DNA Methylation on N6-Adenine Regulates the Hyphal Development during Dimorphism in the Early-Diverging Fungus Mucor lusitanicus
Abstract
1. Introduction
2. Materials and Methods
2.1. Fungal Strains and Culture Conditions
2.2. Disruption of dmt Genes and Generation of Complemented Strains
2.3. Southern Blot
2.4. Experiments of Yeast to Hyphae Transition
2.5. In Vitro Host-Pathogen Interaction Assays
2.6. RNA Extraction and cDNA Synthesis
2.7. RT-qPCR Analysis
2.8. Analysis of 6mA in DNA by HPLC-MS
2.9. Sensitivity Tests
3. Results
3.1. Genes of M. lusitanicus Encoding Putative 6mA Demethylases
3.2. Generation of Single and Double dmt Knockout Mutants in M. lusitanicus
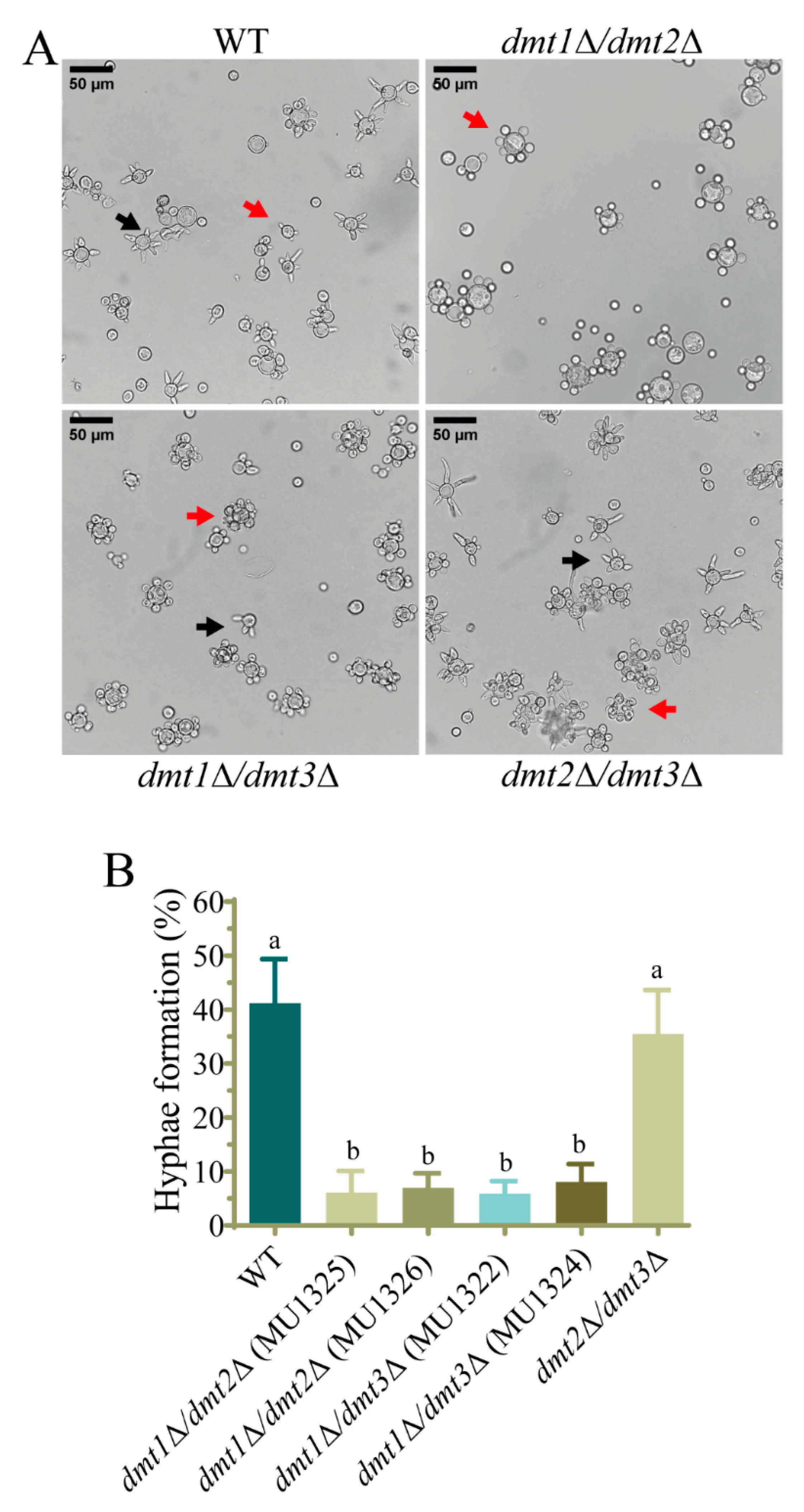
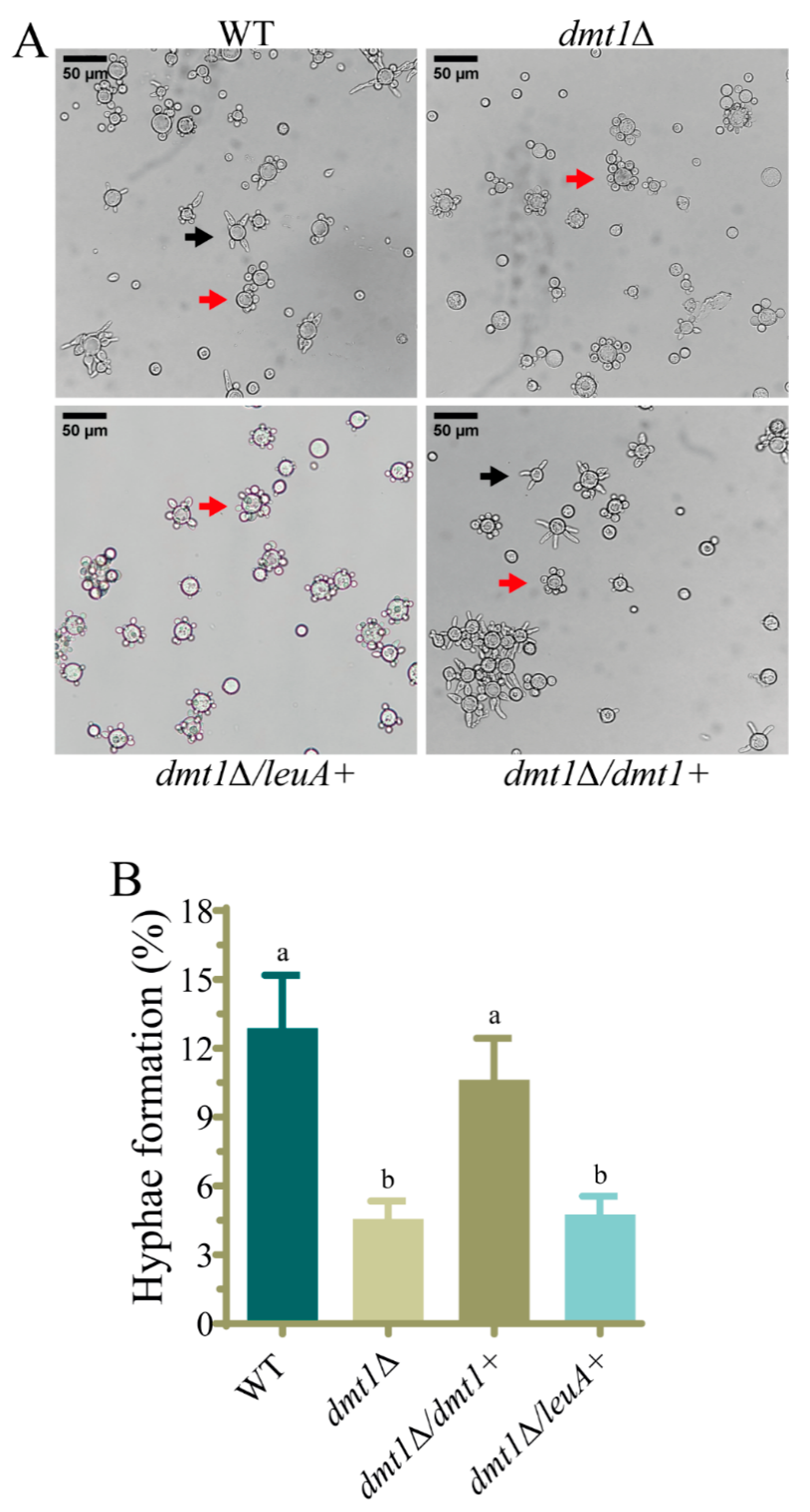
3.3. The dmt1 Is Involved in the Hyphae Formation during the Yeast to Mycelium Transition
3.4. The dmt1 Is Important for the Appropriated pkaR4 Transcription during the Transition from Yeast to Hyphae
3.5. Deletion of dmt1 Alters 6mA Levels during Yeast to Hyphae Transition
3.6. dmt1 Regulates Germination in the Interaction of Spores with Macrophages In Vitro
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Gleissner, B.; Schilling, A.; Anagnostopolous, I.; Siehl, I.; Thiel, E. improved outcome of zygomycosis in patients with hematological diseases? Leuk. Lymphoma 2004, 45, 1351–1360. [Google Scholar] [CrossRef]
- Brown, G.D.; Denning, D.W.; Levitz, S.M. Tackling human fungal infections. Science 2012, 336, 647. [Google Scholar] [CrossRef]
- Binder, U.; Maurer, E.; Lass-Flörl, C. Mucormycosis—From the pathogens to the disease. Clin. Microbiol. Infect. 2014, 20, 60–66. [Google Scholar] [CrossRef]
- Ibrahim, A.S.; Edwards, J.E.; Filler, S.G.; Spellberg, B. Mucormycosis and Entomophthoramycosis (Zygomycosis). In Essentials of Clinical Mycology; Springer: New York, NY, USA, 2011; pp. 265–280. [Google Scholar]
- Veisi, A.; Bagheri, A.; Eshaghi, M.; Rikhtehgar, M.H.; Rezaei Kanavi, M.; Farjad, R. Rhino-orbital mucormycosis during steroid therapy in COVID-19 patients: A case report. Eur. J. Ophthalmol. 2021, 112067212110094. [Google Scholar] [CrossRef] [PubMed]
