Abstract
Biofilm formation during infections with the opportunistic pathogen Aspergillus fumigatus can be very problematic in clinical settings, since it provides the fungal cells with a protective environment. Resistance against drug treatments, immune recognition as well as adaptation to the host environment allows fungal survival in the host. The exact molecular mechanisms behind most processes in the formation of biofilms are unclear. In general, the formation of biofilms can be categorized roughly in a few stages; adhesion, conidial germination and development of hyphae, biofilm maturation and cell dispersion. Fungi in biofilms can adapt to the in-host environment. These adaptations can occur on a level of phenotypic plasticity via gene regulation. However, also more substantial genetic changes of the genome can result in increased resistance and adaptation in the host, enhancing the survival chances of fungi in biofilms. Most research has focused on the development of biofilms. However, to tackle developing microbial resistance and adaptation in biofilms, more insight in mechanisms behind genetic adaptations is required to predict which defense mechanisms can be expected. This can be helpful in the development of novel and more targeted antifungal treatments to combat fungal infections.
1. Introduction
Aspergillus fumigatus (A. fumigatus) is a saprophytic fungus that can normally be found in the soil, where it lives on organic debris. The airborne conidia of this opportunistic fungus are spread abundantly, and hundreds of conidia are inhaled by humans on a daily basis [1]. Inhalation of these conidia by healthy individuals is normally not harmful since they are eliminated by lung innate immune defense systems like mucociliary clearance [2] and alveolar macrophages, the main phagocytic cells of the respiratory tract [3]. However, A. fumigatus can cause infections in the respiratory tract of immunocompromised individuals, such as patients following chemotherapy, receiving transplants and immune-suppressive drugs or those in intensive care [4,5]. In most of these patients, A. fumigatus infection starts in the respiratory tract, resulting in varying clinical diseases such as invasive aspergillosis (IA), allergic bronchopulmonary aspergillosis (ABPA) and aspergilloma [1]. Next to Candida albicans (C. albicans), A. fumigatus is also a major pathogen responsible for nosocomial infections. Whereas Candida species are responsible for 80% of fungal nosocomial infections [6], A. fumigatus is the most common species to occur in infections in the airways of immunocompromised individuals, causing severe and fatal invasive infections [1].
Developments in medical therapy and technology such as organ transplantations accompanied with immunosuppression and new chemotherapeutic agents have increased the survival of patients suffering from severe diseases that previously would be fatal. Due to these advances in medical technology, the number of immunocompromised patients that are vulnerable to nosocomial infections by fungi has also increased [7]. In hospitals, nosocomial aspergillosis has become a severe threat to immunosuppressed patients due to the airborne conidia of A. fumigatus, and where outbreaks of nosocomial aspergillosis can occur due to construction work or deficient ventilation systems [8,9]. Moreover, the implementation of medical devices such as prostheses, catheters, and mechanical heart devices are also a major source of fungal as well as bacterial infections that can be difficult to treat [10,11].
Infection of A. fumigatus can be severe and difficult to treat due to its ability to form a biofilm on surfaces [12]. These biofilms consist of a microbial community that can strongly adhere to each other and to biotic and abiotic surfaces. The microbial community is encased by a polymeric extracellular matrix (ECM), that is composed of primarily of polysaccharides. The ECM functions as a protective sheet and offers a framework for cell cohesion and adhesion to the surface [13]. Moreover, the ECM controls dispersion of cells and acts as a source of nutrients for the cells. The shield of biofilm also possesses defensive characteristics, making the biofilm more tolerant against immune cells and drug treatments [10].
To develop strategies to prevent or treat infections of A. fumigatus, the underlying mechanism of biofilm formation needs to be understood. This paper will give an overview of the current knowledge on A. fumigatus biofilm formation compared to the knowledge available for C. albicans biofilm, which is studied more broadly. First, the different stages and mechanisms of biofilm formation will be described and compared with the known mechanisms of C. albicans. Secondly, the interaction between A. fumigatus and other microorganisms will be highlighted. Finally, the mechanism and effect of genetic adaptation and micro-evolution in A. fumigatus biofilms will be described.
2. Stages of Biofilm Development and Molecular Pathways
The formation of a fungal biofilm starts with the attachment of conidia to the biotic or abiotic surface. In vitro studies using scanning electron microscopy (SEM) and confocal laser scanning microscopy [14,15] showed that the formation of A. fumigatus biofilm can be roughly divided into different stages; (1) adhesion to the surface, (2) conidial germination into hyphae and development, (3) biofilm maturation with development of mycelia, ECM production, hyphal layering networks and formation of channels, and (4) cell dispersion [15]. In C. albicans, the stages of biofilm development are similar to A. fumigatus and comprise adhesion, initiation of biofilm formation, biofilm maturation and dispersion [16].
2.1. Adhesion
Adhesion is considered to be an important step in the process of fungal infection, since it is the first interaction between the fungi and the host. After inhalation, the airborne conidia come in contact with the respiratory tract and adhere onto airway epithelial cells [17]. The initial attraction between the conidia and surface is weak, and to strengthen attachment to the host surface, cell-surface components known as adhesins are used in fungal biofilm formation [15]. In the biofilm formation of C. albicans, fungal adhesion is regulated by members of a class of proteins known as glycosylphosphatidylinositol-dependent cell wall proteins (GPI-CWP). These GPI-CWP proteins include Hwp1, Hwp2 and a group of eight adhesion genes (ALS1-ALS7 and ALS9) that are part of the agglutinin-like sequence (ALS) family. These genes encode for proteins that possess characteristics of adhesin glycoproteins on the cell surface [18,19,20].
In A. fumigatus, electron microscopic and biochemical studies have identified molecules on the outer layer of the cell wall of A. fumigatus conidia. This study showed that the outer layer of the conidia was characterized by a layer of rodlets made of hydrophobins, a family of small hydrophobic proteins on the cell surface [21]. These hydrophobins were identified as RodAp, RodBp, RodCp, RodDp, RodEp and RodFp [15]. It is suggested that the hydrophobic characteristic of these proteins allows for their adhesion to hydrophobic abiotic or biotic surfaces, and that they are involved in the initiation of the biofilm process [22]. In a study where single and multiple hydrophobin-deletion mutants were constructed, the results showed that only RodA is necessary for efficient rodlet formation on the cell surface of conidia and adherence on polystyrene plates [23]. RodA proved to be responsible for hydrophobicity, integrity of the cell wall, conidial production, and susceptibility to external factors such as drugs. The importance of RodA was also observed in another study, where disruption of RodA decreased adherence of conidia to collagen and albumin, although not laminin or pulmonary epithelial cells [17]. The rodlet layer that includes RodA, also disguises the fungal cell wall components β-glucan, chitin, and glycoproteins that can provoke immunogenic reactions. Whereas β-glucan is recognized by receptors such as Dectin-1 in case of interaction of epithelial cells, interaction with ephrin type-A receptor 2 is also involved [24,25]. Another component that is suggested to be involved in the adhesion process of A. fumigatus is sialic acid. This component is part of a family of derivatives of neuraminic acid present on the cell surface of conidia. Removal of the surface sialic acids on the conidial surface resulted in decreased adherence of the conidia to fibronectin, suggesting that conidia adhere to basal lamina proteins via the negatively charged sugars on their surface, which are presumed to be sialic acids [26].
Other cell wall carbohydrate components also showed to mediate fungal adherence. Purified fragments of fibronectin were used to analyze which components on the conidial surface are involved in the process of adherence. The results showed that A. fumigatus conidia preferably bound to the non-glycosylated 40-kDa fragment, containing the glycosaminoglycan binding domain. Negatively charged carbohydrates, such as dextran sulfate and heparin, as well as high-ionic strength buffers, hindered binding of conidia to both fibronectin and basal lamina. This indicated that negatively charged carbohydrates on the cell wall surface of conidia may bind to the glycosaminoglycan binding domain of fibronectin and other basal-lamina proteins. Together, these data suggest that negatively charged carbohydrate moieties on the conidial surface could mediate binding to macromolecules of the host [27].
The developmentally regulating protein MedA was proposed to play an important role as an adhesin and in regulating the adherence and expression of conidiation genes. A. fumigatus mutants deficient in MedA were impaired in adherence to pulmonary epithelial cells, vascular endothelial cells and fibronectin [28]. Next to fungal adhesion to host cells, MedA also manages efficient biofilm formation, since MedA deletion mutants also showed to be decreased in biofilm formation. However, the exact molecular mechanism whereby MedA enhances adherence remains unclear.
At a later stage during biofilm formation, adhesion can also occur on a deeper level in the human host. Growing hyphae can interact with pulmonary epithelial cells, where they adhere to and invade the abluminal surface of vascular endothelial cells to eventually access the vascular compartment. When the hyphae entered the blood vessels, hyphal fragments can be distributed to other sites in the host where they can adhere to the luminal surface of endothelial cells before passing through them and invading into deep tissues. A. fumigatus hyphae can induce host cell damage and death. There, it is likely that the basal lamina within the airways and blood vessels are exposed, and that fungal cells can adhere to basement membrane macromolecules including laminin, fibronectin and collagen [17].
2.2. Conidial Germination and Hyphal Development
After attachment of A. fumigatus conidia to the surface, further colonization occurs through hyphal proliferation [14]. Additionally, as shown in a study by Wasylnka & Moore [29] with two A. fumigatus strains expressing green fluorescent protein, conidia can be internalized by nonprofessional phagocytes in vitro such as epithelial and endothelial cells to germinate at a later stage [30]. This was also observed with conidia of C. albicans, where a study showed that blastoconidia could be internalized by endothelial cells and macrophages in mice [31].
The conidial germination of A. fumigatus involves disruption of the hydrophobic rodlet layer, revealing the inner conidium walls that are composed of polysaccharides, which are hydrophilic cell wall components. During germination, a hydrophobic tip can be found on a single germinating spore. The conidium loses its surface hydrophobicity progressively and the new growth-point exhibits a coexistence of hydrophobic rodlets and hydrophilic polysaccharides [15,22].
After adhesion of the conidia, conidial germination into hyphae begins with the formation of tube-like channels that possess the hydrophobic nature of the cell wall that is suggested to enhance hyphal development [22]. In hyphal development, the adhesive exopolysaccharide galactosaminogalactan (GAG) plays an important role in adherence of A. fumigatus hyphae to cells of the human host and modulating the immune response during infection. GAG is secreted by growing hyphae, where it binds to the surfaces of these hyphae, resulting in a polysaccharide sheath covering the growing organisms [32]. The fungal regulatory protein MedA and developmental transcription factor StuA both affect the formation of adherent biofilms. Disruption of StuA in A. fumigatus strains resulted in reduced adherence to pulmonary epithelial lines and other abiotic substrates [33]. A comparative transcriptome analysis of the ΔmedA and ΔstuA regulatory mutants identified a gene encoding a putative UDP-glucose-epimerase, designated uge3, which was dysregulated in both the ΔstuA and ΔmedA mutants. Disruption of Uge3 resulted in full impairment of GAG synthesis, and markedly decreased adhesion to host cells and biofilm formation. This indicates that GAG is necessary for the efficient attachment of hyphae and biofilm structural integrity [33].
In C. albicans, a proliferation phase is also followed after attachment of the yeast cells, and initiation of filamentation will result in hyphal development, eventually leading to the biofilm development process [34]. In hyphal development, hyphal morphogenesis is important for adhesion maintenance. In this process, the proteins EFG1 and BCR1 are key regulators of several adhesin genes, including most of the ALS gene family, EAP1 and HWP1. A study by McCall et al. [35] showed that adhesion maintenance proteins in C. albicans are expressed at different times during growth phases. Whereas the adhesion maintenance protein Ywp1 is expressed during late biofilm growth, disruption of EFG1 and BCR1 in C. albicans deletion strains demonstrated poor adherence of hyphae in biofilm. This indicates that filamentation is important for further expression of adhesion maintenance proteins. Moreover, the expression of BCR1 is under the regulation of the hyphal regulator Tec1p, where BCR1 is a downstream component of the hyphal regulatory network that couples expression of cell-surface genes to hyphal differentiation. This indicates that hyphal cells are specialized to present adherence components to ensure biofilm structure and integrity [36].
