestivum. Front. Microbiol. 2019, 10, 1437. [Google Scholar] [CrossRef]
- Perlińska-Lenart, U.; Piłsyk, S.; Gryz, E.; Turło, J.; Hilszczańska, D.; Kruszewska, J.S. Identification of bacteria and fungi inhabiting fruiting bodies of Burgundy truffle (Tuber aestivum Vittad.). Arch. Microbiol. 2020, 202, 2727–2738. [Google Scholar] [CrossRef]
- Antony-Babu, S.; Deveau, A.; Van Nostrand, J.D.; Zhou, J.; Le Tacon, F.; Robin, C.; Frey-Klett, P.; Uroz, S. Black truffle-associated bacterial communities during the development and maturation of Tuber melanosporum ascocarps and putative functional roles. Environ. Microbiol. 2014, 16, 2831–2847. [Google Scholar]
- Deveau, A.; Antony-Babu, S.; Le Tacon, F.; Robin, C.; Frey-Klett, P.; Uroz, S. Temporal changes of bacterial communities in the Tuber melanosporum ectomycorrhizosphere during ascocarp development. Mycorrhiza 2016, 26, 389–399. [Google Scholar] [CrossRef] [PubMed]
- Kang, Z.; Zou, J.; Huang, Y.; Zhang, X.; Ye, L.; Zhang, B.; Li, X. Tuber melanosporum shapes nirS-type denitrifying and ammonia-oxidizing bacterial communities in Carya illinoinensis ectomycorrhizosphere soils. PeerJ 2020, 8, e9457. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Li, X.; Wu, C.; Ye, L.; Kang, Z.; Zhang, X. Exogenous nitric oxide and phosphorus stress affect the mycorrhization, plant growth, and associated microbes of Carya illinoinensis seedlings colonized by Tuber indicum. Front. Microbiol. 2019, 10, 2634. [Google Scholar] [CrossRef] [PubMed]
- Liu, D.; Pérez-Moreno, J.; He, X.; Garibay-Orijel, R.; Yu, F. Truffle microbiome is driven by fruit body compartmentalization rather than soils conditioned by different host trees. mSphere 2021, 6, e00039-21. [Google Scholar] [CrossRef]
- Liu, D.; Herrera, M.; Yu, F.; Pèrez-Moreno, J. Provenances originate morphological and microbiome variation of Tuber pseudobrumale in southwestern China despite strong genetic consistency. Mycol. Prog. 2020, 19, 1545–1558. [Google Scholar] [CrossRef]
- Grigoriev, I.V.; Cullen, D.; Goodwin, S.B.; Hibbett, D.; Jeffries, T.W.; Kubicek, C.P.; Kuske, C.; Magnuson, J.K.; Martin, F.; Spatafora, J.W.; et al. Fueling the future with fungal genomics. Mycology 2011, 2, 192–209. [Google Scholar]
- Grigoriev, I.V.; Nikitin, R.; Haridas, S.; Kuo, A.; Ohm, R.; Otillar, R.; Riley, R.; Salamov, A.; Zhao, X.; Korzeniewski, F.; et al. MycoCosm portal: Gearing up for 1000 fungal genomes. Nucleic Acids Res. 2014, 42, D699–D704. [Google Scholar] [CrossRef]
- Zhang, J.; Kobert, K.; Flouri, T.; Stamatakis, A. PEAR: A fast and accurate Illumina Paired-End reAd mergeR. Bioinformatics 2014, 30, 614–620. [Google Scholar] [CrossRef]
- Glöckner, F.O.; Yilmaz, P.; Quast, C.; Gerken, J.; Beccati, A.; Ciuprina, A.; Bruns, G.; Yarza, P.; Peplies, J.; Westram, R. 25 years of serving the community with ribosomal RNA gene reference databases and tools. J. Biotechnol. 2017, 261, 169–176. [Google Scholar] [CrossRef]
- Edgar, R.C. Search and clustering orders of magnitude faster than BLAST. Bioinformatics 2010, 26, 2460–2461. [Google Scholar] [CrossRef]
- Callahan, B.J.; McMurdie, P.J.; Rosen, M.J.; Han, A.W.; Johnson, A.J.A.; Holmes, S.P. DADA2: High-resolution sample inference from Illumina amplicon data. Nat. Methods 2016, 13, 581–583. [Google Scholar] [CrossRef] [PubMed]
- Oksanen, J.; Blanchet, F.; Friendly, M.; Kindt, R.; Legendre, P.; McGlinn, D.; Minchin, P.; O’Hara, R.; Simpson, G.; Solymos, P. Vegan: Community Ecology Package. R Package, version 2.5-2; R Foundation for Statistical Computing: Vienna, Austria, 2018. [Google Scholar]
- Wickham, H. Elegant graphics for data analysis (ggplot2). In Applied Spatial Data Analysis R; Springer: New York, NY, USA, 2009. [Google Scholar]
- McCarthy, D.J.; Chen, Y.; Smyth, G.K. Differential expression analysis of multifactor RNA-Seq experiments with respect to biological variation. Nucleic Acids Res. 2012, 40, 4288–4297. [Google Scholar] [CrossRef]
- Benjamini, Y.; Hochberg, Y. Controlling the false discovery rate: A practical and powerful approach to multiple testing. J. R. Stat. Soc. Ser. B Methodol. 1995, 57, 289–300. [Google Scholar] [CrossRef]
- Olsen, S.R. Estimation of Available Phosphorus in Soils by Extraction with Sodium Bicarbonate. No. 939; US Department of Agriculture: Washington, DC, USA, 1954.