- John, T.M.; Jacob, C.N.; Kontoyiannis, D.P. When uncontrolled diabetes mellitus and severe COVID-19 converge: The perfect storm for mucormycosis. J. Fungi 2021, 7, 298. [Google Scholar] [CrossRef]
- Kauffman, C.A. Zygomycosis: Reemergence of an old pathogen. Clin. Infect. Dis. 2004, 39, 588–590. [Google Scholar] [CrossRef]
- Wagner, L.; Stielow, J.B.; de Hoog, G.S.; Bensch, K.; Schwartze, V.U.; Voigt, K.; Alastruey-Izquierdo, A.; Kurzai, O.; Walther, G. A new species concept for the clinically relevant Mucor circinelloides complex. Persoonia 2020, 44, 67–97. [Google Scholar] [CrossRef] [PubMed]
- Ibrahim, A.S.; Voelz, K. The mucormycete–host interface. Curr. Opin. Microbiol. 2017, 40, 40–45. [Google Scholar] [CrossRef]
- Lax, C.; Pérez-arques, C.; Navarro-mendoza, M.I.; Cánovas-márquez, J.T.; Tahiri, G.; Pérez-ruiz, J.A.; Osorio-concepción, M.; Murcia-flores, L.; Navarro, E.; Garre, V.; et al. Genes, pathways, and mechanisms involved in the virulence of mucorales. Genes 2020, 11, 317. [Google Scholar] [CrossRef]
- Orlowski, M. Mucor dimorphism. Microbiol. Rev. 1991, 55, 234–258. [Google Scholar] [CrossRef] [PubMed]
- Boyce, K.J.; Andrianopoulos, A. Fungal dimorphism: The switch from hyphae to yeast is a specialized morphogenetic adaptation allowing colonization of a host. FEMS Microbiol. Rev. 2015, 39, 797–811. [Google Scholar] [CrossRef]
- Nicolás, F.E.; Murcia, L.; Navarro, E.; Navarro-Mendoza, M.I.; Pérez-Arques, C.; Garre, V. Mucorales species and macrophages. J. Fungi 2020, 6, 94. [Google Scholar] [CrossRef]
- Lee, S.C.; Li, A.; Calo, S.; Heitman, J. Calcineurin plays key roles in the dimorphic transition and virulence of the human pathogenic zygomycete Mucor circinelloides. PLoS Pathog. 2013, 9, e1003625. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.C.; Li, A.; Calo, S.; Inoue, M.; Tonthat, N.K.; Bain, J.M.; Louw, J.; Shinohara, M.L.; Erwig, L.P.; Schumacher, M.A.; et al. Calcineurin orchestrates dimorphic transitions, antifungal drug responses and host-pathogen interactions of the pathogenic mucoralean fungus Mucor circinelloides. Mol. Microbiol. 2015, 97, 844–865. [Google Scholar] [CrossRef]
