Fungal Cell Wall Proteins and Signaling Pathways Form a Cytoprotective Network to Combat Stresses
Abstract
1. Introduction
2. Function of Cell Wall Proteins
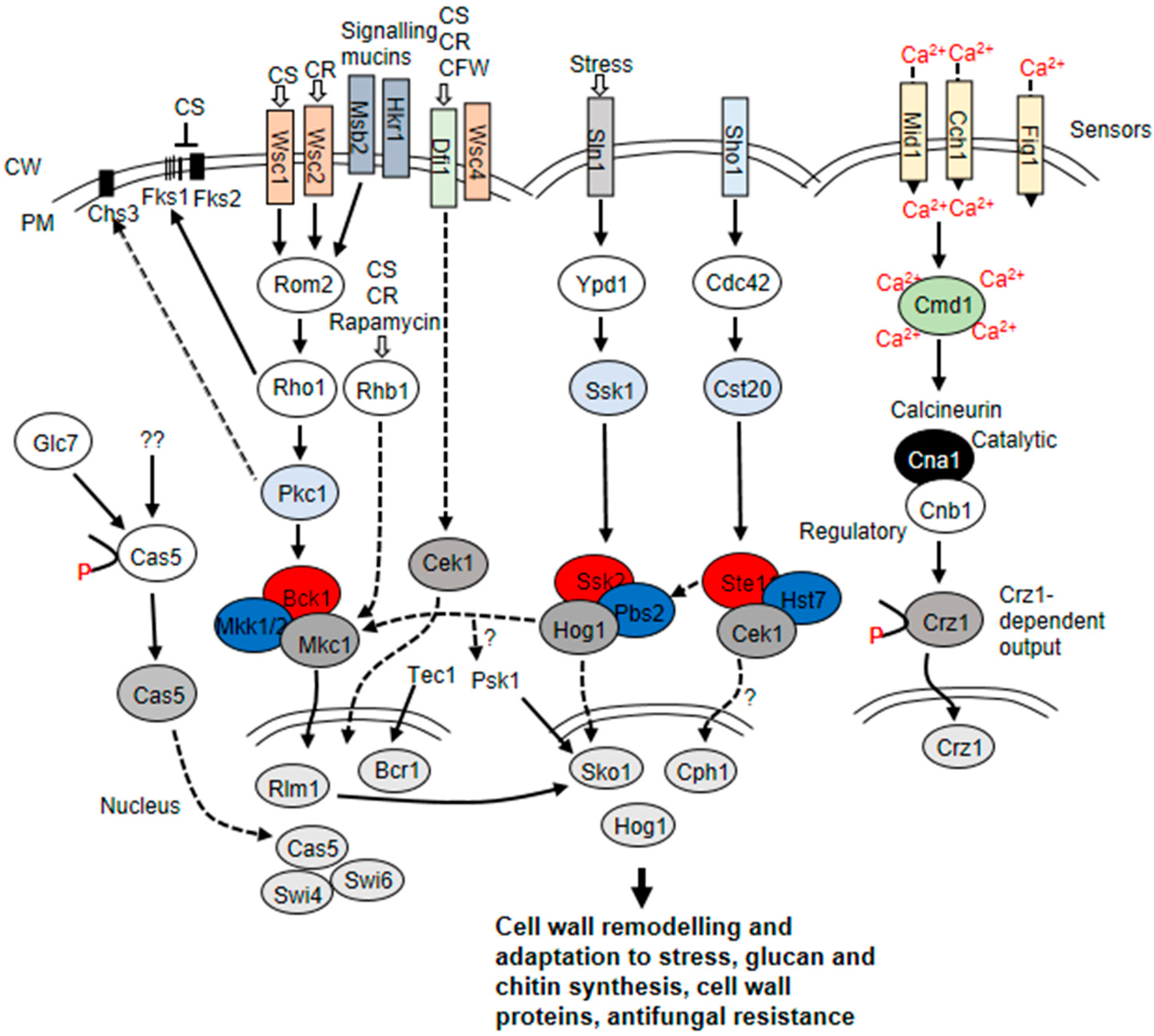
3. Fungal Cell Wall Remodeling and Signaling Pathways That Are Activated in Response to Stress
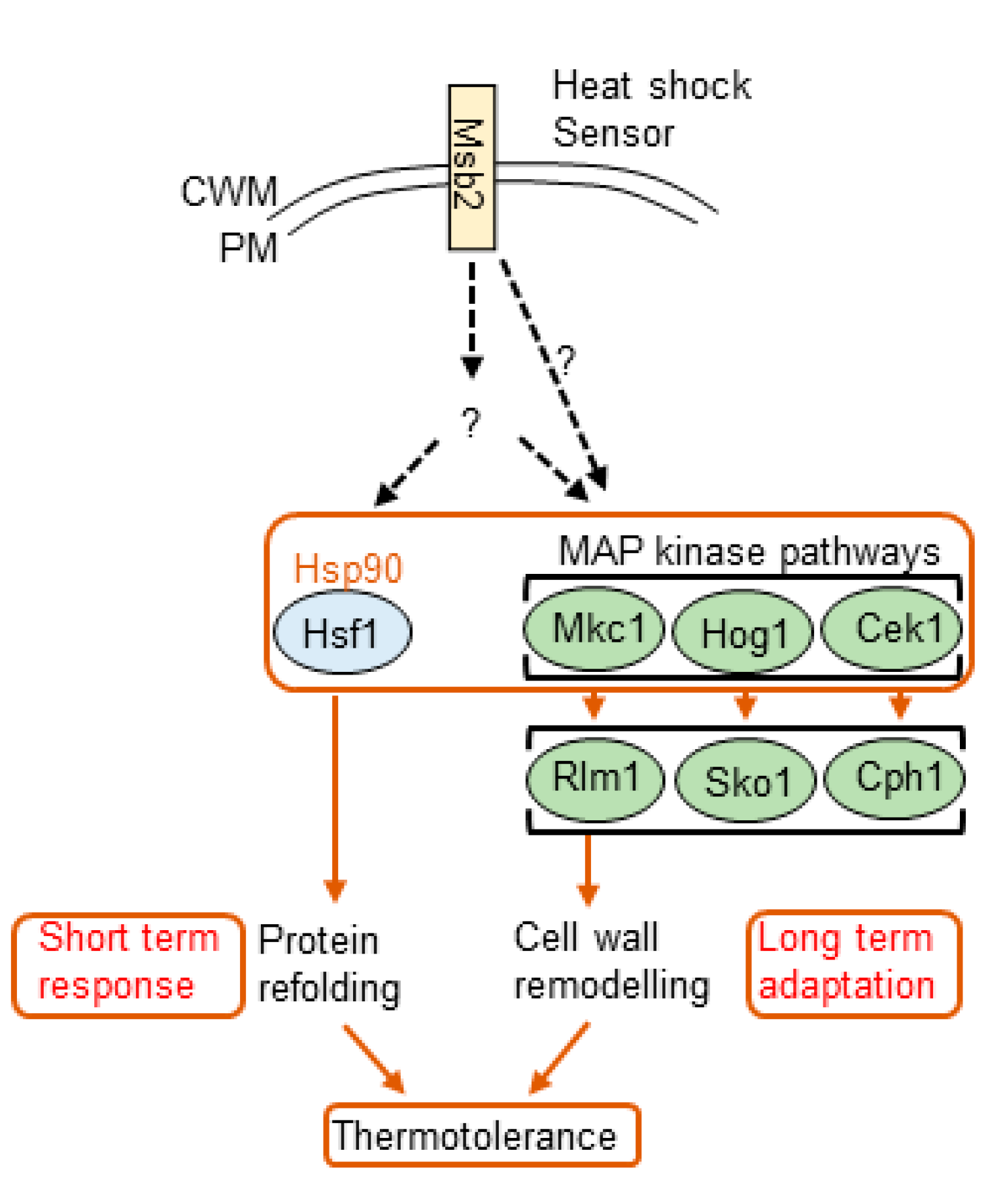
4. Cell Wall Remodeling in Response to Thermal Stress
5. Echinocandin-Induced Cell Wall Remodeling in Yeast
6. Cell Wall Remodeling and Protein Abundance
7. Perspective
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Conflicts of Interest
References
- Brown, G.D.; Denning, D.W.; Gow, N.A.R.; Levitz, S.M.; Netea, M.G.; White, T.C. Hidden killers: Human fungal infections. Sci. Transl. Med. 2012, 4, 165rv13. [Google Scholar] [CrossRef]
- Mayer, F.L.; Wilson, D.; Hube, B. Candida albicans pathogenicity mechanisms. Virulence 2013, 4, 119–128. [Google Scholar] [CrossRef]
- D’Enfert, C.; Kaune, A.K.; Alaban, L.R.; Chakraborty, S.; Cole, N.; Delavy, M.; Kosmala, D.; Marsaux, B.; Frois-Martins, R.; Morelli, M.; et al. The impact of the Fungus-Host-Microbiota interplay upon Candida albicans infections: Current knowledge and new perspectives. FEMS Microbiol. Rev. 2021, 45, 1–55. [Google Scholar]
- Lenardon, M.D.; Sood, P.; Dorfmueller, H.C.; Brown, A.J.; Gow, N.A. Scalar nanostructure of the Candida albicans cell wall; A molecular, cellular and ultrastructural analysis and interpretation. Cell Surf. 2020, 6, 100047. [Google Scholar] [CrossRef]
- Luo, G.; Ibrahim, A.S.; Spellberg, B.; Nobile, C.; Mitchell, A.P.; Fu, Y. Candida albicans Hyr1p confers resistance to neutrophil killing and is a potential vaccine target. J. Infect. Dis. 2010, 201, 1718–1728. [Google Scholar] [CrossRef]
- Edwards, J.E., Jr.; Schwartz, M.M.; Schmidt, C.S.; Sobel, J.D.; Nyirjesy, P.; Schodel, F.; Marchus, E.; Lizakowski, M.; DeMontigny, E.A.; Hoeg, J.; et al. A fungal immunotherapeutic vaccine (NDV-3A) for treatment of recurrent vulvovaginal candidiasis-A Phase 2 randomized, double-blind, placebo-controlled trial. Clin. Infect. Dis. 2018, 66, 1928–1936. [Google Scholar] [CrossRef]
- Spellberg, B.J.; Ibrahim, A.S.; Avanesian, V.; Fu, Y.; Myers, C.; Phan, Q.T.; Filler, S.G.; Yeaman, M.R.; Edwards, J.E., Jr. Efficacy of the anti-Candida rAls3p-N or rAls1p-N vaccines against disseminated and mucosal candidiasis. J. Infect. Dis. 2006, 194, 256–260. [Google Scholar] [CrossRef]
- Klis, F.M.; Sosinska, G.J.; De Groot, P.; Brul, S. Covalently linked cell wall proteins of Candida albicans and their role in fitness and virulence. FEMS Yeast Res. 2009, 9, 1013–1028. [Google Scholar] [CrossRef]
- Klis, F.M.; de Groot, P.; Hellingwerf, K. Molecular organization of the cell wall of Candida albicans. Med. Mycol. 2001, 39 (Suppl. S1), 1–8. [Google Scholar] [CrossRef]
- Sherrington, S.L.; Sorsby, E.; Mahtey, N.; Kumwenda, P.; Lenardon, M.D.; Brown, I.; Ballou, E.R.; Maccallum, D.M.; Hall, R.A. Adaptation of Candida albicans to environmental pH induces cell wall remodeling and enhances innate immune recognition. PLoS Pathog. 2017, 13, e1006403. [Google Scholar] [CrossRef]
- Cheng, S.-C.; van de Veerdonk, F.L.; Lenardon, M.; Stoffels, M.; Plantinga, T.; Smeekens, S.; Rizzetto, L.; Mukaremera, L.; Preechasuth, K.; Cavalieri, D.; et al. The dectin-1/inflammasome pathway is responsible for the induction of protective T-helper 17 responses that discriminate between yeasts and hyphae of Candida albicans. J. Leukoc. Biol. 2011, 90, 357–366. [Google Scholar] [CrossRef]
- Gow, N.A.R.; Latge, J.-P.; Munro, C.A. The fungal cell wall: Structure, biosynthesis, and function. Microbiol. Spectr. 2017, 5, 267–292. [Google Scholar] [CrossRef]
- Kollar, R.; Petrakova, E.; Ashwell, G.; Robbins, P.W.; Cabib, E. Architecture of the yeast cell wall. The linkage between chitin and beta(1-->3)-glucan. J. Biol. Chem. 1995, 270, 1170–1178. [Google Scholar] [CrossRef]
- Gow, N.A.; Hube, B. Importance of the Candida albicans cell wall during commensalism and infection. Curr. Opin. Microbiol. 2012, 15, 406–412. [Google Scholar] [CrossRef]
- Zlotnik, H.; Fernandez, M.P.; Bowers, B.; Cabib, E. Saccharomyces cerevisiae mannoproteins form an external cell wall layer that determines wall porosity. J. Bacteriol. 1984, 159, 1018–1026. [Google Scholar] [CrossRef]