2.3. Biofilm Maturation
After development of the hyphae, the process of biofilm maturation is initiated. During this stage, a biofilm matrix is formed consisting of the extracellular matrix (ECM) that covers the colony surface and binds the cells to form the structural base of the biofilm and glues the hyphae together [12,15]. In A. fumigatus, the structural components of the mature biofilm generally consist of GAG, galactomannan, α-1,3 glucans, monosaccharides, proteins, antigens and lipids, melanin, polyols, and extracellular DNA (eDNA) [13]. Additionally, an expression study showed that there are at least two hydrophobin genes (rodB and rodC) expressed in aerial static mycelium. However, no rodlet proteins were detected in the same ECM [12].
In C. albicans, the biofilm matrix is characterized by a structured mixture of pseudohyphal and hyphal cells encased by an ECM that consists of glycoproteins, carbohydrates, α-mannan and β-1,6-glucan polysaccharides, β-1,3-glucan, lipids and eDNA [16].
The structure of biofilms is dependent of the environmental conditions of the microorganism. Therefore, the biofilm structure can vary and keep reshuffling as they keep on adapting to their environment. For example, the composition of the ECM could vary between aspergillosis pathologies. In a study by Loussert et al. [37] it was shown that an ECM is produced in the lungs at the surface of A. fumigatus hyphae that are present in patients suffering from aspergilloma or IA. The ECM at these different sites showed a different hyphal organization. Light microscopic observations showed that during aspergilloma a ball of strongly agglutinated hyphae without any host cells inside was formed. Additionally, a dense material surrounded the hyphae, and at the periphery of the ball, many blood cells were identified. On the contrary, the hyphae detected in experimental IA were shown to be separated. Moreover, the ECM at these sites contained galactomannan and GAG as observed in vitro. Nevertheless, α-1,3 glucan which was also present in the ECM in vitro was detected only in aspergilloma at the periphery of the ECM. Other components, such as the major antigenic glycoproteins, were present in aspergillosis and IA ECM in vitro but were not observed in the ECM produced in vivo. A possible explanation for this effect is that these antigens are secreted during infection, but do not accumulate as much as seen in vitro, since these antigens were detected in patients with aspergillosis and IA [37].
In fungal biofilm development, the ECM that is formed contributes to the structural base of the biofilm, including the exopolymeric substance (EPS) and network of mycelia. The EPS occurs with a mucous appearance that adheres completely and covers the hyphae, forming a covering sheath over the fungal cells [15]. In the mature biofilm, the immobilized cells function as an ecosystem that includes continuous interactions in the form of cell-cell communication acting as a structured network that holds the cells together. The EPS in the ECM undertakes many important functions such as adhesion, cell aggregation, providing protection against antimicrobial agents and host defense mechanisms, providing a nutrient source, stabilization of the cell community and the exchanging of genetic information [15].
In both A. fumigatus and C. albicans, eDNA is an important component of the ECM, where it contributes to structural integrity and antifungal resistance [38,39,40]. A mechanism by which eDNA can provide biofilm resistance is by changing the extracellular environment. Since eDNA is an anionic macromolecule, this molecule is able to chelate cations such as magnesium ions, resulting in a decline of effective concentration of Mg2+ in the fungal environment. Moreover, certain concentrations of DNA can regulate the induction of the cationic antimicrobial peptide resistance operon, PA3552–PA3559 in Pseudomonas aeruginosa (P. aeruginosa). This indicates that the presence of eDNA can contribute to genomic DNA release and inducible antibiotic resistance [41].
Another role of eDNA is identified as providing the exchange of genetic information by horizontal gene transfer [42]. Several studies showed that plasmid transfer occurs at an increased rate in biofilms compared to planktonic cultures. A study by Hausner & Wuertz [43] quantified the gene transfer process in a simple laboratory-based biofilm system with a strain of Alcaligenes eutrophus as a recipient of a green fluorescent protein-tagged plasmid derivative from Escherichia coli (E. coli). The results of this experiment showed that more transconjugants occurred compared to on a plate count. The transfer of genes in eDNA can contribute to the development of antibiotic resistance in microbial populations.
In A. fumigatus, the eDNA is created by autolysis and derives from fungal cells by the secretion of chitinases by A. fumigatus, favoring its release [39]. Although it is not yet clear how eDNA contributes to antifungal resistance, treatment of A. fumigatus biofilm with DNase resulted in a destabilized integrity of the biofilm and an increased susceptibility to antifungals amphotericin B and caspofungin. Therefore, it is suggested that the presence of eDNA reduces exposure of the target cells to the antifungal drugs [13].
In C. albicans as well as A. fumigatus, a large part of the ECM consists of components of the host when grown in vivo. A previous study showed that in three infection site models of C. albicans, a biofilm matrix was produced containing host components such as hemoglobin, albumin, and alpha globulins, amylase, fibrinogen and keratin. The presence of host components also depended on the biofilm surface (venous catheter, urinary catheter or denture model). Although the precise role of these host components in the biofilms are not yet clear, the attraction of blood cells could be beneficial in the acquisition of iron. Moreover, another explanation could be that the host components are used for the structure of the biofilm to save energy by producing these components themselves [44,45].
2.4. Cell Dispersion
The stage of cell dispersion is generally considered as the terminal stage of biofilm development, also referred to as seeding dispersal [46]. Cell dispersion typically occurs in response to stressful environmental changes, where dissemination allows viable cells to persist in other locations of the host where it can further reproduce [15]. In cell dispersion of A. fumigatus, the part comprising the conidia or hyphae are detached from the biofilm. In the study of González-Ramírez et al. [15], asynchronous biofilm development was observed, in particular at the biofilm-maturation stage when the new conidia were capable of germinating, producing new mycelial growth and hyphal modifications prior to dispersion. In C. albicans, the cells dispersed from a biofilm are yeast cells that originate from the topmost hyphal layers of the biofilm [47]. In C. albicans, the dispersion of round, yeast-form cells are suggested to occur during biofilm development, but larger amounts of yeast cells are dispersed when the biofilm is in a more mature stage. Cell dispersion can lead to novel biofilm formation in the host, possibly leading to systemic infections in the bloodstream or to the spreading of the infection to other parts of the host. Known transcriptional regulators that are involved in C. albicans cell dispersion are Ume6, Nrg1 and Pes1 (also known as Nop7). Also, a component of a chromatin-modifying complex, known as Set3, is required for dispersal. Set3 is suggested to be recruited by the transcriptional regulator Nrg1 [16]. The heat shock protein 90 (Hsp90) is suggested to play a role in cell dispersion of C. albicans. Experiments performed in vitro showed that inactivation of Hsp90 reduced C. albicans biofilm growth and maturation and reduced dispersal of biofilm cells [48]. Additionally, Hsp90 plays a role in the stress response pathways, which is discussed in the next section. If Hsp90 also plays a role in the process of A. fumigatus cell dispersion is not known.
3. Drug Resistance Mechanisms
One of the most important characteristics of the biofilm is to function as a protective shield, making the community more tolerant against physical disruptions and drug treatments. A. fumigatus and C. albicans have several resistance mechanisms to protect them against environmental pressures and the in-host environment. An overview of these mechanisms is given in Table 1. In both A. fumigatus and C. albicans, it is hypothesized that the presence of the ECM reduces drug susceptibility by working as a shield that absorbs the drug and inhibits it from reaching its cellular targets. In a study by Al-Fattani and Douglas [49], a C. albicans biofilm was examined under continuous flow resembling in vivo conditions, and a correlation between the degree of extracellular matrix production and antifungal resistance was found. For A. fumigatus, the formation of biofilm is also suggested to hinder or delay penetration of antifungal agents, but the underlying mechanisms remains unclear [12,40]. Other mechanisms in A. fumigatus and C. albicans to promote drug resistance are the use of multidrug resistance (MDR) pumps, also known as efflux pumps, which transport toxic substrates from the cells into their environment. The expression of MDR pumps in A. fumigatus (AfuMDR) was examined in a study by Rajendran et al. [50], where in an in vivo mouse biofilm model, constitutive upregulation of AfuMDR4 was detected after treatment with voriconazole after 24 h. This suggested that efflux pumps were expressed in A. fumigatus biofilms, and thus contributes to azole resistance. Other known efflux pumps contributing to the transport of toxic components are MDR1, MDR2, and MDR4 [51]. Quorum sensing mechanisms could affect biofilm formation and development of drug resistance in A. fumigatus biofilms, although this has hardly been investigated. In this respect the inhibitory effects of farnesol on A. fumigatus growth by misplacing the tip localized Rho1 and Rho3 proteins and inhibition of cell wall signaling are remarkable [52]. Farnesol was previously shown to reduce the transition to hyphal forms and activated the expression of drug-resistance genes of C. albicans [53]. Furthermore, it was shown that farnesol affected biofilm formation of C. albicans and was a specific modulator of drug efflux pumps of the ABC-multidrug family [54].
For C. albicans, a study by Mukherjee et al. [55], showed that efflux pumps CDR1, CDR2 and MDR1 contribute to azole resistance. However, this effect was only shown in the early-phase biofilms but not the mature biofilm phase, since knock-out mutants showed to be more susceptible to the azole in early-phase biofilms. Disruption of these efflux pumps did not affect biofilm formation. Furthermore, variation in the sterol profile as a possible mechanism of drug resistance was investigated. Sterol analysis showed that changes in the composition of sterol are involved in the intermediate and mature phases of biofilm development in C. albicans.
Next to contributing to biofilm growth, integrity and maintenance, eDNA was found to play a role in promoting drug resistance. As mentioned earlier, degradation of eDNA by DNase resulted in loss of biofilm mass and increased susceptibility to antifungals in A. fumigatus and C. albicans [38,39]. Moreover, eDNA induces conidial surface adhesion in early stages of biofilm development and co-localizes with the polysaccharides of the ECM biofilm, and becomes part of the ECM surrounding the biofilm cells. The eDNA is suggested to enhance the structure of biofilms, but the exact mechanisms behind this effect remains unclear. Therefore, it would be interesting to analyze whether eDNA that originates from neutrophils serves as a crucial nutrient source for the fungal biofilm and is beneficial to biofilm formation in vivo [45].
A resistance characteristic of A. fumigatus and C. albicans biofilm against antimicrobial agents are a special class of protected cells also known as persister cells. These persister cells are hypothesized to be cells that have differentiated into an inactive but protected state and neither grow nor die in the presence of microbicidal antibiotics. Persister cells are dormant variants of regular cells and are randomly formed in biofilms. These cells are found in A. fumigatus and C. albicans biofilms, and hold high tolerance against antifungals [56]. In the study of Lafleur et al. [57], treatment of C. albicans biofilms with two antimicrobial agents, amphotericin B and chlorhexidine, resulted in a biphasic killing pattern, indicating that a subpopulation of highly tolerant cells, termed persisters, existed. The surviving C. albicans persisters were only detected in biofilms, in contrast to planktonic populations. The persister cell hypothesis could explain why biofilms are protected from a wide set of antimicrobial agents. Moreover, reinoculation of the cells that survived the killing of the biofilm by amphotericin B produced a new biofilm with a new subpopulation of persisters. This indicated that C. albicans persister cells are not mutants but phenotypic variants of the wild type [57]. The population of persister cells has been estimated to form about 0.1–10% of all cells in the biofilm and are suggested to survive exposure to antimicrobial agents and reseed the biofilm. Persister cells have been identified in several pathogens such as E. coli, Staphyloccoccus aureus (S. aureus) and P. aeruginosa, where large populations of persisters were identified [58].