- Ocampo, J.; McCormack, B.; Navarro, E.; Moreno, S.; Garre, V.; Rossi, S. Protein kinase A regulatory subunit isoforms regulate growth and differentiation in Mucor circinelloides: Essential role of PKAR4. Eukaryot. Cell 2012, 11, 989–1002. [Google Scholar] [CrossRef]
- Robbins, N.; Leach, M.D.; Cowen, L.E. Lysine Deacetylases Hda1 and Rpd3 Regulate Hsp90 Function thereby Governing Fungal Drug Resistance. Cell Rep. 2012, 2, 878–888. [Google Scholar] [CrossRef] [PubMed]
- Hnisz, D.; Bardet, A.F.; Nobile, C.J.; Petryshyn, A.; Glaser, W.; Schöck, U.; Stark, A.; Kuchler, K. A histone deacetylase adjusts transcription kinetics at coding sequences during Candida albicans morphogenesis. PLoS Genet. 2012, 8, e1003118. [Google Scholar] [CrossRef] [PubMed]
- Nobile, C.J.; Fox, E.P.; Hartooni, N.; Mitchell, K.F.; Hnisz, D.; Andes, D.R.; Kuchler, K.; Johnson, A.D. A histone deacetylase complex mediates biofilm dispersal and drug resistance in Candida albicans. MBio 2014, 5, e01201-14. [Google Scholar] [CrossRef]
- Sedgwick, B.; Bates, P.A.; Paik, J.; Jacobs, S.C.; Lindahl, T. Repair of alkylated DNA: Recent advances. DNA Repair 2007, 6, 429–442. [Google Scholar] [CrossRef] [PubMed]
- Iyer, L.M.; Abhiman, S.; Aravind, L. Natural History of Eukaryotic DNA Methylation Systems. Prog. Mol. Biol. Transl. Sci. 2011, 101, 25–104. [Google Scholar] [CrossRef]
- Bird, A. DNA methylation patterns and epigenetic memory. Genes Dev. 2002, 16, 6–21. [Google Scholar] [CrossRef]
- Greer, E.L.; Blanco, M.A.; Gu, L.; Sendinc, E.; Liu, J.; Aristizábal-Corrales, D.; Hsu, C.-H.; Aravind, L.; He, C.; Shi, Y. DNA Methylation on N6-Adenine in C. elegans. Cell 2015, 161, 868–878. [Google Scholar] [CrossRef]
- Koziol, M.J.; Bradshaw, C.R.; Allen, G.E.; Costa, A.S.H.; Frezza, C.; Gurdon, J.B. Identification of methylated deoxyadenosines in vertebrates reveals diversity in DNA modifications. Nat. Struct. Mol. Biol. 2016, 23, 24–30. [Google Scholar] [CrossRef]
- Mondo, S.J.; Dannebaum, R.O.; Kuo, R.C.; Louie, K.B.; Bewick, A.J.; LaButti, K.; Haridas, S.; Kuo, A.; Salamov, A.; Ahrendt, S.R.; et al. Widespread adenine N6-methylation of active genes in fungi. Nat. Genet. 2017, 49, 964–968. [Google Scholar] [CrossRef]
- Fu, Y.; Luo, G.Z.; Chen, K.; Deng, X.; Yu, M.; Han, D.; Hao, Z.; Liu, J.; Lu, X.; Doré, L.C.; et al. N6-methyldeoxyadenosine marks active transcription start sites in Chlamydomonas. Cell 2015, 161, 879–892. [Google Scholar] [CrossRef]