- Paz-Kagan, T.; DeMalach, N.; Zaady, E.; Shachak, M. Resource redistribution effects on annual plant communities in a runoff harvesting system in dryland. J. Arid Environ. 2019, 171, 103984. [Google Scholar] [CrossRef]
- Rosenberg, E. The family chitinophagaceae. In The Prokaryotes; Springer: Berlin/Heidelberg, Germany, 2014; pp. 493–495. [Google Scholar]
- Bailey, V.L.; Fansler, S.J.; Stegen, J.C.; McCue, L.A. Linking microbial community structure to β-glucosidic function in soil aggregates. ISME J. 2013, 7, 2044–2053. [Google Scholar] [CrossRef] [PubMed]
- Reichenbach, S. The order cytophagales. In The Prokaryotes; Springer: Berlin/Heidelberg, Germany, 2006; pp. 549–590. [Google Scholar]
- Kamat, S.S.; Raushel, F.M. The enzymatic conversion of phosphonates to phosphate by bacteria. Curr. Opin. Chem. Biol. 2013, 17, 589–596. [Google Scholar] [CrossRef] [PubMed]
- Niu, B.; Wang, W.; Yuan, Z.; Sederoff, R.R.; Sederoff, H.; Chiang, V.L.; Borriss, R. Microbial interactions within multiple-strain biological control agents impact soil-borne plant disease. Front. Microbiol. 2020, 11, 585404. [Google Scholar] [CrossRef] [PubMed]
- Sieber, C.M.; Paul, B.G.; Castelle, C.J.; Hu, P.; Tringe, S.G.; Valentine, D.L.; Andersen, G.L.; Banfield, J.F. Unusual metabolism and hypervariation in the genome of a gracilibacterium (Bd1-5) from an oil-degrading community. MBio 2019, 10, e02128–19. [Google Scholar] [CrossRef] [PubMed]
- Rivas, R.; Velázquez, E.; Willems, A.; Vizcaíno, N.; Subba-Rao, N.S.; Mateos, P.F.; Gillis, M.; Dazzo, F.B.; Martínez-Molina, E. A new species of Devosia that forms a unique nitrogen-fixing root-nodule symbiosis with the aquatic legume Neptunia natans (Lf) Druce. Appl. Environ. Microbiol. 2002, 68, 5217–5222. [Google Scholar] [CrossRef] [PubMed]
- Dong, X.; Lv, L.; Wang, W.; Liu, Y.; Yin, C.; Xu, Q.; Yan, H.; Fu, J.; Liu, X. Differences in distribution of potassium-solubilizing bacteria in forest and plantation soils in Myanmar. Int. J. Environ. Res. Public Health 2019, 16, 700. [Google Scholar] [CrossRef] [PubMed]
- Morte, M.A.; Cano, A.; Honrubia, M.; Torres, P. In vitro mycorrhization of micropropagated Helianthemum almeriense plantlets with Terfezia claveryi (desert truffle). Agric. Food Sci. 1994, 3, 309–314. [Google Scholar] [CrossRef]
- Zitouni-Haouar, F.E.; Fortas, Z.; Chevalier, G. Morphological characterization of mycorrhizae formed between three Terfezia species (desert truffles) and several Cistaceae and Aleppo pine. Mycorrhiza 2014, 24, 397–403. [Google Scholar] [CrossRef]
- Badger-Emeka, L.I.; Emeka, P.M.; Aldossari, S.; Khalil, H.E. Terfezia claveryi and Terfezia boudieri extracts: An antimicrobial and molecular assay on clinical isolates associated with eye infections. Pharmacogn. Mag. 2020, 16, 780. [Google Scholar] [CrossRef]
- Benucci, G.M.; Bonito, G.M. The truffle microbiome: Species and geography effects on bacteria associated with fruiting bodies of hypogeous Pezizales. Microb. Ecol. 2016, 72, 4–8. [Google Scholar] [CrossRef]
- Vahdatzadeh, M.; Deveau, A.; Splivallo, R. The role of the microbiome of truffles in aroma formation: A meta-analysis approach. Appl. Environ. Microbiol. 2015, 81, 6946–6952. [Google Scholar] [CrossRef]
- Giorgio, M.; Niccolò, B.G.; Benedetta, T.; Luisa, M.; Leonardo, B.F.; Gregory, B.; Pietro, B.; Alberto, A.; Domizia, D.; Emidio, A. Fungal and bacterial diversity in the Tuber magnatum ecosystem and microbiome. Microb. Ecol. 2022, 2, 1–4. [Google Scholar] [CrossRef] [PubMed]