Certain stress pathways can be induced in the biofilm to enhance antimicrobial resistance. In C. albicans, the component calcineurin, a Ca2+ calmodulin-activated serine/threonine-specific protein phosphatase, plays an important role in homeostasis, morphogenesis, virulence, and was also found to play a role in resistance development. Disruption of this component was shown to improve the efficacy of fluconazole on C. albicans biofilm [59]. Next to its role in cell dispersion, the heat shock protein Hsp90 is also involved in another C. albicans stress response pathway. Hsp90 has been shown to enable the emergence and maintenance of drug resistance in planktonic conditions by stabilizing the protein phosphatase calcineurin and MAPK Mkc1. Next to reduced biofilm growth, maturation and cell dispersion, inactivation of Hsp90 also impairs resistance of C. albicans biofilm to antifungal drugs such as azoles [48]. Also in A. fumigatus, inhibition of Hsp90 in vitro was shown to improve the efficacy of antifungals such as azoles and echinocandins against biofilms. This implicated that Hsp90 acts as a regulator in drug resistance [48].
An additional component involved in the stress response pathway includes the MAPK MKc1p, an important component of the signal transduction pathway that is triggered by cell wall stress. Inactivation of this protein resulted in defective hyphal formation and the development of biofilm and resistance to fluconazole in C. albicans mutants [60].


Table 1.
Drug resistance mechanisms in A. fumigatus and C. albicans biofilm.
Table 1.
Drug resistance mechanisms in A. fumigatus and C. albicans biofilm.
| A. fumigatus | C. albicans | References | |
|---|---|---|---|
| ECM | Reducing drug susceptibility by preventing the drug from reaching their cellular target | Reducing drug susceptibility by preventing the drug from reaching their cellular target | [12,49] |
| Efflux pumps | Upregulation of efflux pumps genes such as AfuMDR4, MDR1, MDR2, MDR4 | CDR1, CDR2 and MDR1 contributing to azole resistance | [40,51] |
| eDNA | Promotes resistance against amphotericin B and caspofungins | Promoting resistance to amphotericin B and echinocandins | [39,40] |
| Persister cells | Dormant drug tolerant cells developed during biofilm formation that can serve as an inoculum for new biofilms | Dormant drug tolerant cells developed during biofilm formation that can serve as an inoculum for new biofilms | [40,56] |
| Induced stress response pathway | HSP90 pathway | MAPK-, HSP90- and calcineurin pathway to develop azole resistance | [40,48] |
4. Interaction with Host Immune Systems
Next to drug resistance mechanisms, another essential characteristic of the biofilm is the ability to evade or affect the immune systems of the host. Whereas A. fumigatus and C. albicans share similar mechanisms in drug resistance, the mechanisms to evade host immune systems are more different.
In A. fumigatus, the exopolysaccharide GAG is secreted by the growing hyphae and plays several roles in the interaction between the host and pathogen (Figure 1). It functions as an adhesin and interferes with immune recognition of the host by reducing exposure of the cell wall components like β-1,3 glucans that are considered important fungal pathogen-associated molecular patterns (PAMPs) recognized by the innate immune system [32]. The β-1,3 glucans are increasingly exposed as conidia swell and begin to germinate, but are covered again during the production of GAG during hyphal growth. Although it is likely that there are more PAMPS that are covered by GAG, these are not identified yet. GAG also interferes with neutrophil-mediated immunity to A. fumigatus, where it induces neutrophil apoptosis by triggering neutrophils to produce reactive oxygen species (ROS). Moreover, GAG also impedes neutrophils through extracellular killing by neutrophil extracellular traps (NETs). Additionally, GAG is able to induce immunomodulatory effects by modulating the cytokine production and T-cell responses [32].

Figure 1.
Potential roles of GAG during fungal infection.
The extracellular matrix of C. albicans biofilms are crucial in the protection against the host immune cells. For example, ROS and NETs are less triggered by C. albicans biofilms compared to planktonic cells [61,62]. Time lapse imaging showed impaired NET release during growth of C. albicans biofilms. This revealed that NET inhibition depended on an intact ECM since genetic or physical disruption of this component resulted in NET release. This showed that C. albicans biofilms can hinder neutrophil response through an inhibitory pathway induced by the ECM [61]. Additionally, the polysaccharide β-1,3 glucan in the biofilm matrix is suggested to prevent drugs from reaching their targets, but also to reduce neutrophil activation. In a study by Xie et al. [63] it was shown that the presence of extracellular polysaccharides such as β-glucans can inhibit neutrophil antibiofilm function. Early biofilms were coated with glucanase-treated matrix or laminarin, which showed that some β-glucans interfere with neutrophil–biofilm interactions that activate an ROS response. Although components on the surface of the biofilm matrix can interfere with the pattern recognition receptors in the host immune cells, the exact mechanisms underlying the interaction between biofilms and the host immune systems still need to be investigated further.
5. Interaction between A. fumigatus and Other Micro-Organisms
In nature, biofilms are generally composed of several micro-organisms. Co-infections of interacting microorganisms of varying species can form an ecosystem of pathogens leading to worsened disease in the patient. For example, co-infection of C. albicans with other pathogens such as E. coli can enhance the adhesion of C. albicans in bladder mucosa. Similarly, pneumonia caused by P. aeruginosa infection is more likely to occur in individuals already infected by C. albicans [64]. Co-existence of bacteria and fungi in the human host can result in co-aggregation and form mixed-species biofilm. This can lead to an even more harmful infection in the human host than infection of a single pathogen.
Whereas interaction of C. albicans with other microorganism species are more widely studied [16], interaction of A. fumigatus with other microbial species is less described, except for interaction with P. aeruginosa and S. aureus. Interaction between A. fumigatus and S. aureus is observed in the eye disease keratitis. However, this interaction is scarcely studied, but analysis of the mixed biofilm showed structural changes of both microbes and an antibiosis effect of S. aureus on A. fumigatus was observed and which strongly affected hyphal development [65]. More information is available regarding the interaction between A. fumigatus and P. aeruginosa and which will be described in the following sections. In patients suffering from cystic fibrosis (CF), A. fumigatus and P. aeruginosa are the species that are found most commonly [66,67]. Co-colonization of these opportunistic pathogens can lead to both mutual inhibition and promotion of their growth in biofilm.
5.1. Inhibitory Mechanisms of P. aeruginosa on A. Fumigatus
In vitro, growth of A. fumigatus can be inhibited by P. aeruginosa by the effect of phenazines, a nitrogen-containing heterocyclic compound secreted by P. aeruginosa during growth. This compound is able to affect the modification of multiple host cellular response mechanisms, and is considered an important antimicrobial factor against bacteria, fungi or mammalian cells, and is involved in the interactions with eukaryotic hosts and host tissues [68]. The P. aeruginosa phenazines are involved in electron shuttling, redox chemistry, and the promotion of P. aeruginosa biofilm growth through toxic superoxide generation and signaling. Phenazines include five secreted molecules; 5-N-methyl-1-hydroxyphenazine (PYO), 1-hydroxyphenazine (1-HP), phenazine-1-carboxamide (PCN), phenazine-1-carboxylic acid (PCA) and dirhamnolipids (diRhls) [69]. The secreted phenazines inhibit A. fumigatus growth by generating reactive oxygen species (ROS) and reactive nitrogen species (RNS). Whereas PYO, 1-HP, PCN and PCA inhibit A. fumigatus growth through RNS/ROS, DiRhls can inhibit A. fumigatus biofilm growth by blocking the β-1,3 glucan synthase activity in A. fumigatus [70]. An overview of different interaction mechanisms between these two pathogens is described by Zhao & Yu, [69]. An important note is that the studies that show the inhibitory effect of phenazines on A. fumigatus were performed in vitro. To analyze if the same effects are also present in vivo, further research is needed [69].
One of the key mechanisms of A. fumigatus inhibition by P. aeruginosa involves the competition for iron. To survive, both microbes are dependent on iron in their environment. P. aeruginosa produces the siderophore pyoverdine that is able to chelate iron and deny this molecule to A. fumigatus. In turn, A. fumigatus uses hydroxamate siderophores (sidA) to defend itself against these antifungal effects of P. aeruginosa [71]. Another study showed that the Pseudomonas quinolone signal (PQS), a quorum sensing molecule, is also able to chelate iron and delivers iron to the P. aeruginosa cell membrane using siderophores [72]. The bacteriophage Pf4 produced by P. aeruginosa also binds iron and thereby causes an iron deficiency for A. fumigatus. However, inhibition by Pf4 was shown to be more effective in the developing stage of A. fumigatus biofilm than in biofilm formation [73]. Small colony variants (SCVs) of P. aeruginosa are slow growing sub-populations of bacteria with different phenotypic traits such as an atypical colony morphology and different biochemical characteristics. Therefore, they are more difficult to identify clinically. These SCVs are more adapted to persist in mammalian cells and are less vulnerable to antimicrobial agents than the wild type colonies. They can cause recurrent or latent infections in case of an emerging protective environment of the host cell [74]. The SCVs formed by P. aeruginosa are involved in the inhibition of A. fumigatus biofilm formation. This inhibition is related to the pyoverdine pathway [75].
5.2. Inhibiting Mechanisms of A. fumigatus on P. aeruginosa
A. fumigatus has developed mechanisms to withstand the growth-inhibitory effects of P. aeruginosa. For example, the secreted bacterial phenazines can be modified by A. fumigatus metabolically and decrease their inhibitory effect. The phenazine PCA can be modified to 1-HP,1-methoxyphenazine and phenazine-1-sulfate. As described previously, 1-HP has an inhibitory effect on A. fumigatus. However, 1-HP is also able to induce the production of the A. fumigatus siderophores TAFC and FsC. The phenazines PCA and PYO produced by P. aeruginosa can be converted by A. fumigatus to phenazine dimers, which have a decreased inhibitory effect on A. fumigatus biofilm growth. PCA also plays a role in the iron acquisition of P. aeruginosa and conversion of PCA can lead to reduced biofilm formation. PYO is a QS molecule and affects transcriptional regulation and induces biofilm formation. Therefore, conversion of PYO might negatively affect the QS regulation system [69].
Gene expression and proteomic analyses showed that in a mature biofilm, specialized genes for the production of gliotoxin are expressed [76]. Gliotoxin secreted by A. fumigatus is an immuno-evasive toxin that plays an important role in mediating A. fumigatus-associated colonization. Production of gliotoxin is upregulated during mycelial growth in A. fumigatus and in biofilms. The increased biofilm production of A. fumigatus also helps to inhibit P. aeruginosa growth. The hyphae present in the A. fumigatus biofilm are more mature with a different morphology, and it is suggested that these mature hyphae are less susceptible to the cytotoxic molecules produced by P. aeruginosa compared to germinating conidia and germlings. The increased sensitivity of the latter for P. aeruginosa virulence factors was proposed to be due to a difference in the metabolic activity which is uniform in germlings but more localized to apical regions in mature hyphae [77].
5.3. Mutualism between P. aeruginosa and A. fumigatus
P. aeruginosa can promote the growth of A. fumigatus under certain conditions. Although secreted phenazines of P. aeruginosa inhibit growth of A. fumigatus, sub-bacteriostatic concentrations of phenazines can promote growth in A. fumigatus under specific circumstances. For example, the sub-bacteriostatic phenazines PYO, PCA and PCN can promote A. fumigatus growth by enhancing iron uptake. The redox function of 1-HP which is able to chelate iron and deprive A. fumigatus of iron can also promote A. fumigatus growth [78].
Volatile organic compounds (VOCs) secreted by P. aeruginosa such as volatile sulfur compounds (VSCs) can promote A. fumigatus growth in vitro [79]. Via genetic approaches and gas chromatography mass spectrometry-mediated volatile analysis, it was shown that A. fumigatus assimilates volatile sulfur compounds (VSCs) via cysteine synthase or homocysteine-synthase. This process is necessary for the use of VSCs as sulfur sources, since P. aeruginosa-derived VSCs trigger growth of A. fumigatus wild-type, but not of a mutant defective in cysteine synthase and homocysteine-synthase on sulfur-limiting media [79]. However, in this study a substrate was used that resembles the lung environment, indicating that this volatile-based synergy also occurs in the human lungs, although further research is still needed to confirm this. Also, the mechanistic basis of such cross-talk and its physiological relevance during co-infection remains unknown.