- Beh, L.Y.; Debelouchina, G.T.; Clay, D.M.; Thompson, R.E.; Lindblad, K.A.; Hutton, E.R.; Bracht, J.R.; Sebra, R.P.; Muir, T.W.; Landweber, L.F. Identification of a DNA N6-adenine methyltransferase complex and its impact on chromatin organization. Cell 2019, 177, 1781–1796.e25. [Google Scholar] [CrossRef] [PubMed]
- Luo, G.Z.; Hao, Z.; Luo, L.; Shen, M.; Sparvoli, D.; Zheng, Y.; Zhang, Z.; Weng, X.; Chen, K.; Cui, Q.; et al. N 6-methyldeoxyadenosine directs nucleosome positioning in Tetrahymena DNA. Genome Biol. 2018, 19, 200. [Google Scholar] [CrossRef] [PubMed]
- O’Brown, Z.K.; Greer, E.L. N6-methyladenine: A conserved and dynamic DNA mark. Adv. Exp. Med. Biol. 2016, 945, 213–246. [Google Scholar] [CrossRef] [PubMed]
- Duncan, T.; Trewick, S.C.; Koivisto, P.; Bates, P.A.; Lindahl, T.; Sedgwick, B. Reversal of DNA alkylation damage by two human dioxygenases. Proc. Natl. Acad. Sci. USA 2002, 99, 16660–16665. [Google Scholar] [CrossRef] [PubMed]
- Fedeles, B.I.; Singh, V.; Delaney, J.C.; Li, D.; Essigmann, J.M. The AlkB Family of Fe(II)/α-Ketoglutarate-dependent Dioxygenases: Repairing Nucleic Acid Alkylation Damage and Beyond. J. Biol. Chem. 2015, 290, 20734–20742. [Google Scholar] [CrossRef] [PubMed]
- Kamat, S.S.; Fan, H.; Sauder, J.M.; Burley, S.K.; Shoichet, B.K.; Sali, A.; Raushel, F.M. Enzymatic deamination of the epigenetic base N-6-methyladenine. J. Am. Chem. Soc. 2011, 133, 2080–2083. [Google Scholar] [CrossRef] [PubMed]
- Chen, K.; Luo, G.Z.; He, C. High-resolution mapping of n6-methyladenosine in transcriptome and genome using a photo-crosslinking-assisted strategy. Methods Enzymol. 2015, 560, 161–185. [Google Scholar] [CrossRef] [PubMed]
- Van den Born, E.; Bekkelund, A.; Moen, M.N.; Omelchenko, M.V.; Klungland, A.; Falnes, P. Bioinformatics and functional analysis define four distinct groups of AlkB DNA-dioxygenases in bacteria. Nucleic Acids Res. 2009, 37, 7124–7136. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Aas, P.A.; Otterlei, M.; Falnes, P.; Vågbø, C.B.; Skorpen, F.; Akbari, M.; Sundheim, O.; Bjørås, M.; Slupphaug, G.; Seeberg, E.; et al. Human and bacterial oxidative demethylases repair alkylation damage in both RNA and DNA. Nature 2003, 421, 859–863. [Google Scholar] [CrossRef]