- Gow, N.A.; Netea, M.G.; Munro, C.A.; Ferwerda, G.; Bates, S.; Mora-Montes, H.M.; Walker, L.; Jansen, T.; Jacobs, L.; Tsoni, V.; et al. Immune recognition of Candida albicans beta-glucan by dectin-1. J. Infect. Dis. 2007, 196, 1565–1571. [Google Scholar] [CrossRef]
- Munro, C.; Richard, M.L. The cell wall: Glycoproteins, remodeling, and regulation. In Candida and Candidiasis, 2nd ed.; Calderone, R.A., Clancy, C.J., Eds.; ASM Press: Washington, DC, USA, 2012; pp. 197–223. [Google Scholar]
- De Groot, P.W.; Ram, A.F.; Klis, F.M. Features and functions of covalently linked proteins in fungal cell walls. Fungal Genet. Biol. 2005, 42, 657–675. [Google Scholar] [CrossRef]
- Ibe, C.; Walker, L.A.; Gow, N.; Munro, C.A. Unlocking the therapeutic potential of the fungal cell wall: Clinical implications and drug resistance. In Candida albicans: Cellular and Molecular Biology; Prasad, R., Ed.; Springer International Publishing: London, UK, 2017; pp. 313–346. [Google Scholar] [CrossRef]
- Orlean, P. Architecture and biosynthesis of the Saccharomyces cerevisiae cell wall. Genetics 2012, 192, 775–818. [Google Scholar] [CrossRef]
- Bowman, S.M.; Free, S.J. The structure and synthesis of the fungal cell wall. BioEssays 2006, 28, 799–808. [Google Scholar] [CrossRef]
- Ferguson, M.A. The structure, biosynthesis and functions of glycosylphosphatidylinositol anchors, and the contributions of trypanosome research. J. Cell Sci. 1999, 112 Pt 17, 2799–2809. [Google Scholar] [CrossRef]
- Kinoshita, T.; Inoue, N. Dissecting and manipulating the pathway for glycosylphosphatidylinositol-anchor biosynthesis. Curr. Opin. Chem. Biol. 2000, 4, 632–638. [Google Scholar] [CrossRef]
- Richard, M.L.; Plaine, A. Comprehensive analysis of glycosylphosphatidylinositol-anchored proteins in Candida albicans. Eukaryot. Cell 2007, 6, 119–133. [Google Scholar] [CrossRef]
- Ruiz-Herrera, J.; Elorza, M.V.; Valentin, E.; Sentandreu, R. Molecular organization of the cell wall of Candida albicans and its relation to pathogenicity. FEMS Yeast Res. 2006, 6, 14–29. [Google Scholar] [CrossRef]
- Kapteyn, J.C.; Van Egmond, P.; Sievi, E.; Ende, H.V.D.; Makarow, M.; Klis, F.M. The contribution of the O-glycosylated protein Pir2p/Hsp150 to the construction of the yeast cell wall in wild-type cells and beta1,6-glucan-deficient mutants. Mol. Microbiol. 1999, 31, 1835–1844. [Google Scholar] [CrossRef]
- Zhao, X.; Daniels, K.J.; Oh, S.-H.; Green, C.B.; Yeater, K.M.; Soll, D.R.; Hoyer, L.L. Candida albicans Als3p is required for wild-type biofilm formation on silicone elastomer surfaces. Microbiology 2006, 152, 2287–2299. [Google Scholar] [CrossRef]
- Zhao, X.; Oh, S.-H.; Yeater, K.M.; Hoyer, L.L. Analysis of the Candida albicans Als2p and Als4p adhesins suggests the potential for compensatory function within the Als family. Microbiology 2005, 151, 1619–1630. [Google Scholar] [CrossRef]
- Hoyer, L.L.; Green, C.B.; Oh, S.H.; Zhao, X. Discovering the secrets of the Candida albicans agglutinin-like sequence (ALS) gene family—A sticky pursuit. Med. Mycol. 2008, 46, 1–15. [Google Scholar] [CrossRef]
- Almeida, R.S.; Brunke, S.; Albrecht, A.; Thewes, S.; Laue, M.; Edwards, J.E., Jr.; Filler, S.G.; Hube, B. The hyphal-associated adhesin and invasin Als3 of Candida albicans mediates iron acquisition from host ferritin. PLoS Pathog. 2008, 4, e1000217. [Google Scholar] [CrossRef]
- Phan, Q.T.; Myers, C.L.; Fu, Y.; Sheppard, D.C.; Yeaman, M.R.; Welch, W.H.; Ibrahim, A.S.; Edwards, J.E., Jr.; Filler, S.G. Als3 Is a Candida albicans invasin that binds to cadherins and induces endocytosis by host cells. PLoS Biol. 2007, 5, e64. [Google Scholar] [CrossRef]
- Laforce-Nesbitt, S.S.; Sullivan, M.A.; Hoyer, L.L.; Bliss, J.M. Inhibition of Candida albicans adhesion by recombinant human antibody single-chain variable fragment specific for Als3p. FEMS Immunol. Med. Microbiol. 2008, 54, 195–202. [Google Scholar] [CrossRef]
- Nobile, C.; Schneider, H.A.; Nett, J.E.; Sheppard, D.C.; Filler, S.G.; Andes, D.; Mitchell, A.P. Complementary adhesin function in C. albicans biofilm formation. Curr. Biol. 2008, 18, 1017–1024. [Google Scholar] [CrossRef] [PubMed]