Under certain conditions, P. aeruginosa contributes to the colonization and growth of A. fumigatus in CF patients. Therefore, it is possible that both pathogens work together to interfere with host immune factors, resulting in increased infection and decreased pulmonary function. Moreover, cytotoxic elastase produced by P. aeruginosa increases in the presence of A. fumigatus, leading to damage of the human lung epithelial cells, decreasing lung function and enhancing progression of the disease [80]. However, their relationship becomes more competitive with growth and the change of environmental circumstances. In a low Fe environment, P. aeruginosa inhibits A. fumigatus via several mechanisms. In an environment with high iron levels, P. aeruginosa enhances A. fumigatus growth [72]. In the presence of co-infection, P. aeruginosa and A. fumigatus do not exist in isolation; instead, they affect each other and maintain a changing synergistic relationship.
6. Genetic Evolution and Adaptation
In order to keep surviving in the human host, pathogenic bacteria, yeast and fungi have to adapt to the in-host environment. Adaptation to the in-host environment can occur through several mechanisms. One of these mechanisms involves epigenetic changes, where the genetic code of DNA is unchanged but the activation of certain genes is modified. Another strategy to enhance the survival chances of the pathogen is through genetic adaptation, which can be defined as the acquisition of heritable modifications. Genetic adaptation in biofilms can be a severe clinical issue, since the resistant biofilms can enhance their resistance even further through additional mutations. Genetic diversification is often more observed in biofilm populations compared to planktonic cell cultures [81]. For example, in the bacterium Mycobacterium tuberculosis (M. tuberculosis), the formed biofilms showed distinct characteristics compared to planktonic cultures. Whereas exposure to isoniazid and rifampicin lead to cell death in planktonic cultures, biofilms exposed to the antibiotics had a high proportion of cells (around 10%) that survived incubation with isoniazid treatment [82]. This example suggests that genetic adaptation is more likely to occur in cells of biofilms. To tackle the problem of these adaptive mutations that seem to occur at a higher rate in biofilms, studies should reveal the underlying mechanism behind genetic adaptation. This could provide new insight in the evolutionary predictability of molecular changes in certain conditions e.g., in CF patients and help to develop novel strategies against A. fumigatus antifungal resistance and infection.
6.1. Primary and Acquired Resistance Mechanisms
Fungal pathogens can use several mechanisms to evolve resistance against antifungal drugs. These adapted resistance mechanisms can be divided into primary and acquired resistance, and an overview of these mechanisms were described previously [80]. The primary resistance mechanisms involve the structure of the formed biofilm that inhibits fungal drug susceptibility, target incompatibility through amino acid substitutions, stress response signaling, overexpression of efflux pumps that act as drug transporters, and alterations in cell wall permeability that could prevent antifungal drugs from reaching their targets [83].
The acquired resistance mechanisms include overexpression of the drug target and alterations in cellular pathways. Other acquired resistance mechanisms can overlap with the mechanisms of the primary resistance pathways. These include amino acid substitutions leading to reduced drug binding, signaling through stress response pathways, and upregulation of efflux pumps. Moreover, acquired resistance mechanisms can be accelerated by several factors that enhance genetic adaptation. One of the factors that can enhance genetic adaptation includes genetic plasticity, the presence of hypermutator strains, or environmental pressures, such as sudden exposure to antifungals, that can lead to strains that are more resistant to fungicides. The combination of these primary and acquired mechanisms can result in the selection of resistant microorganisms [83].
Cells in biofilms are able to adapt to the in-host environment and stresses in multiple ways. The altering of gene expression is a strategy where the cells are able to change the phenotypic traits in favor of survival. Another mechanism to enhance beneficial traits is through genetic adaptation, where mutations or changes in gene expression may lead to more beneficial traits that enhance the survival chances of the pathogenic fungi. Mutations may result in point mutations or can change larger parts of the genome. An overview of these genetic adaptation mechanisms will be presented in the following sections.
6.2. Phenotypic Plasticity
Biofilms are able to quickly adapt to environmental changes or stresses. One of these mechanisms is the ability to rapidly change their phenotypic traits to enhance their survival chances. An example is the upregulation of efflux pumps as a result of the presence of antifungal agents. Multiple studies that were also mentioned earlier showed that efflux pumps, such as AfuMDR4, MDR1, MDR2, and MDR4, were upregulated in A. fumigatus biofilms when exposed to concentrations of antifungals [50,51]. Moreover, in a study where A. fumigatus biofilm was exposed to antimicrobial agents, it was shown that the fungus was able to adjust its transcriptome rapidly within 60 min of exposure to itraconazole [84]. This suggests that when A. fumigatus is challenged with a new and stressful environment, the mechanisms of phenotypic plasticity allow for the adjustment of its physiology to enable growth and reproduction.
Phenotypic plasticity can also be triggered in cases of environmental changes in the host. Since efficient adhesion and colonization of pathogens onto surfaces are crucial steps in the process of biofilm formation, it is possible that biofilms are flexible in adapting to a variety of surfaces. A study by Shemesh et al. [85] analyzed the molecular conditions during biofilm growth of Streptococcus mutans (S. mutans) on different dental surfaces. Growth analyses showed that the bacteria formed a smoother and thicker biofilm on a hydroxyapatite surface compared to the other tested surfaces. The differentially expressed genes of S. mutans were analyzed through transcriptional profiling. This revealed widely based changes in the patterns of gene expression during development of the S. mutans biofilm on various surfaces. These results indicated that the physiological state of bacteria can be influenced by the type of substance of attachment. In another study, separate isolates of P. aeruginosa were exposed to the same environmental pressures, which resulted in the production of similar biofilm phenotypes. This observed independent evolution of particular biofilm phenotypic traits in non-related clinical isolates showed that parallel evolution in P. aeruginosa can occur in adaptation to comparable environments [86].
The same effect of parallel evolution was observed in isolates of Burkholderia cenocepacia (B. cenocepacia). In these experiments, shared selection pressure drove parallel evolution across distinct environments. The B. cenocepacia populations evolved for 90 days under all combinations of high or low carbon availability with selection for either planktonic or biofilm modes of growth. They found that genetic parallelism between low-carbon biofilm and low-carbon planktonic populations was very low despite shared selection for growth under low-carbon conditions, suggesting that evolution in low-carbon environments could develop stronger trade-offs between the growth of biofilm and planktonic cultures. However, they also found that a population’s fitness in a specific environment was positively correlated with the genetic similarity between that population and the populations that evolved in that specific environment. This showed that in general, evolution in similar environments led to higher levels of genetic parallelism [87].
6.3. Genomic Plasticity
Genomic plasticity allows microorganisms to adapt to environmental stresses and in-host defense mechanisms. In most cases, changes are brought to the DNA. An example of this is through horizontal gene transfer (HGT), where genetic material is transferred between microorganisms, and for example contributes to antibiotic resistance development. HGT has been detected in the genomes of A. fumigatus, A. nidulans and A flavus using the phyletic distribution-based bioinformatics software called HGT-Finder. These analyses support the hypothesis that secondary metabolite gene clusters evolved via HGT [88].
A study by Valdes et al. [89] investigated whether A. fumigatus was also able to acquire mutations in the case of long-term exposure to the sino-nasal environment of dogs. Through whole genome sequencing, single-nucleotide polymorphisms (SNPs) were identified in isolates from dogs suffering from sino-nasal aspergillosis. They found that in a total of 28 isolates contained on average around 45,000 non-synonymous (ns) SNPs, but six isolates from a total of three dogs contained approximately 2.5-fold more mutations. The SNP isolates formed a closely related subclade, despite the fact that they originated from 3 genotypes. This indicated that the six isolates had acquired similar genetic changes. Part of these ns-SNPs were shared, which gives the impression that they resulted from pressure in the host. The variants containing 2.5-fold more SNPs might have an increased mutation rate which could be the result of defects in DNA repair, since many mutations actually were detected in DNA-repair machinery. Together, the results of this study indicated that the mixture of variants of A. fumigatus found at the sino-nasal mucosal surface in the dog is a result of in-host adaptation.
In another study where the adaptive resistance mechanisms were analyzed, thirteen A. fumigatus strains were isolated from a patient suffering from recurring IA over a period of two years. In all strains identical microsatellite genotypes were observed and were considered isogenic. Through whole genome comparisons they identified non-synonymous single nucleotide polymorphisms that potentially play a role in adaptation in the host. Further experiments analyzing growth, virulence, and conidiation in these strains demonstrated that A. fumigatus in this patient had enhanced azole resistance mechanisms and additional phenotypes that promoted fungal resistance [90].
Other forms of adaptation strategies of A. fumigatus and C. albicans lie in their reproduction strategies [91,92]. A. fumigatus is able to reproduce through asexual reproduction, sexual reproduction, and parasexual reproduction. Genomic changes can occur during the three reproduction processes. During asexual reproduction, conidiophores that are produced by hyphae can produce a great number of spores. Through every mitotic phase, there is a probability of a mutation. However, since the number of mitotic divisions is so large during asexual production, asexual spores can contribute to a large number of spontaneous mutations that can be beneficial for azole resistance mutations when exposed to azoles [93].
During sexual reproduction, genetic variation can occur through recombination during the meiosis process, where the genotypes of the parental strains of opposite mating types are reshuffled. In case of azole pressure, more resistant phenotypes can occur through natural selection. However, sexual production can take several months to complete, and was only demonstrated under controlled laboratory conditions. Sexual production is unlikely to occur in the human host due to required conditions [92,94].
The third reproduction strategy of A. fumigatus involves parasexual recombination. This strategy also enhances genetic diversity through the reshuffling of genes that can result in genetically different but compatible hyphae. Whereas asexual and sexual reproduction are known to occur in many microorganisms, parasexual recombination is more considered as a strategy of the fungus to generate genetic diversity than a reproduction mode. During parasexual recombination, the fungal hyphae fuse together, allowing the nuclei to fuse as well. This can result in a diploid phase in which mitotic recombination can occur, before changing back to the initial haploid stage. This form of reproduction strategy can also result in genetic variation and enabling the production of genotypes that are more adapted to the changing in-host environment [92,95].
6.4. Hypermutation
Microorganisms that undergo mutations at a high rate, also known as hypermutator strains, can be very efficient in genetically adapting to environmental stresses. These mutations can be caused by errors in DNA replication or chromosome segregation, which can result in changes in whole chromosomes or chromosome segments [96].
In a study by Mena et al. [97], it was demonstrated that P. aeruginosa strains in CF patients suffering from chronic respiratory infection (CRI) experience substantial genetic adaptation. In a collection of isolates, the role of hypermutation was examined in this process, since a study by Oliver et al. [98] found that one of the trademarks of CRI is the widespread occurrence of DNA mismatch repair in (MMR) system-deficient mutator strains. They found mutations in 34 genes in the study collection of 90 P. aeruginosa isolates, but these mutations were not homogeneous. The mutations were significantly concentrated in the mutator lineages, which represented 17% of the isolates. On an interesting note, they found that increased accumulation of mutations in mutator lineages was not a consequence of overrepresentation of mutations in genes involved in antimicrobial resistance, which was the only adaptive trait that was connected so far to hypermutation in CF patients. This demonstrated that hypermutation in DNA repair systems also plays a major role in P. aeruginosa genome evolution and adaptation during CRI [97].
Another mechanism that was found to possibly contribute to genetic adaptation in biofilms is the appearance of coding tandem repeats. Genes containing multiple coding minisatellite- and short tandem repeats are highly dynamic components of genomes. This variation in intragenic repeat number supplies the functional diversity of cell surface antigens. Through this mechanism, pathogens are able to adapt rapidly to environmental pressures and avoid the host immune system [99]. In a study by Levdansky et al. [100], the genome of A. fumigatus was analyzed for open reading frames (ORFs) containing tandem repeat regions, and the variability in the length of these regions in different isolates was determined. They found that expansion and contraction of coding tandem repeat regions occurred in A. fumigatus genes, and that a significant proportion of those genes undergoing expansion and contraction of coding repeat regions encode putative cell surface proteins. Deletion of one of these genes, Afu3g08990, which encodes the CspA protein localized at the cell wall, results in rapid germination and decreased adherence to ECM, suggesting that it plays a role in defining cell surface properties. Moreover, in a study by Fan et al. [101] disruption of the cspA gene resulted in an alteration of the conidial surface, a decrease in biofilm formation and less resistance to antifungal drugs. Also, internalization by lung epithelial cells was shown to be increased in the ΔcspA mutant, which suggests that CspA not only contributes in preserving the integrity of the cell wall, but also plays a role in biofilm establishment, response to antifungal drugs, and the uptake of A. fumigatus.