- Korvald, H.; Mølstad Moe, A.M.; Cederkvist, F.H.; Thiede, B.; Laerdahl, J.K.; Bjørås, M.; Alseth, I. Schizosaccharomyces pombe Ofd2 is a nuclear 2-oxoglutarate and iron dependent dioxygenase interacting with histones. PLoS ONE 2011, 6, e25188. [Google Scholar] [CrossRef]
- Meza, T.J.; Moen, M.N.; Vågbo, C.B.; Krokan, H.E.; Klungland, A.; Grini, P.E.; Falnes, P.O. The DNA dioxygenase ALKBH2 protects Arabidopsis thaliana against methylation damage. Nucleic Acids Res. 2012, 40, 6620–6631. [Google Scholar] [CrossRef]
- Tsujikawa, K.; Koike, K.; Kitae, K.; Shinkawa, A.; Arima, H.; Suzuki, T.; Tsuchiya, M.; Makino, Y.; Furukawa, T.; Konishi, N.; et al. Expression and sub-cellular localization of human ABH family molecules. J. Cell. Mol. Med. 2007, 11, 1105–1116. [Google Scholar] [CrossRef]
- Ougland, R.; Rognes, T.; Klungland, A.; Larsen, E. Non-homologous functions of the AlkB homologs. J. Mol. Cell Biol. 2015, 7, 494–504. [Google Scholar] [CrossRef]
- Sundheim, O.; Vågbø, C.B.; Bjørås, M.; Sousa, M.M.L.; Talstad, V.; Aas, P.A.; Drabløs, F.; Krokan, H.E.; Tainer, J.A.; Slupphaug, G. Human ABH3 structure and key residues for oxidative demethylation to reverse DNA/RNA damage. EMBO J. 2006, 25, 3389–3397. [Google Scholar] [CrossRef]
- Liu, Y.; Yuan, Q.; Xie, L. The AlkB Family of Fe (II)/Alpha-ketoglutarate-dependent dioxygenases modulates embryogenesis through epigenetic regulation. Curr. Stem Cell Res. Ther. 2018, 13, 136–143. [Google Scholar] [CrossRef]
- Ma, C.-J.; Ding, J.-H.; Ye, T.-T.; Yuan, B.-F.; Feng, Y.-Q. AlkB homologue 1 demethylates N3-methylcytidine in mRNA of mammals. ACS Chem. Biol. 2019, 14, 1418–1425. [Google Scholar] [CrossRef]
- Wu, T.P.; Wang, T.; Seetin, M.G.; Lai, Y.; Zhu, S.; Lin, K.; Liu, Y.; Byrum, S.D.; Mackintosh, S.G.; Zhong, M.; et al. DNA methylation on N6-adenine in mammalian embryonic stem cells. Nature 2016, 532, 329–333. [Google Scholar] [CrossRef] [PubMed]
- Nigam, R.; Babu, K.R.; Ghosh, T.; Kumari, B.; Akula, D.; Rath, S.N.; Das, P.; Anindya, R.; Khan, F.A. Indenone derivatives as inhibitor of human DNA dealkylation repair enzyme AlkBH3. Bioorg. Med. Chem. 2018, 26, 4100–4112. [Google Scholar] [CrossRef] [PubMed]