- Nobile, C.; Andes, D.; Nett, J.E.; Smith, F.J.; Yue, F.; Phan, Q.-T.; Edwards, J.E., Jr.; Filler, S.G.; Mitchell, A.P. Critical role of Bcr1-dependent adhesins in C. albicans biofilm formation In Vitro and In Vivo. PLoS Pathog. 2006, 2, e63. [Google Scholar] [CrossRef] [PubMed]
- Nobile, C.; Mitchell, A.P. Regulation of cell-surface genes and biofilm formation by the C. albicans transcription factor Bcr1p. Curr. Biol. 2005, 15, 1150–1155. [Google Scholar] [CrossRef]
- Nobile, C.; Nett, J.E.; Andes, D.; Mitchell, A.P. Function of Candida albicans adhesin Hwp1 in biofilm formation. Eukaryot. Cell 2006, 5, 1604–1610. [Google Scholar] [CrossRef]
- Ruben, S.; Garbe, E.; Mogavero, S.; Albrecht-Eckardt, D.; Hellwig, D.; Häder, A.; Krüger, T.; Gerth, K.; Jacobsen, I.D.; Elshafee, O.; et al. Ahr1 and Tup1 contribute to the transcriptional control of virulence-associated genes in Candida albicans. mBio 2020, 11, e00206-20. [Google Scholar] [CrossRef] [PubMed]
- Sosinska, G.J.; De Groot, P.; De Mattos, M.J.T.; Dekker, H.L.; de Koster, C.; Hellingwerf, K.J.; Klis, F.M. Hypoxic conditions and iron restriction affect the cell-wall proteome of Candida albicans grown under vagina-simulative conditions. Microbiology 2008, 154 Pt 2, 510–520. [Google Scholar] [CrossRef]
- Staab, J.F.; Bradway, S.D.; Fidel, P.L.; Sundstrom, P. Adhesive and mammalian transglutaminase substrate properties of Candida albicans. HwpScience 1999, 283, 1535–1538. [Google Scholar] [CrossRef]
- Xin, H.; Dziadek, S.; Bundle, D.R.; Cutler, J.E. Synthetic glycopeptide vaccines combining beta-mannan and peptide epitopes induce protection against candidiasis. Proc. Natl. Acad. Sci. USA 2008, 105, 13526–13531. [Google Scholar] [CrossRef]
- Lain, A.; Elguezabal, N.; Brena, S.; Garcia-Ruiz, J.C.; Del Palacio, A.; Moragues, M.D.; Pontón, J. Diagnosis of invasive candidiasis by enzyme-linked immunosorbent assay using the N-terminal fragment of Candida albicans hyphal wall protein-1. BMC Microbiol. 2007, 7, 35. [Google Scholar] [CrossRef]
- Hayek, P.; Dib, L.; Yazbeck, P.; Beyrouthy, B.; Khalaf, R.A. Characterization of Hwp2, a Candida albicans putative GPI-anchored cell wall protein necessary for invasive growth. Microbiol. Res. 2010, 165, 250–258. [Google Scholar] [CrossRef]
- Younes, S.; Bahnan, W.; Dimassi, H.; Khalaf, R.A. The Candida albicans Hwp2 is necessary for proper adhesion, biofilm formation and oxidative stress tolerance. Microbiol. Res. 2011, 166, 430–436. [Google Scholar] [CrossRef] [PubMed]
- McCall, A.D.; Pathirana, R.; Prabhakar, A.; Cullen, P.J.; Edgerton, M. Candida albicans biofilm development is governed by cooperative attachment and adhesion maintenance proteins. NPJ Biofilms Microbiomes 2019, 5, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Fonzi, W.A. PHR1 and PHR2 of Candida albicans encode putative glycosidases required for proper cross-linking of beta-1,3- and beta-1,6-glucans. J. Bacteriol. 1999, 181, 7070–7079. [Google Scholar] [CrossRef] [PubMed]
- Mouyna, I.; Fontaine, T.; Vai, M.; Monod, M.; Fonzi, W.A.; Diaquin, M.; Popolo, L.; Hartland, R.P.; Latgé, J.-P. Glycosylphosphatidylinositol-anchored glucanosyltransferases play an active role in the biosynthesis of the fungal cell wall. J. Biol. Chem. 2000, 275, 14882–14889. [Google Scholar] [CrossRef] [PubMed]
- Kovacova, K.; Degani, G.; Stratilová, E.; Farkaš, V.; Popolo, L. Catalytic properties of Phr family members of cell wall glucan remodeling enzymes: Implications for the adaptation of Candida albicans to ambient pH. FEMS Yeast Res. 2015, 15, fou011. [Google Scholar] [CrossRef]
- Eckert, S.E.; Heinz, W.J.; Zakikhany, K.; Thewes, S.; Haynes, K.; Hube, B.; Mühlschlegel, F.A. PGA4, a GAS homologue from Candida albicans, is up-regulated early in infection processes. Fungal Genet. Biol. 2007, 44, 368–377. [Google Scholar] [CrossRef]
- Calderon, J.; Zavrel, M.; Ragni, E.; Fonzi, W.A.; Rupp, S.; Popolo, L. PHR1, a pH-regulated gene of Candida albicans encoding a glucan-remodeling enzyme, is required for adhesion and invasion. Microbiology 2010, 156, 2484–2494. [Google Scholar] [CrossRef]
- Cabib, E.; Farkas, V.; Kosík, O.; Blanco, N.; Arroyo, J.; McPhie, P. Assembly of the yeast cell wall: Crh1p AND Crh2p act as transglycosylases in vivo and in vitro. J. Biol. Chem. 2008, 283, 29859–29872. [Google Scholar] [CrossRef]