- Sharma, M.P.; Grover, M.; Chourasiya, D.; Bharti, A.; Agnihotri, R.; Maheshwari, H.S.; Pareek, A.; Buyer, J.S.; Sharma, S.K.; Schütz, L.; et al. Deciphering the role of trehalose in tripartite symbiosis among rhizobia, arbuscular mycorrhizal fungi, and legumes for enhancing abiotic stress tolerance in crop plants. Front. Microbiol. 2020, 11, 509919. [Google Scholar] [CrossRef] [PubMed]
- Rahimlou, S.; Bahram, M.; Tedersoo, L. Phylogenomics reveals the evolution of root nodulating alpha-and beta-Proteobacteria (rhizobia). Microbiol. Res. 2021, 21, 126788. [Google Scholar] [CrossRef] [PubMed]
- Schulze, J.; Pöschel, G. Bacterial inoculation of maize affects carbon allocation to roots and carbon turnover in the rhizosphere. Plant Soil 2004, 267, 235–241. [Google Scholar] [CrossRef]
- Deveau, A.; Palin, B.; Delaruelle, C.; Peter, M.; Kohler, A.; Pierrat, J.C.; Sarniguet, A.; Garbaye, J.; Martin, F.; Frey-Klett, P. The mycorrhiza helper Pseudomonas fluorescens BBc6R8 has a specific priming effect on the growth, morphology and gene expression of the ectomycorrhizal fungus Laccaria bicolor S238N. New Phytol. 2007, 175, 743–755. [Google Scholar] [CrossRef] [PubMed]
- Wurst, S.; Vender, V.; Rillig, M.C. Testing for allelopathic effects in plant competition: Does activated carbon disrupt plant symbioses? Plant Ecol. 2010, 211, 19–26. [Google Scholar] [CrossRef]
- Glick, B.R. The enhancement of plant growth by free-living bacteria. Can. J. Microbiol. 1995, 41, 109–117. [Google Scholar] [CrossRef]
- Mehta, S.; Nautiyal, C.S. An efficient method for qualitative screening of phosphate-solubilizing bacteria. Curr. Microbiol. 2001, 43, 51–56. [Google Scholar] [CrossRef]
- Sbrana, C.; Bagnoli, G.; Bedini, S.; Filippi, C.; Giovannetti, M.; Nuti, M.P. Adhesion to hyphal matrix and antifungal activity of Pseudomonas strains isolated from Tuber borchii ascocarps. Can. J. Microbiol. 2000, 46, 259–268. [Google Scholar] [CrossRef]
- Li, Q.; Yan, L.; Ye, L.; Zhou, J.; Zhang, B.; Peng, W.; Zhang, X.; Li, X. Chinese black truffle (Tuber indicum) alters the ectomycorrhizosphere and endoectomycosphere microbiome and metabolic profiles of the host tree Quercus aliena. Front. Microbiol. 2018, 9, 2202. [Google Scholar] [CrossRef] [PubMed]
- Belimov, A.A.; Safronova, V.I.; Sergeyeva, T.A.; Egorova, T.N.; Matveyeva, V.A.; Tsyganov, V.E.; Borisov, A.Y.; Tikhonovich, I.A.; Kluge, C.; Preisfeld, A.; et al. Characterization of plant growth promoting rhizobacteria isolated from polluted soils and containing 1-aminocyclopropane-1-carboxylate deaminase. Can. J. Microbiol. 2001, 47, 642–652. [Google Scholar] [CrossRef]
- Dupré Ch Chevalier, G.; Morizet, J.; Leblevenec, L. Influence de l’azote et du phosphore sur la mycorhization de Quercus pubescens Willd. par Tuber melanosporum Vitt. en conditions contrôlées. Les Mycorhizes: Biologie et utilisation. Colloq. l’INRA 1982, 13, 147–153. [Google Scholar]
- Sourzat, P. Comment cultiver la truffière? In Guide Pratique de Trufficulture; Lycée Professionnel Agricole et Viticole de Cahors: Le Montat, France, 2002; pp. 65–94. [Google Scholar]
- Treseder, K. A meta-analysis of mycorrhizal responses to nitrogen, phosphorus and atmospheric CO2 in field studies. New Phytol. 2004, 164, 347–355. [Google Scholar] [CrossRef] [PubMed]