- Pilžys, T.; Marcinkowski, M.; Kukwa, W.; Garbicz, D.; Dylewska, M.; Ferenc, K.; Mieczkowski, A.; Kukwa, A.; Migacz, E.; Wołosz, D.; et al. ALKBH overexpression in head and neck cancer: Potential target for novel anticancer therapy. Sci. Rep. 2019, 9, 13249. [Google Scholar] [CrossRef] [PubMed]
- Nordstrand, L.M.; Svärd, J.; Larsen, E.; Nilsen, A.; Ougland, R.; Furu, K.; Lien, G.F.; Rognes, T.; Namekawa, S.H.; Lee, J.T.; et al. Mice lacking Alkbh1 display sex-ratio distortion and unilateral eye defects. PLoS ONE 2010, 5, e13827. [Google Scholar] [CrossRef] [PubMed]
- Kawarada, L.; Suzuki, T.; Ohira, T.; Hirata, S.; Miyauchi, K.; Suzuki, T. ALKBH1 is an RNA dioxygenase responsible for cytoplasmic and mitochondrial tRNA modifications. Nucleic Acids Res. 2017, 45, 7401–7415. [Google Scholar] [CrossRef]
- Yuan Wang, S.I.; Mao, H.I.; Shibuya, H.I.; Uzawa, S.I.; Klapholz, Z.O.; Wesenberg, S.; ShinID, N.; Saito, T.T.; Gao, J.; Meyer, B.J.; et al. The demethylase NMAD-1 regulates DNA replication and repair in the Caenorhabditis elegans germline. PLoS Genet. 2019, 15, e1008252. [Google Scholar] [CrossRef]
- Korvald, H.; Falnes, P.; Laerdahl, J.K.; Bjørås, M.; Alseth, I. The Schizosaccharomyces pombe AlkB homolog Abh1 exhibits AP lyase activity but no demethylase activity. DNA Repair 2012, 11, 453–462. [Google Scholar] [CrossRef]
- Schipper, M.A.A. On Mucor circinelloides, Mucor racemosus and Related Species. Stud. Mycol. 1976, 12, 1–40. [Google Scholar] [CrossRef]
- Navarro-Mendoza, M.I.; Pérez-Arques, C.; Panchal, S.; Nicolás, F.E.; Mondo, S.J.; Ganguly, P.; Pangilinan, J.; Grigoriev, I.V.; Heitman, J.; Sanyal, K.; et al. Early diverging fungus Mucor circinelloides lacks centromeric histone CENP-A and displays a mosaic of point and regional centromeres. Curr. Biol. 2019, 29, 3791–3802.e6. [Google Scholar] [CrossRef] [PubMed]
- Nicolás, F.E.; de Haro, J.P.; Torres-Martínez, S.; Ruiz-Vázquez, R.M. Mutants defective in a Mucor circinelloides dicer-like gene are not compromised in siRNA silencing but display developmental defects. Fungal Genet. Biol. 2007, 44, 504–516. [Google Scholar] [CrossRef]