DNA modification through aneuploidy is another described mechanism where the fungal genome obtains a different chromosome number than the wildtype [102]. Aneuploidy is also detected in other fungal pathogens isolated from patients and the environmental setting, which suggests that variation in chromosome organization is a familiar mechanism of pathogenic fungi to quickly generate genetic diversity in the case of environmental stresses, such as exposure to antifungals [91]. Large changes in chromosomes in cases of antimicrobial pressure have also been observed in C. albicans, where truncation or fragmentation of chromosomes have been reported for isolates from the clinical and laboratory setting. In an isolate of C. albicans from a bone marrow transplant patient treated with fluconazole, truncations in the (Chr5) chromosome were observed [103]. Also, processes such as variation in copy numbers during replication and loss of heterozygosity, a cross chromosomal event where one of the alleles at the locus can get lost, may result in genomic changes and can stimulate antimicrobial resistance and virulence. However, the underlying mechanism is not fully clear [104].
7. Transcriptomic Studies of Biofilms
Together with gene deletion analysis and morphological studies, biofilms produced by A. fumigatus have been studied to a very limited extent in in vitro and in vivo model systems with microarray and RNA-sequencing (Table 2). A comparison between in vitro biofilms using microarray, RNA-sequencing and proteomic analysis was already described by Muszkieta et al. [105] and Beauvais & Latgé [106]. In these studies, gene expression was compared between A. fumigatus biofilms grown on solid agar medium (BF) versus submerged planktonic growth in liquid medium conditions (PL). These studies clearly indicated that aerial growth on agar medium represented a BF-like state with characteristics like reduced susceptibility for antifungals, the presence of an extracellular matrix, and changes in metabolism that were more associated with virulence. In contrast, the submerged growth system (PL) was shown to be a poor in vitro disease model. Furthermore, RNA-sequencing was shown to be a much more sensitive technique, and only around 49% of the differently expressed genes identified in microarrays were also detected in RNA-sequencing analysis. The proteomic analysis provided very limited data that could be connected to gene-expression analysis [105]. Currently, proteomic technology is much more advanced, however no new proteomic analysis of A. fumigatus biofilms has been described yet.
Among the important genes that were found to be upregulated in the datasets of the studies described in Table 2 were the ones involved in production of gliotoxin (GliG, GliK) and melanin (Arb1, Arp2). Gliotoxin, for example, is an important molecule in the regulation of the host response via several mechanisms, for example the induction of apoptosis in leukocytes and the inhibition of T-cell and B-cells response [107]. Interestingly, expression of the gliotoxin and hexadehydro-astechrome clusters was also detected in mature biofilms in dogs suffering from sino-nasal aspergillosis (SNA) [89], underscoring the important role of these secondary metabolites for A. fumigatus growing in biofilm.
Stress response is another crucial factor for the establishment of a successful biofilm since the fungus encounters stress in the host environment (e.g., oxidative stress, low oxygen and nutritional immunity). Mechanisms to cope with such challenges can be expected to be upregulated. For example, superoxide dismutase (SODs) are proteins that detoxify harmful reactive oxygen species (ROS), and a well-known immunoreactive SOD from A. fumigatus is SodA (Afu5g09240), which was upregulated in 24 h and 48 h biofilm [76], Interestingly, however, two SOD proteins, SodB (Afu4g11580), and SodM (Afu1g14550) were downregulated in 24 h and 48 h biofilms, indicating a condition dependent expression of the superoxide dismutases of A. fumigatus. Such a pattern is not new and was also reported previously in a study comparing the gene expression of conidia, swollen conidia, germinated conidia and mycelia [108]. Here it was shown that SodA and SodB were highly expressed in resting conidia, and SodM only on mycelium.
Transcription factors play a major role in the resistance to stress as well. It has been shown in Aspergillus nidulans that the transcription factor AtfA plays a critical role in the induction of many different stress-responsive genes. Its influence stretches from responses towards osmotic, heat shock- and oxidative-stress [109,110]. The importance of this gene in A. fumigatus biofilm is underlined by the relative high expression levels shown in all studies indicated in Table 2. In in vivo gene expression studies in mice, a role for the transcription factor PacC in development of invasive aspergillosis was described. Bronchoalveolar lavage of infected mice in a time course experiment up to 16 h post infection followed by micro-array analysis revealed insight into the early steps in fungal development which can also be regarded as initial steps of biofilm formation. PacC mutants revealed a non-invasive phenotype and were shown to remain localized at the surface of lung epithelial cells.

Table 2.
Overview of gene expression studies of A. fumigatus biofilms. Whole genome expression studies of A. fumigatus biofilm growth were used to cross-reference with the PHI database [111]. No modifications of expression data or reanalysis of raw data was made in the case of Bertuzzi et al. 2014 and Gibbons et al. 2012 [112,113]. The data marked with bullet points was used to describe which genes were up- or down-regulated in that specific study. Expression thresholds for differential expression were kept as reported by the authors. Additionally, names of the categories used for Figure 2 are depicted.
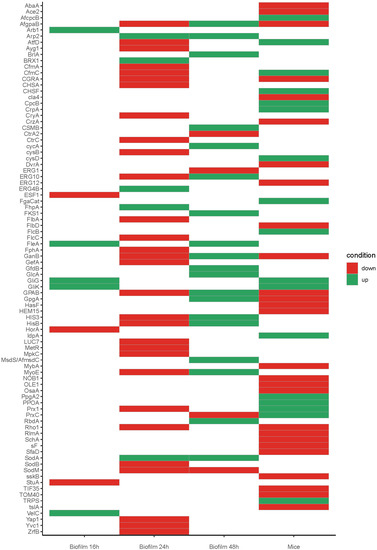
Figure 2.
Heatmap showing the expression profiles of selected genes that are known to be involved in pathogenesis as reported by PHI database [111], expression and thresholds for differential expression was kept as reported by the authors, for mice and biofilm data, some conditions were grouped into one condition were: Mice up = Early up + late up. Mice down = Early down + late down. Biofilm 16 up = Biofilm unique (only expressed in Biofilm) 2-fold up in Biofilm. Biofilm 16 down = 2-fold down in Biofilm (Table 2). Full expression values are on Supplementary Material Table S1.
Table 2.
Overview of gene expression studies of A. fumigatus biofilms. Whole genome expression studies of A. fumigatus biofilm growth were used to cross-reference with the PHI database [111]. No modifications of expression data or reanalysis of raw data was made in the case of Bertuzzi et al. 2014 and Gibbons et al. 2012 [112,113]. The data marked with bullet points was used to describe which genes were up- or down-regulated in that specific study. Expression thresholds for differential expression were kept as reported by the authors. Additionally, names of the categories used for Figure 2 are depicted.
| Objective | Model | Object of Analysis | Time Point/Series of Analysis after Growth | Expression Threshold for Differential Expression, and Selected Categories | |
|---|---|---|---|---|---|
| Bertuzzi et al. (2014) [112] | Transcriptional profiling of ΔpacC ATCC mutants | In vivo | Bronchoalveolar lavage of infected mice (in vivo) | 4, 8, 12 and 16 h of WT and ΔpacC ATCC | Log2 ratios ≥ +/− 1.5 relative to ungerminated spores (only WT) Early expressed: (differentially expressed at any or all of 4, 8 and 12 h post-infection Late expressed: (differentially expressed at either or both of 12 and 16 h post-infection
|
| Bruns et al. (2010) [76] | Proteome and transcriptome analysis of PL versus BF | In vitro | Planktonic and biofilm-grown A. fumigatus mycelium | 24 and 48 h |
|
| Gibbons et al. (2012) [113] | Gene expression analysis of BF versus PL | In vitro | Fungal tissue from planktonic and biofilm grown A. fumigatus | 16 h | Log2 ratio between Biofilm and planktonic RPKM values.
|
Another transcription factor known to play a role on stress response is Yap1. This transcription factor specifically regulates genes with defensive functions towards reactive oxygen intermediates (ROIs). ROIs are produced by alveolar macrophages, which have been shown to kill conidia. These alveolar macrophages are the most common immune cell that fight A. fumigatus in the lungs [3,114]. Yap1 was found to be down regulated in 24 h [76].
A. fumigatus growing in the host and biofilm will encounter nutritional immunity stress as a result of deprivation of nutrients, and zinc deprivation is one of them [112]. In the study of Bertuzzi et al. [112] it was observed that the zinc transporter ZrfB (Afu2g03860) was downregulated under neutral-alkaline conditions via PacC (Afu3g11970), suggesting that A. fumigatus biofilms are neutral to alkaline. Interestingly, mature biofilms in dogs suffering from sino-nasal aspergillosis (SNA) have an elevated pH of around 8 [89]. In agreement with this, it was found that the ZrfC (Afu4g09560) zinc transporter was also expressed in biofilms in SNA, which is required for zinc acquisition under alkaline conditions [115].
We compared the in vivo whole gene expression studies in the mice of Bertuzzi et al. [112] with the transcriptomic studies of in vitro formed biofilms [76,113] and analyzed which differentially expressed genes that are known to be involved in pathogenesis (as reported in the PHI database) were shared in these three studies (Figure 2). Remarkably few genes were found to be shared between all conditions. For example, the dataset of Gibbons et al. [113] shared upregulation of genes involved in the gliotoxin cluster (GliG and GliK) with the dataset of Bertuzzi et al. [112], and upregulation of the lectin FleA with the 48 h biofilm condition of Bruns et al. [76]. Although this analysis was made on a small set of genes, it shows that future work needs a standard biofilm assay to study biofilm formation and pathogenesis. Furthermore, the results emphasize the great plasticity of this fungus to adapt to several conditions.
8. Conclusions and Future Directions
Since the increased developments in medical treatments, patients suffering from severe diseases now have higher chances of survival compared to years before. However, these developments in the medical field also led to an increase of immunosuppressed patients in hospitals. These patients are more vulnerable to nosocomial infection caused by opportunistic pathogens such as C. albicans and A. fumigatus. Candida spp. can cause diseases such as urinary tract infections, hospital pneumonia, and biofilm formation on the surface of medical devices. A. fumigatus is responsible for diseases such as invasive aspergillosis, and can also infect the human host by biofilm growth on implanted medical devices. These infections are an increasing problem in the clinical setting. Infection of these pathogens are often harder to treat in the case of biofilm formation, since biofilms often possess strong antibiotic resistance.
This review first covered the classical view of biofilm development, focusing on the different stages of biofilm formation that can be generally divided into adhesion of the conidia to host-cells, conidial germination and development of hyphae, maturation of the biofilm, and cell dispersion. Compared to A. fumigatus, biofilm formation in C. albicans is studied more broadly. Biofilm growth of A. fumigatus is slower compared to C. albicans, and studies regarding A. fumigatus biofilm seem to be performed more often in vitro than in vivo compared to C. albicans studies. Following A. fumigatus isolates more often in a clinical setting could help to gain more insights in the underlying mechanisms of A. fumigatus biofilm formation.
The reason biofilms have such a strong antimicrobial resistance is due to the ability to rapidly adapt to their (in-host) environment. The most efficient mechanisms behind this adjustment to environmental stresses are linked to genetic adaptation. The strategy of genetic adaptation can occur on different levels. Alterations can occur in the form of phenotypic traits where certain genes can be upregulated such as efflux pumps, but in essence the fungal genome remains unchanged.
On a deeper level of genetic adaptation, modifications can be made in the genome through point mutations, horizontal gene transfer, and errors in DNA repair systems leading to mutations that may result in beneficial traits for the survival of the fungi. Another moment in the lifecycle of the fungus that is sensitive for probable mutations is during the reproduction process. Through asexual, sexual, and parasexual reproduction, spontaneous mutations can occur that may result in helpful characteristics in antifungal resistance. At last, through processes such as hypermutation in strains, loss of heterozygosity, severe errors in DNA mismatch repair, aneuploidy and copy number variation, large, or even whole parts of the chromosome can be altered, increasing the mutation rate in these strains and therefore the chance of novel beneficial traits.