- Pardini, G.; De Groot, P.; Coste, A.; Karababa, M.; Klis, F.M.; de Koster, C.; Sanglard, D. The CRH family coding for cell wall Glycosylphosphatidylinositol proteins with a predicted transglycosidase domain affects cell wall organization and virulence of Candida albicans. J. Biol. Chem. 2006, 281, 40399–40411. [Google Scholar] [CrossRef]
- Ene, I.V.; Walker, L.A.; Schiavone, M.; Lee, K.K.; Martin-Yken, H.; Dague, E.; Gow, N.A.R.; Munro, C.A.; Brown, A.J.P. Cell wall remodeling enzymes modulate fungal cell wall elasticity and osmotic stress resistance. mBio 2015, 6, e00986-15. [Google Scholar] [CrossRef]
- Karababa, M.; Valentino, E.; Pardini, G.; Coste, A.T.; Bille, J.; Sanglard, D. CRZ1, a target of the calcineurin pathway in Candida albicans. Mol. Microbiol. 2006, 59, 1429–1451. [Google Scholar] [CrossRef] [PubMed]
- Bruno, V.M.; Kalachikov, S.; Subaran, R.; Nobile, C.J.; Kyratsous, C.; Mitchell, A.P. Control of the C. albicans cell wall damage response by transcriptional regulator Cas5. PLoS Pathog. 2006, 2, e21. [Google Scholar] [CrossRef] [PubMed]
- Ao, J.; Chinnici, J.L.; Maddi, A.; Free, S.J. The N-linked outer chain mannans and the Dfg5p and Dcw1p Endo-α-1,6-Mannanases are needed for incorporation of Candida albicans glycoproteins into the cell wall. Eukaryot. Cell 2015, 14, 792–803. [Google Scholar] [CrossRef] [PubMed]
- Mancuso, R.; Chinnici, J.; Tsou, C.; Busarajan, S.; Munnangi, R.; Maddi, A. Functions of Candida albicans cell wall glycosidases Dfg5p and Dcw1p in biofilm formation and HOG MAPK pathway. PeerJ 2018, 6, e5685. [Google Scholar] [CrossRef] [PubMed]
- Butler, G.; Rasmussen, M.D.; Lin, M.F.; Santos, M.; Sakthikumar, S.; Munro, C.; Rheinbay, E.; Grabherr, M.; Forche, A.; Reedy, J.L.; et al. Evolution of pathogenicity and sexual reproduction in eight Candida genomes. Nature 2009, 459, 657–662. [Google Scholar] [CrossRef]
- De Boer, A.D.; de Groot, P.W.; Weindl, G.; Schaller, M.; Riedel, D.; Diez-Orejas, R.; Klis, F.M.; de Koster, C.G.; Dekker, H.L.; Gross, U.; et al. The Candida albicans cell wall protein Rhd3/Pga29 is abundant in the yeast form and contributes to virulence. Yeast 2010, 27, 611–624. [Google Scholar] [CrossRef] [PubMed]
- Castillo, L.; Martínez, A.I.; Garcerá, A.; Garcia-Martinez, J.; Ruiz-Herrera, J.; Valentín, E.; Sentandreu, R. Genomic response programs of Candida albicans following protoplasting and regeneration. Fungal Genet. Biol. 2006, 43, 124–134. [Google Scholar] [CrossRef]
- Plaine, A.; Walker, L.; Da Costa, G.; Mora-Montes, H.M.; McKinnon, A.; Gow, N.; Gaillardin, C.; Munro, C.; Richard, M.L. Functional analysis of Candida albicans GPI-anchored proteins: Roles in cell wall integrity and caspofungin sensitivity. Fungal Genet. Biol. 2008, 45, 1404–1414. [Google Scholar] [CrossRef]
- Frohner, I.E.; Bourgeois, C.; Yatsyk, K.; Majer, O.; Kuchler, K. Candida albicans cell surface superoxide dismutases degrade host-derived reactive oxygen species to escape innate immune surveillance. Mol. Microbiol. 2009, 71, 240–252. [Google Scholar] [CrossRef]
- Martchenko, M.; Alarco, A.-M.; Harcus, D.; Whiteway, M. Superoxide dismutases in Candida albicans: Transcriptional regulation and functional characterization of the hyphal-induced SOD5 gene. Mol. Biol. Cell 2004, 15, 456–467. [Google Scholar] [CrossRef]
- Albrecht, A.; Felk, A.; Pichova, I.; Naglik, J.; Schaller, M.; De Groot, P.; MacCallum, D.; Odds, F.C.; Schäfer, W.; Klis, F.; et al. Glycosylphosphatidylinositol-anchored proteases of Candida albicans target proteins necessary for both cellular processes and host-pathogen interactions. J. Biol. Chem. 2006, 281, 688–694. [Google Scholar] [CrossRef] [PubMed]
- Moreno-Ruiz, E.; Ortu, G.; De Groot, P.; Cottier, F.; Loussert, C.; Prévost, M.-C.; De Koster, C.; Klis, F.M.; Goyard, S.; D’Enfert, C. The GPI-modified proteins Pga59 and Pga62 of Candida albicans are required for cell wall integrity. Microbiology 2009, 155, 2004–2020. [Google Scholar] [CrossRef] [PubMed]