- Gutiérrez, A.; López-García, S.; Garre, V. High reliability transformation of the basal fungus Mucor circinelloides by electroporation. J. Microbiol. Methods 2011, 84, 442–446. [Google Scholar] [CrossRef]
- Vellanki, S.; Navarro-Mendoza, M.I.; Garcia, A.; Murcia, L.; Perez-Arques, C.; Garre, V.; Nicolas, F.E.; Lee, S.C. Mucor circinelloides: Growth, maintenance, and genetic manipulation. Curr. Protoc. Microbiol. 2018, 49, e53. [Google Scholar] [CrossRef]
- Nicolás, F.E.; Navarro-Mendoza, M.I.; Pérez-Arques, C.; López-García, S.; Navarro, E.; Torres-Martínez, S.; Garre, V. Molecular tools for carotenogenesis analysis in the mucoral Mucor circinelloides. In Methods in Molecular Biology; Humana Press Inc.: New York, NY, USA, 2018; Volume 1852, pp. 221–237. [Google Scholar]
- Rodríguez-Frómeta, R.A.; Gutiérrez, A.; Torres-Martínez, S.; Garre, V. Malic enzyme activity is not the only bottleneck for lipid accumulation in the oleaginous fungus Mucor circinelloides. Appl. Microbiol. Biotechnol. 2013, 97, 3063–3072. [Google Scholar] [CrossRef]
- Pérez-Arques, C.; Navarro-Mendoza, M.I.; Murcia, L.; Lax, C.; Martínez-García, P.; Heitman, J.; Nicolás, F.E.; Garrea, V. Mucor circinelloides thrives inside the phagosome through an atf-mediated germination pathway. MBio 2019, 10, e02765-18. [Google Scholar] [CrossRef] [PubMed]
- Valle-Maldonado, M.I.; Jácome-Galarza, I.E.; Gutiérrez-Corona, F.; Ramírez-Díaz, M.I.; Campos-García, J.; Meza-Carmen, V. Selection of reference genes for quantitative real time RT-PCR during dimorphism in the zygomycete Mucor circinelloides. Mol. Biol. Rep. 2015, 42, 705–711. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Jia, G.; Fu, Y.; Zhao, X.; Dai, Q.; Zheng, G.; Yang, Y.; Yi, C.; Lindahl, T.; Pan, T.; Yang, Y.G.; et al. N6-Methyladenosine in nuclear RNA is a major substrate of the obesity-associated FTO. Nat. Chem. Biol. 2011, 7, 885–887. [Google Scholar] [CrossRef] [PubMed]
- Trewick, S.C.; Henshaw, T.F.; Hausinger, R.P.; Lindahl, T.; Sedgwick, B. Oxidative demethylation by Escherichia coli AlkB directly reverts DNA base damage. Nature 2002, 419, 174–178. [Google Scholar] [CrossRef] [PubMed]