Most research concerning A. fumigatus biofilm formation is focused on the underlying mechanisms in the biofilm development process. However, the problem in treating A. fumigatus infections lies in the ability to quickly develop antifungal resistance. Therefore, more research is needed to understand the strategies that A. fumigatus uses to increase its genetic diversity during infection in the environment of the human host. A strategy to investigate genetic adaptation could include performing large scale analysis with in depth sequencing methods of isolates from patients. This way, genomic modifications, large or small, can be analyzed and could give a better view of the A. fumigatus mutations that occur randomly or perhaps in a more controlled manner when exposed to antifungal drugs. Perhaps specific genes could be discovered that are related to genomic plasticity.
With this information, genomic changes leading to resistance could be detected easier, or even predict which specific antifungal treatments can be used best to enhance treatment efficiency. With such strategies, more targeted treatments for aspergillosis and other A. fumigatus related diseases could be developed and improved. In order to understand the molecular processes of biofilm formation, standard biofilm assays are required for future work which will allow more in-depth gene expression studies. It will be challenging to develop such assays that reflect in vivo biofilm growth.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/jof8010048/s1, Table S1: Biofilm expression data.
Author Contributions
Conceptualization, H.d.C.; formal analysis, I.V. and J.v.N.; writing—original draft preparation, E.S.; writing—review and editing, I.V., J.v.N. and H.d.C.; supervision, H.d.C. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Latgé, J.-P. Aspergillus fumigatus and Aspergillosis. Clin. Microbiol. Rev. 1999, 12, 310–350. [Google Scholar] [CrossRef] [Green Version]
- Nawroth, J.C.; van der Does, A.M.; Ryan, A.; Kanso, E. Multiscale mechanics of mucociliary clearance in the lung. Philos. Trans. R. Soc. B Biol. Sci. 2020, 375, 20190160. [Google Scholar] [CrossRef] [PubMed]
- Philippe, B.; Ibrahim-Granet, O.; Prévost, M.C.; Gougerot-Pocidalo, M.A.; Perez, M.S.; Van der Meeren, A.; Latgé, J.P. Killing of Aspergillus fumigatus by Alveolar Macrophages Is Mediated by Reactive Oxidant Intermediates. Infect. Immun. 2003, 71, 3034–3042. [Google Scholar] [CrossRef] [Green Version]
- Young, R.C.; Bennett, J.E.; Vogel, C.L.; Carbone, P.P.; Devita, V.T. The Spectrum of the Disease in 98 Patients. Medicine 1970, 49, 147–173. [Google Scholar] [CrossRef]
- Zilberberg, M.D.; Shorr, A.F. Fungal Infections in the ICU. Infect. Dis. Clin. N. Am. 2009, 23, 625–642. [Google Scholar] [CrossRef] [PubMed]
- Pendleton, K.M.; Huffnagle, G.B.; Dickson, R.P. The significance of Candida in the human respiratory tract: Our evolving understanding. Pathog. Dis. 2017, 75. [Google Scholar] [CrossRef] [Green Version]
- Fridkin, S.K.; Jarvis, W.R. Epidemiology of nosocomial fungal infections. Clin. Microbiol. Rev. 1996, 9, 499–511. [Google Scholar] [CrossRef] [PubMed]
- Chazalet, V.; Debeaupuis, J.P.; Sarfati, J.; Lortholary, J.; Ribaud, P.; Shah, P.; Cornet, M.; Thien, H.V.; Gluckman, E.; Brücker, G.; et al. Molecular Typing of Environmental and Patient Isolates of Aspergillus fumigatus from Various Hospital Settings. J. Clin. Microbiol. 1998, 36, 1494–1500. [Google Scholar] [CrossRef] [Green Version]
- Vonberg, R.-P.; Gastmeier, P. Nosocomial aspergillosis in outbreak settings. J. Hosp. Infect. 2006, 63, 246–254. [Google Scholar] [CrossRef]
- Ramage, G.; Rajendran, R.; Gutierrez-Correa, M.; Jones, B.; Williams, C. Aspergillus biofilms: Clinical and industrial significance. FEMS Microbiol. Lett. 2011, 324, 89–97. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Desai, J.; Mitchell, A.P.; Andes, D. Fungal Biofilms, Drug Resistance, and Recurrent Infection. Cold Spring Harb. Perspect. Med. 2014, 4, a019729. [Google Scholar] [CrossRef] [Green Version]
- Beauvais, A.; Schmidt, C.; Guadagnini, S.; Roux, P.; Perret, E.; Henry, C.; Paris, S.; Mallet, A.; Prévost, M.-C.; Latgé, J.P. An extracellular matrix glues together the aerial-grown hyphae of Aspergillus fumigatus. Cell. Microbiol. 2007, 9, 1588–1600. [Google Scholar] [CrossRef] [PubMed]
- Mitchell, K.F.; Zarnowski, R.; Andes, D.R. Fungal Super Glue: The Biofilm Matrix and Its Composition, Assembly, and Functions. PLOS Pathog. 2016, 12, e1005828. [Google Scholar] [CrossRef] [Green Version]
- Singhal, D.; Baker, L.; Wormald, P.-J.; Tan, L. Aspergillus FumigatusBiofilm on Primary Human Sinonasal Epithelial Culture. Am. J. Rhinol. Allergy 2011, 25, 219–225. [Google Scholar] [CrossRef]
- González-Ramírez, A.I.; Ramírez-Granillo, A.; Medina-Canales, M.G.; Rodríguez-Tovar, A.V.; Martínez-Rivera, M.A. Analysis and description of the stages of Aspergillus fumigatus biofilm formation using scanning electron microscopy. BMC Microbiol. 2016, 16, 243. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lohse, M.B.; Gulati, M.; Johnson, A.D.; Nobile, C.J. Development and regulation of single- and multi-species Candida albicans biofilms. Nat. Rev. Microbiol. 2018, 16, 19–31. [Google Scholar] [CrossRef] [Green Version]
- Sheppard, D.C. Molecular mechanism of Aspergillus fumigatus adherence to host constituents. Curr. Opin. Microbiol. 2011, 14, 375–379. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hoyer, L.L. The ALS gene family of Candida albicans. Trends Microbiol. 2001, 9, 176–180. [Google Scholar] [CrossRef]
- Sundstrom, P. Adhesion in Candida spp. Cell. Microbiol. 2002, 4, 461–469. [Google Scholar] [CrossRef]
- Younes, S.; Bahnan, W.; Dimassi, H.; Khalaf, R.A. The Candida albicans Hwp2 is necessary for proper adhesion, biofilm formation and oxidative stress tolerance. Microbiol. Res. 2011, 166, 430–436. [Google Scholar] [CrossRef]
- Thau, N.; Monod, M.; Crestani, B.; Rolland, C.; Tronchin, G.; Latgé, J.P.; Paris, S. rodletless mutants of Aspergillus fumigatus. Infect. Immun. 1994, 62, 4380–4388. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dague, E.; Alsteens, D.; Latgé, J.-P.; Dufrêne, Y.F. High-Resolution Cell Surface Dynamics of Germinating Aspergillus fumigatus Conidia. Biophys. J. 2008, 94, 656–660. [Google Scholar] [CrossRef] [Green Version]
- Valsecchi, I.; Dupres, V.; Stephen-Victor, E.; Guijarro, J.I.; Gibbons, J.; Beau, R.; Bayry, J.; Coppee, J.-Y.; Lafont, F.; Latgé, J.-P.; et al. Role of Hydrophobins in Aspergillus fumigatus. J. Fungi 2017, 4, 2. [Google Scholar] [CrossRef] [Green Version]
- Latgé, J.-P.; Beauvais, A. Functional duality of the cell wall. Curr. Opin. Microbiol. 2014, 20, 111–117. [Google Scholar] [CrossRef] [PubMed]
- Keizer, E.M.; Wösten, H.A.B.; De Cock, H. EphA2-Dependent Internalization of A. fumigatus Conidia in A549 Lung Cells Is Modulated by DHN-Melanin. Front. Microbiol. 2020, 11, 534118. [Google Scholar] [CrossRef]
- Warwas, M.L.; Watson, J.N.; Bennet, A.J.; Moore, M.M. Structure and role of sialic acids on the surface of Aspergillus fumigatus conidiospores. Glycobiology 2007, 17, 401–410. [Google Scholar] [CrossRef] [Green Version]
- Wasylnka, J.A.; Moore, M.M. Adhesion of Aspergillus Species to Extracellular Matrix Proteins: Evidence for Involvement of Negatively Charged Carbohydrates on the Conidial Surface. Infect. Immun. 2000, 68, 3377–3384. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gravelat, F.N.; Ejzykowicz, D.E.; Chiang, L.Y.; Chabot, J.C.; Urb, M.; Macdonald, K.D.; Al-Bader, N.; Filler, S.G.; Sheppard, D.C. Aspergillus fumigatusMedA governs adherence, host cell interactions and virulence. Cell. Microbiol. 2010, 12, 473–488. [Google Scholar] [CrossRef] [Green Version]
- Wasylnka, J.A.; Moore, M.M. Uptake of Aspergillus fumigatus Conidia by Phagocytic and Nonphagocytic Cells In Vitro: Quantitation Using Strains Expressing Green Fluorescent Protein. Infect. Immun. 2002, 70, 3156–3163. [Google Scholar] [CrossRef] [Green Version]
- Filler, S.G.; Sheppard, D.C. Fungal Invasion of Normally Non-Phagocytic Host Cells. PLoS Pathog. 2006, 2, e129. [Google Scholar] [CrossRef] [PubMed]
- Kaposzta, R.; Marodi, L.; Hollinshead, M.; Gordon, S.; da Silva, R. Rapid recruitment of late endosomes and lysosomes in mouse macrophages ingesting Candida albicans. J. Cell Sci. 1999, 112, 3237–3248. [Google Scholar] [CrossRef]
- Speth, C.; Rambach, G.; Lass-Flörl, C.; Howell, P.L.; Sheppard, D.C. Galactosaminogalactan (GAG) and its multiple roles in Aspergillus pathogenesis. Virulence 2019, 10, 976–983. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gravelat, F.; Beauvais, A.; Liu, H.; Lee, M.J.; Snarr, B.D.; Chen, D.; Xu, W.; Kravtsov, I.; Hoareau, C.M.Q.; Vanier, G.; et al. Aspergillus Galactosaminogalactan Mediates Adherence to Host Constituents and Conceals Hyphal β-Glucan from the Immune System. PLoS Pathog. 2013, 9, e1003575. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wall, G.; Montelongo-Jauregui, D.; Bonifacio, B.V.; Lopez-Ribot, J.L.; Uppuluri, P. Candida albicans biofilm growth and dispersal: Contributions to pathogenesis. Curr. Opin. Microbiol. 2019, 52, 1–6. [Google Scholar] [CrossRef]
- McCall, A.D.; Pathirana, R.; Prabhakar, A.; Cullen, P.J.; Edgerton, M. Candida albicans biofilm development is governed by cooperative attachment and adhesion maintenance proteins. NPJ Biofilms Microbiomes 2019, 5, 1–12. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nobile, C.; Mitchell, A.P. Regulation of Cell-Surface Genes and Biofilm Formation by the C. albicans Transcription Factor Bcr1p. Curr. Biol. 2005, 15, 1150–1155. [Google Scholar] [CrossRef] [Green Version]