- Perez, A.; Pedros, B.; Murgui, A.; Casanova, M.; Lopez-Ribot, J.L.; Martinez, J.P. Biofilm formation by Candida albicans mutants for genes coding fungal proteins exhibiting the eight-cysteine-containing CFEM domain. FEMS Yeast Res. 2006, 6, 1074–1084. [Google Scholar] [CrossRef] [PubMed]
- Braun, B.R.; Head, W.S.; Wang, M.X.; Johnson, A.D. Identification and characterization of tup1-regulated genes in Candida albicans. Genetics 2000, 156, 31–44. [Google Scholar] [CrossRef] [PubMed]
- Weissman, Z.; Kornitzer, D. A family of Candida cell surface haem-binding proteins involved in haemin and haemoglobin-iron utilization. Mol. Microbiol. 2004, 53, 1209–1220. [Google Scholar] [CrossRef] [PubMed]
- Kuznets, G.; Vigonsky, E.; Weissman, Z.; Lalli, D.; Gildor, T.; Kauffman, S.J.; Turano, P.; Becker, J.; Lewinson, O.; Kornitzer, D. A relay network of extracellular heme-binding proteins drives, C. albicans iron acquisition from hemoglobin. PLOS Pathog. 2014, 10, e1004407. [Google Scholar] [CrossRef] [PubMed]
- Martínez, A.I.; Castillo, L.; Garcerá, A.; Elorza, M.V.; Valentín, E.; Sentandreu, R. Role of Pir1 in the construction of the Candida albicans cell wall. Microbiology 2004, 150, 3151–3161. [Google Scholar] [CrossRef] [PubMed]
- Pietrella, D.; Lupo, P.; Rachini, A.; Sandini, S.; Ciervo, A.; Perito, S.; Bistoni, F.; Vecchiarelli, A. A Candida albicans mannoprotein deprived of its mannan moiety is efficiently taken up and processed by human dendritic cells and induces T-cell activation without stimulating proinflammatory cytokine production. Infect. Immun. 2008, 76, 4359–4367. [Google Scholar] [CrossRef]
- Willaert, R.G. Adhesins of Yeasts: Protein Structure and Interactions. J. Fungi 2018, 4, 119. [Google Scholar] [CrossRef]
- Munro, C.A. Chitin and glucan, the yin and yang of the fungal cell wall, implications for antifungal drug discovery and therapy. Adv. Appl. Microbiol. 2013, 83, 145–172. [Google Scholar] [CrossRef]
- Ene, I.V.; Heilmann, C.J.; Sorgo, A.G.; Walker, L.A.; de Koster, C.G.; Munro, C.A.; Klis, F.M.; Brown, A.J.P. Carbon source-induced reprogramming of the cell wall proteome and secretome modulates the adherence and drug resistance of the fungal pathogen Candida albicans. Proteomics 2012, 12, 3164–3179. [Google Scholar] [CrossRef] [PubMed]
- Rolli, E.; Ragni, E.; Calderon, J.; Porello, S.; Fascio, U.; Popolo, L. Immobilization of the glycosylphosphatidylinositol-anchored gas1 protein into the chitin ring and septum is required for proper morphogenesis in yeast. Mol. Biol. Cell 2009, 20, 4856–4870. [Google Scholar] [CrossRef]
- Sarthy, A.V.; McGonigal, T.; Coen, M.; Frost, D.J.; Meulbroek, J.A.; Goldman, R.C. Phenotype in Candida albicans of a disruption of the BGL2 gene encoding a 1,3-β-glucosyltransferase. Microbiology 1997, 143, 367–376. [Google Scholar] [CrossRef] [PubMed]
- Cantarel, B.L.; Coutinho, P.M.; Rancurel, C.; Bernard, T.; Lombard, V.; Henrissat, B. The Carbohydrate-Active EnZymes database (CAZy): An expert resource for Glycogenomics. Nucleic Acids Res. 2009, 37, D233–D238. [Google Scholar] [CrossRef] [PubMed]
- Klis, F.M.; Boorsma, A.; De Groot, P. Cell wall construction in Saccharomyces cerevisiae. Yeast 2006, 23, 185–202. [Google Scholar] [CrossRef] [PubMed]
- Klis, F.M.; Brul, S.; De Groot, P.W.J. Covalently linked wall proteins in ascomycetous fungi. Yeast 2010, 27, 489–493. [Google Scholar] [CrossRef]
- Dranginis, A.M.; Rauceo, J.M.; Coronado, J.E.; Lipke, P.N. A Biochemical guide to yeast adhesins: Glycoproteins for social and antisocial occasions. Microbiol. Mol. Biol. Rev. 2007, 71, 282–294. [Google Scholar] [CrossRef]
- Kapteyn, J.C.; Hoyer, L.; Hecht, J.E.; Muller, W.H.; Andel, A.; Verkleij, A.J.; Makarow, M.; Ende, H.V.D.; Klis, F.M. The cell wall architecture of Candida albicans wild-type cells and cell wall-defective mutants. Mol. Microbiol. 2000, 35, 601–611. [Google Scholar] [CrossRef]
- García-Sánchez, S.; Aubert, S.; Iraqui, I.; Janbon, G.; Ghigo, J.-M.; D’Enfert, C. Candida albicans Biofilms: A Developmental State Associated With Specific and Stable Gene Expression Patterns. Eukaryot. Cell 2004, 3, 536–545. [Google Scholar] [CrossRef]
- Garcerá, A.; Martínez, A.I.; Castillo, L.; Elorza, M.V.; Sentandreu, R.; Valentín, E. Identification and study of a Candida albicans protein homologous to Saccharomyces cerevisiae Ssr1p, an internal cell-wall protein. Microbiology 2003, 149 Pt 8, 2137–2145. [Google Scholar] [CrossRef][Green Version]