- Falnes, P.; Johansen, R.F.; Seeberg, E. AlkB-mediated oxidative demethylation reverses DNA damage in Escherichia coli. Nature 2002, 419, 178–182. [Google Scholar] [CrossRef]
- Velayos, A.; Eslava, A.P.; Iturriaga, E.A. A bifunctional enzyme with lycopene cyclase and phytoene synthase activities is encoded by the carRP gene of Mucor circinelloides. Eur. J. Biochem. 2000, 267, 5509–5519. [Google Scholar] [CrossRef] [PubMed]
- Yi, C.; He, C. DNA repair by reversal of DNA damage. Cold Spring Harb. Perspect. Biol. 2013, 5, 12575–12576. [Google Scholar] [CrossRef] [PubMed]
- Kataoka, H.; Yamamoto, Y.; Sekiguchi, M. A new gene (alkB) of Escherichia coli that controls sensitivity to methyl methane sulfonate. J. Bacteriol. 1983, 153, 1301–1307. [Google Scholar] [CrossRef] [PubMed]
- Gauthier, G.; Klein, B.S. Insights into fungal morphogenesis and immune evasion: Fungal conidia, when situated in mammalian lungs, may switch from mold to pathogenic yeasts or spore-forming spherules. Microbe 2008, 3, 416–423. [Google Scholar] [CrossRef]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Osorio-Concepción, M.; Lax, C.; Navarro, E.; Nicolás, F.E.; Garre, V. DNA Methylation on N6-Adenine Regulates the Hyphal Development during Dimorphism in the Early-Diverging Fungus Mucor lusitanicus. J. Fungi 2021, 7, 738. https://doi.org/10.3390/jof7090738
Osorio-Concepción M, Lax C, Navarro E, Nicolás FE, Garre V. DNA Methylation on N6-Adenine Regulates the Hyphal Development during Dimorphism in the Early-Diverging Fungus Mucor lusitanicus. Journal of Fungi. 2021; 7(9):738. https://doi.org/10.3390/jof7090738
Chicago/Turabian StyleOsorio-Concepción, Macario, Carlos Lax, Eusebio Navarro, Francisco E. Nicolás, and Victoriano Garre. 2021. "DNA Methylation on N6-Adenine Regulates the Hyphal Development during Dimorphism in the Early-Diverging Fungus Mucor lusitanicus" Journal of Fungi 7, no. 9: 738. https://doi.org/10.3390/jof7090738
APA StyleOsorio-Concepción, M., Lax, C., Navarro, E., Nicolás, F. E., & Garre, V. (2021). DNA Methylation on N6-Adenine Regulates the Hyphal Development during Dimorphism in the Early-Diverging Fungus Mucor lusitanicus. Journal of Fungi, 7(9), 738. https://doi.org/10.3390/jof7090738