- Loussert, C.; Schmitt, C.; Prevost, M.-C.; Balloy, V.; Fadel, E.; Philippe, B.; Kauffmann-Lacroix, C.; Latgé, J.P.; Beauvais, A. In vivobiofilm composition ofAspergillus fumigatus. Cell. Microbiol. 2010, 12, 405–410. [Google Scholar] [CrossRef]
- Martins, M.; Uppuluri, P.; Thomas, D.P.; Cleary, I.; Henriques, M.; Lopez-Ribot, J.; Oliveira, R. Presence of extracellular DNA in the Candida albicans biofilm matrix and its contribution to biofilms. Mycopathologia 2009, 169, 323–331. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rajendran, R.; Williams, C.; Lappin, D.F.; Millington, O.; Martins, M.; Ramage, G. Extracellular DNA Release Acts as an Antifungal Resistance Mechanism in Mature Aspergillus fumigatus Biofilms. Eukaryot. Cell 2013, 12, 420–429. [Google Scholar] [CrossRef] [Green Version]
- Taff, H.T.; Mitchell, K.F.; Edward, J.A.; Andes, D.R. Mechanisms ofCandidabiofilm drug resistance. Future Microbiol. 2013, 8, 1325–1337. [Google Scholar] [CrossRef] [Green Version]
- Mulcahy, H.; Charron-Mazenod, L.; Lewenza, S. Extracellular DNA Chelates Cations and Induces Antibiotic Resistance in Pseudomonas aeruginosa Biofilms. PLoS Pathog. 2008, 4, e1000213. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Molin, S.; Tolker-Nielsen, T. Gene transfer occurs with enhanced efficiency in biofilms and induces enhanced stabilisation of the biofilm structure. Curr. Opin. Biotechnol. 2003, 14, 255–261. [Google Scholar] [CrossRef]
- Hausner, M.; Wuertz, S. High rates of conjugation in bacterial biofilms as determined by quantitative in situ analysis. Appl. Environ. Microbiol. 1999, 65, 3710–3713. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nett, J.E.; Zarnowski, R.; Cabezas-Olcoz, J.; Brooks, E.G.; Bernhardt, J.; Marchillo, K.; Mosher, D.F.; Andes, D.R. Host Contributions to Construction of Three Device-Associated Candida albicans Biofilms. Infect. Immun. 2015, 83, 4630–4638. [Google Scholar] [CrossRef] [Green Version]
- Shopova, I.; Bruns, S.M.; Thywissen, A.; Kniemeyer, O.; Brakhage, A.A.; Hillmann, F. Extrinsic extracellular DNA leads to biofilm formation and colocalizes with matrix polysaccharides in the human pathogenic fungus Aspergillus fumigatus. Front. Microbiol. 2013, 4, 141. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rumbaugh, K.P.; Sauer, K. Biofilm dispersion. Nat. Rev. Genet. 2020, 18, 571–586. [Google Scholar] [CrossRef]
- Uppuluri, P.; Acosta-Zaldívar, M.; Anderson, M.Z.; Dunn, M.J.; Berman, J.; Ribot, J.L.L.; Köhler, J.R. Candida albicans Dispersed Cells Are Developmentally Distinct from Biofilm and Planktonic Cells. mBio 2018, 9, e01338-18. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Robbins, N.; Uppuluri, P.; Nett, J.; Rajendran, R.; Ramage, G.; Lopez-Ribot, J.; Andes, D.; Cowen, L.E. Hsp90 Governs Dispersion and Drug Resistance of Fungal Biofilms. PLoS Pathog. 2011, 7, e1002257. [Google Scholar] [CrossRef] [PubMed]
- Al-Fattani, M.A.; Douglas, L.J. Biofilm matrix of Candida albicans and Candida tropicalis: Chemical composition and role in drug resistance. J. Med. Microbiol. 2006, 55, 999–1008. [Google Scholar] [CrossRef] [PubMed]
- Rajendran, R.; Mowat, E.; McCulloch, E.; Lappin, D.F.; Jones, B.; Lang, S.; Majithiya, J.B.; Warn, P.; Williams, C.; Ramage, G. Azole Resistance of Aspergillus fumigatus Biofilms Is Partly Associated with Efflux Pump Activity. Antimicrob. Agents Chemother. 2011, 55, 2092–2097. [Google Scholar] [CrossRef] [Green Version]
- Fanning, S.; Mitchell, A.P. Fungal Biofilms. PLoS Pathog. 2012, 8, e1002585. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dichtl, K.; Ebel, F.; Dirr, F.; Routier, F.H.; Heesemann, J.; Wagener, J. Farnesol misplaces tip-localized Rho proteins and inhibits cell wall integrity signalling in Aspergillus fumigatus. Mol. Microbiol. 2010, 76, 1191–1204. [Google Scholar] [CrossRef]
- Enjalbert, B.; Whiteway, M. Release from Quorum-Sensing Molecules Triggers Hyphal Formation during Candida albicans Resumption of Growth. Eukaryot. Cell 2005, 4, 1203–1210. [Google Scholar] [CrossRef] [Green Version]
- Sharma, M.; Prasad, R. The Quorum-Sensing Molecule Farnesol Is a Modulator of Drug Efflux Mediated by ABC Multidrug Transporters and Synergizes with Drugs in Candida albicans. Antimicrob. Agents Chemother. 2011, 55, 4834–4843. [Google Scholar] [CrossRef] [Green Version]
- Mukherjee, P.K.; Chandra, J.; Kuhn, D.M.; Ghannoum, M.A. Mechanism of Fluconazole Resistance in Candida albicans Biofilms: Phase-Specific Role of Efflux Pumps and Membrane Sterols. Infect. Immun. 2003, 71, 4333–4340. [Google Scholar] [CrossRef] [Green Version]
- Roberts, M.E.; Stewart, P. Modelling protection from antimicrobial agents in biofilms through the formation of persister cells. Microbiology 2005, 151, 75–80. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- LaFleur, M.D.; Kumamoto, C.A.; Lewis, K. Candida albicans Biofilms Produce Antifungal-Tolerant Persister Cells. Antimicrob. Agents Chemother. 2006, 50, 3839–3846. [Google Scholar] [CrossRef] [Green Version]
- Keren, I.; Kaldalu, N.; Spoering, A.; Wang, Y.; Lewis, K. Persister cells and tolerance to antimicrobials. FEMS Microbiol. Lett. 2004, 230, 13–18. [Google Scholar] [CrossRef] [Green Version]
- Uppuluri, P.; Nett, J.; Heitman, J.; Andes, D. Synergistic Effect of Calcineurin Inhibitors and Fluconazole against Candida albicans Biofilms. Antimicrob. Agents Chemother. 2008, 52, 1127–1132. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kumamoto, C.A. A contact-activated kinase signals Candida albicans invasive growth and biofilm development. Proc. Natl. Acad. Sci. USA 2005, 102, 5576–5581. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Johnson, C.J.; Cabezas-Olcoz, J.; Kernien, J.F.; Wang, S.X.; Beebe, D.J.; Huttenlocher, A.; Ansari, H.; Nett, J.E. The Extracellular Matrix of Candida albicans Biofilms Impairs Formation of Neutrophil Extracellular Traps. PLoS Pathog. 2016, 12, e1005884. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kernien, J.F.; Snarr, B.D.; Sheppard, D.C.; Nett, J.E. The Interface between Fungal Biofilms and Innate Immunity. Front. Immunol. 2018, 8, 1968. [Google Scholar] [CrossRef] [Green Version]
- Xie, Z.; Thompson, A.; Sobue, T.; Kashleva, H.; Xu, H.; Vasilakos, J.; Dongari-Bagtzoglou, A. Candida albicans Biofilms Do Not Trigger Reactive Oxygen Species and Evade Neutrophil Killing. J. Infect. Dis. 2012, 206, 1936–1945. [Google Scholar] [CrossRef] [Green Version]
- Morales, D.; Hogan, D.A. Candida albicans Interactions with Bacteria in the Context of Human Health and Disease. PLoS Pathog. 2010, 6, e1000886. [Google Scholar] [CrossRef] [PubMed]
- Granillo, A.R.; Canales, M.G.M.; Espíndola, M.E.S.; Rivera, M.A.M.; de Lucio, V.M.B.; Tovar, A.V.R. Antibiosis interaction of Staphylococccus aureus on Aspergillus fumigatus assessed in vitro by mixed biofilm formation. BMC Microbiol. 2015, 15, 33. [Google Scholar] [CrossRef] [Green Version]
- Williams, H.; Davies, J. Basic science for the chest physician:Pseudomonas aeruginosaand the cystic fibrosis airway. Thorax 2012, 67, 465–467. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sabino, R.; Ferreira, J.A.; Moss, R.B.; Valente, J.; Veríssimo, C.; Carolino, E.; Clemons, K.V.; Everson, C.; Banaei, N.; Penner, J.; et al. Molecular epidemiology of Aspergillus collected from cystic fibrosis patients. J. Cyst. Fibros. 2014, 14, 474–481. [Google Scholar] [CrossRef] [Green Version]
- Pierson, L.S.; Pierson, E.A. Metabolism and function of phenazines in bacteria: Impacts on the behavior of bacteria in the environment and biotechnological processes. Appl. Microbiol. Biotechnol. 2010, 86, 1659–1670. [Google Scholar] [CrossRef] [Green Version]
- Zhao, J.; Yu, W. Interaction betweenPseudomonas aeruginosaandAspergillus fumigatusin cystic fibrosis. PeerJ 2018, 6, e5931. [Google Scholar] [CrossRef] [Green Version]
- Briard, B.; Rasoldier, V.; Bomme, P.; Elaouad, N.; Guerreiro, C.; Chassagne, P.; Muszkieta, L.; Latgé, J.-P.; Mulard, L.; Beauvais, A. Dirhamnolipids secreted from Pseudomonas aeruginosa modify anjpegungal susceptibility of Aspergillus fumigatus by inhibiting β1,3 glucan synthase activity. ISME J. 2017, 11, 1578–1591. [Google Scholar] [CrossRef]
- Sass, G.; Ansari, S.R.; Dietl, A.-M.; Déziel, E.; Haas, H.; Stevens, D.A. Intermicrobial interaction: Aspergillus fumigatus siderophores protect against competition by Pseudomonas aeruginosa. PLoS ONE 2019, 14, e0216085. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nazik, H.; Sass, G.; Ansari, S.R.; Ertekin, R.; Haas, H.; Déziel, E.; Stevens, D.A. Novel intermicrobial molecular interaction: Pseudomonas aeruginosa Quinolone Signal (PQS) modulates Aspergillus fumigatus response to iron. Microbiology 2020, 166, 44–55. [Google Scholar] [CrossRef] [PubMed]
- Penner, J.C.; Ferreira, J.A.G.; Secor, P.R.; Sweere, J.; Birukova, M.K.; Joubert, L.-M.; Haagensen, J.A.J.; Garcia, O.; Malkovskiy, A.V.; Kaber, G.; et al. Pf4 bacteriophage produced by Pseudomonas aeruginosa inhibits Aspergillus fumigatus metabolism via iron sequestration. Microbiology 2016, 162, 1583–1594. [Google Scholar] [CrossRef]
- Proctor, R.A.; Von Eiff, C.; Kahl, B.; Becker, K.; McNamara, P.; Herrmann, M.; Peters, G. Small colony variants: A pathogenic form of bacteria that facilitates persistent and recurrent infections. Nat. Rev. Genet. 2006, 4, 295–305. [Google Scholar] [CrossRef]
- Mooij, M.J.; Drenkard, E.; Llamas, M.A.; Vandenbroucke-Grauls, C.M.J.E.; Savelkoul, P.H.M.; Ausubel, F.M.; Bitter, W. Characterization of the integrated filamentous phage Pf5 and its involvement in small-colony formation. Microbiology 2007, 153, 1790–1798. [Google Scholar] [CrossRef] [Green Version]
- Bruns, S.; Seidler, M.; Albrecht, D.; Salvenmoser, S.; Remme, N.; Hertweck, C.; Brakhage, A.A.; Kniemeyer, O.; Müller, F.-M.C. Functional genomic profiling of Aspergillus fumigatus biofilm reveals enhanced production of the mycotoxin gliotoxin. Proteomics 2010, 10, 3097–3107. [Google Scholar] [CrossRef]