- Li, F.; Palecek, S.P. EAP1, a Candida albicans gene involved in binding human epithelial cells. Eukaryot. Cell 2003, 2, 1266–1273. [Google Scholar] [CrossRef] [PubMed]
- Weissman, Z.; Shemer, R.; Conibear, E.; Kornitzer, D. An endocytic mechanism for haemoglobin-iron acquisition in Candida albicans. Mol. Microbiol. 2008, 69, 201–217. [Google Scholar] [CrossRef] [PubMed]
- Munro, C. Candida albican cell wall mediated virulence. The Yeast Handbook Series; In Pathogenic Yeast; Ashbee, R.H., Bignell, E.M., Eds.; Springer: Berlin/Heidelberg, Germany, 2010; pp. 69–95. [Google Scholar]
- Gaur, N.K.; Klotz, S.A. Accessibility of the peptide backbone of protein ligands is a key specificity determinant in Candida albicans SRS adherence. Microbiology 2004, 150, 277–284. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Sheppard, D.C.; Yeaman, M.R.; Welch, W.H.; Phan, Q.T.; Fu, Y.; Ibrahim, A.S.; Filler, S.G.; Zhang, M.; Waring, A.J.; Edwards, J.E. Functional and structural diversity in the Als protein family of Candida albicans. J. Biol. Chem. 2004, 279, 30480–30489. [Google Scholar] [CrossRef]
- Calderone, R.A.; Fonzi, W.A. Virulence factors of Candida albicans. Trends Microbiol. 2001, 9, 327–335. [Google Scholar] [CrossRef]
- Staab, J.F.; Datta, K.; Rhee, P. Niche-specific requirement for hyphal wall protein 1 in virulence of Candida albicans. PLoS ONE 2013, 8, e80842. [Google Scholar] [CrossRef]
- Naglik, J.R.; Moyes, D.L.; Wachtler, B.; Hube, B. Candida albicans interactions with epithelial cells and mucosal immunity. Microbes Infect. 2011, 13, 963–976. [Google Scholar] [CrossRef]
- Zhu, W.; Filler, S.G. Interactions of Candida albicans with epithelial cells. Cell Microbiol. 2010, 12, 273–282. [Google Scholar] [CrossRef]
- Wächtler, B.; Citiulo, F.; Jablonowski, N.; Förster, S.; Dalle, F.; Schaller, M.; Wilson, D.; Hube, B. Candida albicans-epithelial interactions: Dissecting the roles of active penetration, induced endocytosis and host factors on the infection process. PLoS ONE 2012, 7, e36952. [Google Scholar] [CrossRef]
- Naglik, J.R.; Challacombe, S.J.; Hube, B. Candida albicans secreted aspartyl proteinases in virulence and pathogenesis. Microbiol. Mol. Biol. Rev. 2003, 67, 400–428. [Google Scholar] [CrossRef]
- Taylor, B.N.; Hannemann, H.; Sehnal, M.; Biesemeier, A.; Schweizer, A.; Röllinghoff, M.; Schröppel, K. Induction of SAP7 correlates with virulence in an intravenous infection model of candidiasis but not in a vaginal infection model in mice. Infect. Immun. 2005, 73, 7061–7063. [Google Scholar] [CrossRef] [PubMed]
- Hube, B.; Naglik, J. Candida albicans proteinases: Resolving the mystery of a gene family. Microbiology 2001, 147 Pt 8, 1997–2005. [Google Scholar] [CrossRef]
- Schild, L.; Heyken, A.; de Groot, P.W.; Hiller, E.; Mock, M.; de Koster, C.; Horn, U.; Rupp, S.; Hube, B. Proteolytic cleavage of covalently linked cell wall proteins by Candida albicans Sap9 and Sap10. Eukaryot. Cell. 2011, 10, 98–109. [Google Scholar] [CrossRef] [PubMed]
- Ramage, G.; Saville, S.P.; Thomas, D.P.; López-Ribot, J.L. Candida Biofilms: An Update. Eukaryot. Cell 2005, 4, 633–638. [Google Scholar] [CrossRef] [PubMed]
- Nobile, C.J.; Nett, J.E.; Hernday, A.D.; Homann, O.R.; Deneault, J.S.; Nantel, A.; Andes, D.R.; Johnson, A.D.; Mitchell, A.P. Biofilm matrix regulation by Candida albicans Zap1. PLoS Biol. 2009, 7, e1000133. [Google Scholar] [CrossRef]
- Green, C.B.; Cheng, G.; Chandra, J.; Mukherjee, P.; Ghannoum, M.A.; Hoyer, L.L. RT-PCR detection of Candida albicans ALS gene expression in the reconstituted human epithelium (RHE) model of oral candidiasis and in model biofilms. Microbiology 2004, 150 Pt 2, 267–275. [Google Scholar] [CrossRef][Green Version]
- Brown, A.J.P.; Budge, S.; Kaloriti, D.; Tillmann, A.; Jacobsen, M.D.; Yin, Z.; Ene, I.V.; Bohovych, I.; Sandai, D.; Kastora, S.; et al. Stress adaptation in a pathogenic fungus. J. Exp. Biol. 2014, 217, 144–155. [Google Scholar] [CrossRef] [PubMed]
- Walker, L.A.; Munro, C.; de Bruijn, I.; Lenardon, M.D.; McKinnon, A.D.; Gow, N.A.R. Stimulation of chitin synthesis rescues Candida albicans from echinocandins. PLoS Pathog. 2008, 4, e1000040. [Google Scholar] [CrossRef]