- Suz, L.M.; Martín, M.P.; Fischer, C.R.; Bonet, J.A.; Colinas, C. Can NPK fertilizers enhance seedling growth and mycorrhizal status of Tuber melanosporum-inoculated Quercus ilex seedlings? Mycorrhiza 2010, 20, 349–360. [Google Scholar] [CrossRef]
- Morte, A.; Navarro-Ródenas, A.; Nicolás, E. Physiological parameters of desert truffle mycorrhizal Helianthemum almeriense plants cultivated in orchards under water deficit conditions. Symbiosis 2010, 52, 133–139. [Google Scholar] [CrossRef]
- Aasfar, A.; Bargaz, A.; Yaakoubi, K.; Hilali, A.; Bennis, I.; Zeroual, Y.; Meftah Kadmiri, I. Nitrogen fixing Azotobacter species as potential soil biological enhancers for crop nutrition and yield stability. Front. Microbiol. 2021, 12, 628379. [Google Scholar] [CrossRef] [PubMed]
- Timmusk, S.; Behers, L.; Muthoni, J.; Muraya, A.; Aronsson, A.C. Perspectives and challenges of microbial application for crop improvement. Front. Plant Sci. 2017, 8, 49. [Google Scholar] [CrossRef] [PubMed]
- Selosse, M.A.; Schneider-Maunoury, L.; Taschen, E.; Rousset, F.; Richard, F. Black truffle, a hermaphrodite with forced unisexual behaviour. Trends Microbiol. 2017, 25, 784–787. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Li, J.M.; Tang, Y.J.; Xing, Y.M.; Qiao, P.; Li, Y.; Liu, P.G.; Guo, S.X. Chinese black truffle-associated bacterial communities of Tuber indicum from different geographical regions with nitrogen fixing bioactivity. Front. Microbiol. 2019, 10, 2515. [Google Scholar] [CrossRef]
- Robbins, C.; Thiergart, T.; Hacquard, S.; Garrido-Oter, R.; Gans, W.; Peiter, E.; Schulze-Lefert, P.; Spaepen, S. Root-associated bacterial and fungal community profiles of Arabidopsis thaliana are robust across contrasting soil p levels. Phytobiomes 2018, 2, 24–34. [Google Scholar] [CrossRef]

| Nitrite and Ammonia | 2017 | 2018 | 2019 | |||
|---|---|---|---|---|---|---|
| NO2− | NH4− | NO2− | NH4+ | NO2− | NH4+ | |
| (mg/kg) | (mg/kg) | (mg/kg) | ||||
| Control-Soil | N.D. | 2.3 | 0.73 | - | 0.66 | 4.2 |
| Mycorrhizal-Soil | N.D. | 2.4 | 0.63 | - | 0.73 | 12.7 |
| Potassium | K+ (mg/L) | K+ (mg/L) | K+ (mg/L) | |||
| Control-Soil | 0.58 | 8.4 | 8.3 | |||
| Mycorrhizal-Soil | 0.41 | 9.7 | 20.8 | |||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Satish, L.; Barak, H.; Keren, G.; Yehezkel, G.; Kushmaro, A.; Ben-Dov, E.; Kagan-Zur, V.; Barak, Z.; Sitrit, Y. The Microbiome Structure of the Symbiosis between the Desert Truffle Terfezia boudieri and Its Host Plant Helianthemum sessiliflorum. J. Fungi 2022, 8, 1062. https://doi.org/10.3390/jof8101062
Satish L, Barak H, Keren G, Yehezkel G, Kushmaro A, Ben-Dov E, Kagan-Zur V, Barak Z, Sitrit Y. The Microbiome Structure of the Symbiosis between the Desert Truffle Terfezia boudieri and Its Host Plant Helianthemum sessiliflorum. Journal of Fungi. 2022; 8(10):1062. https://doi.org/10.3390/jof8101062
Chicago/Turabian StyleSatish, Lakkakula, Hana Barak, Guy Keren, Galit Yehezkel, Ariel Kushmaro, Eitan Ben-Dov, Varda Kagan-Zur, Ze’ev Barak, and Yaron Sitrit. 2022. "The Microbiome Structure of the Symbiosis between the Desert Truffle Terfezia boudieri and Its Host Plant Helianthemum sessiliflorum" Journal of Fungi 8, no. 10: 1062. https://doi.org/10.3390/jof8101062
APA StyleSatish, L., Barak, H., Keren, G., Yehezkel, G., Kushmaro, A., Ben-Dov, E., Kagan-Zur, V., Barak, Z., & Sitrit, Y. (2022). The Microbiome Structure of the Symbiosis between the Desert Truffle Terfezia boudieri and Its Host Plant Helianthemum sessiliflorum. Journal of Fungi, 8(10), 1062. https://doi.org/10.3390/jof8101062