- Manavathu, E.K.; Vager, D.L.; Vazquez, J.A. Development and antimicrobial susceptibility studies of in vitro monomicrobial and polymicrobial biofilm models with Aspergillus fumigatus and Pseudomonas aeruginosa. BMC Microbiol. 2014, 14, 53. [Google Scholar] [CrossRef] [Green Version]
- Briard, B.; Bomme, P.; Lechner, B.; Mislin, G.L.A.; Lair, V.; Prévost, M.-C.; Latgé, J.-P.; Haas, H.; Beauvais, A. Pseudomonas aeruginosa manipulates redox and iron homeostasis of its microbiota partner Aspergillus fumigatus via phenazines. Sci. Rep. 2015, 25, e106. [Google Scholar] [CrossRef] [Green Version]
- Scott, J.; Sueiro-Olivares, M.; Ahmed, W.; Heddergott, C.; Zhao, C.; Thomas, R.; Bromley, M.; Latgé, J.-P.; Krappmann, S.; Fowler, S.; et al. Pseudomonas aeruginosa-Derived Volatile Sulfur Compounds Promote Distal Aspergillus fumigatus Growth and a Synergistic Pathogen-Pathogen Interaction That Increases Pathogenicity in Co-infection. Front. Microbiol. 2019, 10, 2311. [Google Scholar] [CrossRef] [PubMed]
- Smith, K.; Rajendran, R.; Kerr, S.; Lappin, D.F.; Mackay, W.; Williams, C.; Ramage, G. Aspergillus fumigatusenhances elastase production inPseudomonas aeruginosaco-cultures. Med. Mycol. 2015, 53, 645–655. [Google Scholar] [CrossRef] [Green Version]
- Steenackers, H.P.; Parijs, I.; Foster, K.; Vanderleyden, J. Experimental evolution in biofilm populations. FEMS Microbiol. Rev. 2016, 40, 373–397. [Google Scholar] [CrossRef] [Green Version]
- Richards, J.P.; Cai, W.; Zill, N.A.; Zhang, W.; Ojha, A.K. Adaptation of Mycobacterium tuberculosis to biofilm growth is genetically linked to drug tolerance. Antimicrob. Agents Chemother. 2019, 63, e01213–e01219. [Google Scholar] [CrossRef] [Green Version]
- Revie, N.M.; Iyer, K.R.; Robbins, N.; Cowen, L.E. Antifungal drug resistance: Evolution, mechanisms and impact. Curr. Opin. Microbiol. 2018, 45, 70–76. [Google Scholar] [CrossRef]
- Hokken, M.W.J.; Zoll, J.; Coolen, J.; Zwaan, B.; Verweij, P.; Melchers, W.J.G. Phenotypic plasticity and the evolution of azole resistance in Aspergillus fumigatus; an expression profile of clinical isolates upon exposure to itraconazole. BMC Genom. 2019, 20, 28. [Google Scholar] [CrossRef]
- Shemesh, M.; Tam, A.; Aharoni, R.; Steinberg, D. Genetic adaptation of Streptococcus mutans during biofilm formation on different types of surfaces. BMC Microbiol. 2010, 10, 51. [Google Scholar] [CrossRef] [Green Version]
- Thöming, J.G.; Tomasch, J.; Preusse, M.; Koska, M.; Grahl, N.; Pohl, S.; Willger, S.D.; Kaever, V.; Müsken, M.; Häussler, S. Parallel evolutionary paths to produce more than one Pseudomonas aeruginosa biofilm phenotype. NPJ Biofilms Microbiomes 2020, 6, 2. [Google Scholar] [CrossRef] [PubMed]
- Turner, C.B.; Marshall, C.W.; Cooper, V.S. Parallel genetic adaptation across environments differing in mode of growth or resource availability. Evol. Lett. 2018, 2, 355–367. [Google Scholar] [CrossRef]
- Nguyen, M.; Ekstrom, A.; Li, X.; Yin, Y. HGT-Finder: A New Tool for Horizontal Gene Transfer Finding and Application to Aspergillus genomes. Toxins 2015, 7, 4035–4053. [Google Scholar] [CrossRef] [Green Version]
- Valdes, I.D.; De Ruijter, A.B.P.H.; Torres, C.J.; Breuker, J.C.A.; Wösten, H.A.B.; De Cock, H. The sino-nasal warzone: Transcriptomic and genomic studies on sino-nasal aspergillosis in dogs. NPJ Biofilms Microbiomes 2020, 6, 51. [Google Scholar] [CrossRef] [PubMed]
- Ballard, E.; Melchers, W.J.; Zoll, J.; Brown, A.J.; Verweij, P.; Warris, A. In-host microevolution of Aspergillus fumigatus: A phenotypic and genotypic analysis. Fungal Genet. Biol. 2018, 113, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Selmecki, A.; Forche, A.; Berman, J. Genomic Plasticity of the Human Fungal Pathogen Candida albicans. Eukaryot. Cell 2010, 9, 991–1008. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Verweij, P.E.; Zhang, J.; Debets, A.J.M.; Meis, J.F.; van de Veerdonk, F.L.; Schoustra, S.E.; Zwaan, B.; Melchers, W.J.G. In-host adaptation and acquired triazole resistance in Aspergillus fumigatus: A dilemma for clinical management. Lancet Infect. Dis. 2016, 16, e251–e260. [Google Scholar] [CrossRef]
- Zhang, J.; Debets, A.J.M.; Verweij, P.; Melchers, W.; Zwaan, B.; Schoustra, S.E. Asexual sporulation facilitates adaptation: The emergence of azole resistance inAspergillus fumigatus. Evolution 2015, 69, 2573–2586. [Google Scholar] [CrossRef] [PubMed]
- Dyer, P.S.; O’Gorman, C. Sexual development and cryptic sexuality in fungi: Insights fromAspergillusspecies. FEMS Microbiol. Rev. 2012, 36, 165–192. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pontecorvo, G. The Parasexual Cycle in Fungi. Annu. Rev. Microbiol. 1956, 10, 393–400. [Google Scholar] [CrossRef]
- Barton, R.C.; Gull, K. Isolation, characterization, and genetic analysis of monosomic, aneuploid mutants of Candida albicans. Mol. Microbiol. 1992, 6, 171–177. [Google Scholar] [CrossRef]
- Mena, A.; Smith, E.E.; Burns, J.L.; Speert, D.P.; Moskowitz, S.M.; Perez, J.L.; Oliver, A. Genetic Adaptation of Pseudomonas aeruginosa to the Airways of Cystic Fibrosis Patients Is Catalyzed by Hypermutation. J. Bacteriol. 2008, 190, 7910–7917. [Google Scholar] [CrossRef] [Green Version]
- Oliver, A.; Cantón, R.; Campo, P.; Baquero, F.; Blázquez, J. High Frequency of Hypermutable Pseudomonas aeruginosa in Cystic Fibrosis Lung Infection. Science 2000, 288, 1251–1253. [Google Scholar] [CrossRef] [PubMed]
- Verstrepen, K.; Jansen, A.; Lewitter, F.; Fink, G.R. Intragenic tandem repeats generate functional variability. Nat. Genet. 2005, 37, 986–990. [Google Scholar] [CrossRef] [Green Version]
- Levdansky, E.; Romano, J.; Shadkchan, Y.; Sharon, H.; Verstrepen, K.J.; Fink, G.R.; Osherov, N. Coding Tandem Repeats Generate Diversity in Aspergillus fumigatus Genes. Eukaryot. Cell 2007, 6, 1380–1391. [Google Scholar] [CrossRef] [Green Version]
- Fan, Z.; Li, Z.; Xu, Z.; Li, H.; Li, L.; Ning, C.; Ma, L.; Xie, X.; Wang, G.; Yu, H. cspAInfluences Biofilm Formation and Drug Resistance in Pathogenic FungusAspergillus fumigatus. BioMed Res. Int. 2015, 2015, 960357. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kwon-Chung, K.J.; Chang, Y.C. Aneuploidy and Drug Resistance in Pathogenic Fungi. PLoS Pathog. 2012, 8, e1003022. [Google Scholar] [CrossRef]
- Selmecki, A.; Gerami-Nejad, M.; Paulson, C.; Forche, A.; Berman, J. An isochromosome confers drug resistance in vivo by amplification of two genes, ERG11 and TAC1. Mol. Microbiol. 2008, 68, 624–641. [Google Scholar] [CrossRef]
- Todd, R.T.; Wikoff, T.D.; Forche, A.; Selmecki, A. Genome plasticity in Candida albicans is driven by long repeat sequences. eLife 2019, 8, e45954. [Google Scholar] [CrossRef] [PubMed]
- Muszkieta, L.; Beauvais, A.; Pähtz, V.; Gibbons, J.G.; Leberre, V.A.; Beau, R.; Shibuya, K.; Rokas, A.; Francois, J.M.; Kniemeyyer, O.; et al. Investigation of Aspergillus fumigatus biofilm formation by various “omics” approaches. Front. Microbiol. 2013, 4, 13. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Beauvais, A.; Latgé, J.-P. Aspergillus Biofilm In Vitro and In Vivo. In Microbial Biofilms; ASM Press: Washington, DC, USA, 2015; pp. 149–161. [Google Scholar] [CrossRef]
- Gayathri, L.; Akbarsha, M.A.; Ruckmani, K. In vitro study on aspects of molecular mechanisms underlying invasive aspergillosis caused by gliotoxin and fumagillin, alone and in combination. Sci. Rep. 2020, 10, 14473. [Google Scholar] [CrossRef]
- Lambou, K.; Lamarre, C.; Beau, R.; Dufour, N.; Latge, J.-P. Functional analysis of the superoxide dismutase family inAspergillus fumigatus. Mol. Microbiol. 2010, 75, 910–923. [Google Scholar] [CrossRef] [PubMed]
- Hagiwara, D.; Suzuki, S.; Kamei, K.; Gonoi, T.; Kawamoto, S. The role of AtfA and HOG MAPK pathway in stress tolerance in conidia of Aspergillus fumigatus. Fungal Genet. Biol. 2014, 73, 138–149. [Google Scholar] [CrossRef] [PubMed]
- Silva, L.P.; Horta, M.A.C.; Goldman, G.H. Genetic Interactions Between Aspergillus fumigatus Basic Leucine Zipper (bZIP) Transcription Factors AtfA, AtfB, AtfC, and AtfD. Front. Fungal Biol. 2021, 2, 1. [Google Scholar] [CrossRef]
- Urban, M.; Cuzick, A.; Seager, J.; Wood, V.; Rutherford, K.; Venkatesh, S.Y.; De Silva, N.; Martinez, M.C.; Pedro, H.; Yates, A.D.; et al. PHI-base: The pathogen–host interactions database. Nucleic Acids Res. 2019, 48, D613–D620. [Google Scholar] [CrossRef]
- Bertuzzi, M.; Schrettl, M.; Alcazar-Fuoli, L.; Cairns, T.; Muñoz, A.; Walker, L.A.; Herbst, S.; Safari, M.; Cheverton, A.M.; Chen, D.; et al. The pH-Responsive PacC Transcription Factor of Aspergillus fumigatus Governs Epithelial Entry and Tissue Invasion during Pulmonary Aspergillosis. PLoS Pathog. 2014, 10, e1004413. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gibbons, J.G.; Beauvais, A.; Beau, R.; McGary, K.L.; Latgé, J.-P.; Rokas, A. Global Transcriptome Changes Underlying Colony Growth in the Opportunistic Human Pathogen Aspergillus fumigatus. Eukaryot. Cell 2012, 11, 68–78. [Google Scholar] [CrossRef] [Green Version]
- Lessing, F.; Kniemeyer, O.; Wozniok, I.; Loeffler, J.; Kurzai, O.; Haertl, A.; Brakhage, A.A. The Aspergillus fumigatus Transcriptional Regulator AfYap1 Represents the Major Regulator for Defense against Reactive Oxygen Intermediates but Is Dispensable for Pathogenicity in an Intranasal Mouse Infection Model. Eukaryot. Cell 2007, 6, 2290–2302. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Amich, J.; Leal, F.; Calera, J.A. Repression of the acid ZrfA/ZrfB zinc-uptake system of Aspergillus fumigatus mediated by PacC under neutral, zinc-limiting conditions. Int. Microbiol. 2009, 12, 39–47. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