- Heinisch, J.J.; Dupres, V.; Wilk, S.; Jendretzki, A.; Dufrene, Y.F. Single-molecule atomic force microscopy reveals clustering of the yeast plasma-membrane sensor Wsc1. PLoS ONE 2010, 5, e11104. [Google Scholar] [CrossRef]
- Rodicio, R.R.; Heinisch, J.J. Together we are strong-cell wall integrity sensors in yeasts. Yeast 2010, 27, 531–540. [Google Scholar] [CrossRef] [PubMed]
- Levin, D.E. Regulation of cell wall biogenesis in Saccharomyces cerevisiae: The cell wall integrity signaling pathway. Genetics 2011, 189, 1145–1175. [Google Scholar] [CrossRef] [PubMed]
- Dupres, V.; Alsteens, D.; Wilk, S.; Hansen, B.; Heinisch, J.J.; Dufrêne, Y. The yeast Wsc1 cell surface sensor behaves like a nanospring In Vivo. Nat. Chem. Biol. 2009, 5, 857–862. [Google Scholar] [CrossRef]
- Philip, B.; Levin, D.E. Wsc1 and Mid2 are cell surface sensors for cell wall integrity signaling that act through Rom2, a Guanine Nucleotide Exchange Factor for Rho1. Mol. Cell. Biol. 2001, 21, 271–280. [Google Scholar] [CrossRef] [PubMed]
- Rajavel, M.; Philip, B.; Buehrer, B.M.; Errede, B.; Levin, D.E. Mid2 is a putative sensor for cell integrity signaling in Saccharomyces cerevisiae. Mol. Cell. Biol. 1999, 19, 3969–3976. [Google Scholar] [CrossRef]
- Lara-Aguilar, V.; Rueda, C.; García-Barbazán, I.; Varona, S.; Monzón, S.; Jiménez, P.; Cuesta, I.; Zaballos, Á.; Zaragoza, Ó. Adaptation of the emerging pathogenic yeast Candida auris to high caspofungin concentrations correlates with cell wall changes. Virulence 2021, 12, 1400–1417. [Google Scholar] [CrossRef] [PubMed]
- Kapteyn, J.C.; Ram, A.F.; Groos, E.M.; Kollar, R.; Montijn, R.C.; Ende, H.V.D.; Llobell, A.; Cabib, E.; Klis, F.M. Altered extent of cross-linking of beta1,6-glucosylated mannoproteins to chitin in Saccharomyces cerevisiae mutants with reduced cell wall beta1,3-glucan content. J. Bacteriol. 1997, 179, 6279–6284. [Google Scholar] [CrossRef]
- Munro, C.A.; Selvaggini, S.; de Bruijn, I.; Walker, L.; Lenardon, M.D.; Gerssen, B.; Milne, S.; Brown, A.J.; Gow, N. The PKC, HOG and Ca2+ signalling pathways co-ordinately regulate chitin synthesis in Candida albicans. Mol. Microbiol. 2007, 63, 1399–1413. [Google Scholar] [CrossRef]
- Ene, I.V.; Adya, A.K.; Wehmeier, S.; Brand, A.C.; MacCallum, D.M.; Gow, N.A.; Brown, A.J.P. Host carbon sources modulate cell wall architecture, drug resistance and virulence in a fungal pathogen. Cell Microbiol. 2012, 14, 1319–1335. [Google Scholar] [CrossRef]
- Monge, R.A.; Román, E.; Nombela, C.; Pla, J. The MAP kinase signal transduction network in Candida albicans. Microbiology 2006, 152 Pt 4, 905–912. [Google Scholar] [CrossRef]
- Saraswat, D.; Kumar, R.; Pande, T.; Edgerton, M.; Cullen, P.J. Signalling mucin Msb2 Regulates adaptation to thermal stress in Candida albicans. Mol. Microbiol. 2016, 100, 425–441. [Google Scholar] [CrossRef]
- Walker, L.A.; Gow, N.; Munro, C.A. Fungal echinocandin resistance. Fungal Genet. Biol. 2010, 47, 117–126. [Google Scholar] [CrossRef] [PubMed]
- Navarro-García, F.; Eisman, B.; Fiuza, S.; Nombela, C.; Pla, J. The MAP kinase Mkc1p is activated under different stress conditions in Candida albicans. Microbiology 2005, 151 Pt 8, 2737–2749. [Google Scholar] [CrossRef]
- Bermejo, C.; Rodríguez, E.; Garcia, R.; Peña, J.M.R.; de la Concepción, M.L.R.; Rivas, C.; Arias, P.; Nombela, C.; Posas, F.; Arroyo, J. The Sequential Activation of the Yeast HOG and SLT2 Pathways is required for cell survival to cell wall stress. Mol. Biol. Cell 2008, 19, 1113–1124. [Google Scholar] [CrossRef] [PubMed]
- Eisman, B.; Alonso-Monge, R.; Román, E.; Arana, D.; Nombela, C.; Pla, J. The Cek1 and Hog1 Mitogen-Activated Protein Kinases Play Complementary Roles in Cell wall biogenesis and chlamydospore formation in the fungal pathogen Candida albicans. Eukaryot. Cell 2006, 5, 347–358. [Google Scholar] [CrossRef] [PubMed]
- Diez-Orejas, R.; Molero, G.; Navarro-Garcia, F.; Pla, J.; Nombela, C.; Sanchez-Perez, M. Reduced virulence of Candida albicans MKC1 mutants: A role for mitogen-activated protein kinase in pathogenesis. Infect Immun. 1997, 65, 833–837. [Google Scholar] [CrossRef] [PubMed]
- Csank, C.; Schröppel, K.; Leberer, E.; Harcus, D.; Mohamed, O.; Meloche, S.; Thomas, D.Y.; Whiteway, M. Roles of the Candida albicans mitogen-activated protein kinase homolog, Cek1p, in hyphal development and systemic candidiasis. Infect. Immun. 1998, 66, 2713–2721. [Google Scholar] [CrossRef] [PubMed]
- Alonso-Monge, R.; Navarro-Garcia, F.; Molero, G.; Diez-Orejas, R.; Gustin, M.; Pla, J.; Sanchez, M.; Nombela, C. Role of the mitogen-activated protein kinase Hog1p in morphogenesis and virulence of Candida albicans. J. Bacteriol. 1999, 181, 3058–3068. [Google Scholar] [CrossRef] [PubMed]
- Tsao, C.-C.; Chen, Y.-T.; Lan, C.-Y. A small G protein Rhb1 and a GTPase-activating protein Tsc2 involved in nitrogen starvation-induced morphogenesis and cell wall integrity of Candida albicans. Fungal Genet. Biol. 2009, 46, 126–136. [Google Scholar] [CrossRef] [PubMed]
- Xie, J.L.; Qin, L.; Miao, Z.; Grys, B.T.; Diaz, J.D.L.C.; Ting, K.; Krieger, J.R.; Tong, J.; Tan, K.; Leach, M.D.; et al. The Candida albicans transcription factor Cas5 couples stress responses, drug resistance and cell cycle regulation. Nat. Commun. 2017, 8, 1–18. [Google Scholar] [CrossRef]
- Xiong, K.; Su, C.; Sun, Q.; Lu, Y. Efg1 and Cas5 orchestrate cell wall damage response to caspofungin in Candida albicans. Antimicrob. Agents Chemother. 2021, 65, e01584-20. [Google Scholar] [CrossRef] [PubMed]
- Gregori, C.; Glaser, W.; Frohner, I.E.; Reinoso-Martin, C.; Rupp, S.; Schuller, C.; Kuchler, K. Efg1 Controls caspofungin-induced cell aggregation of Candida albicans through the adhesin Als1. Eukaryot Cell. 2011, 10, 1694–1704. [Google Scholar] [CrossRef] [PubMed]
- Zucchi, P.C.; Davis, T.R.; Kumamoto, C.A. A Candida albicans cell wall-linked protein promotes invasive filamentation into semi-solid medium. Mol. Microbiol. 2010, 76, 733–748. [Google Scholar] [CrossRef] [PubMed]
- Dichtl, K.; Samantaray, S.; Wagener, J. Cell wall integrity signalling in human pathogenic fungi. Cell. Microbiol. 2016, 18, 1228–1238. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Kohler, J.; Fink, G.R. Suppression of hyphal formation in Candida albicans by mutation of a STE12 homolog. Science 1994, 266, 1723–1726. [Google Scholar] [CrossRef]
- Silva, Y.B.D.; Vaz, C.; Pereira, J.; Carneiro, C.; Nogueira, E.; Correia, A.; Carreto, L.; Silva, S.; Faustino, A.; Pais, C.; et al. Participation of Candida albicans transcription factor RLM1 in cell wall biogenesis and virulence. PLoS ONE 2014, 9, e86270. [Google Scholar] [CrossRef]
- Valdivia, R.H.; Schekman, R. The yeasts Rho1p and Pkc1p regulate the transport of chitin synthase III (Chs3p) from internal stores to the plasma membrane. Proc. Natl. Acad. Sci. USA 2003, 100, 10287–10292. [Google Scholar] [CrossRef]
- Lenardon, M.D.; Munro, C.; Gow, N.A. Chitin synthesis and fungal pathogenesis. Curr. Opin. Microbiol. 2010, 13, 416–423. [Google Scholar] [CrossRef]
- Lenardon, M.D.; Lesiak, I.; Munro, C.; Gow, N.A.R. Dissection of the Candida albicans class I chitin synthase promoters. Mol. Genet. Genom. 2009, 281, 459–471. [Google Scholar] [CrossRef] [PubMed]
- Kamada, Y.; Jung, U.S.; Piotrowski, J.; Levin, D. The protein kinase C-activated MAP kinase pathway of Saccharomyces cerevisiae mediates a novel aspect of the heat shock response. Genes Dev. 1995, 9, 1559–1571. [Google Scholar] [CrossRef]
- Rauceo, J.M.; Blankenship, J.R.; Fanning, S.; Hamaker, J.J.; Deneault, J.S.; Smith, F.J.; Nantel, A.; Mitchell, A.P.; Bloom, K. Regulation of the Candida albicans cell wall damage response by transcription factor Sko1 and PAS kinase Psk1. Mol. Biol. Cell. 2008, 19, 2741–2751. [Google Scholar]
- Heredia, M.Y.; Ikeh, M.A.C.; Gunasekaran, D.; Conrad, K.A.; Filimonava, S.; Marotta, D.H.; Nobile, C.J.; Rauceo, J.M. An expanded cell wall damage signaling network is comprised of the transcription factors Rlm1 and Sko1 in Candida albicans. PLoS Genet. 2020, 16, e1008908. [Google Scholar] [CrossRef]
- Chamilos, G.; Nobile, C.J.; Bruno, V.M.; Lewis, R.E.; Mitchell, A.P.; Kontoyiannis, D.P. Candida albicans Cas5, a regulator of cell wall integrity, is required for virulence in murine and toll mutant fly models. J. Infect. Dis. 2009, 200, 152–157. [Google Scholar] [CrossRef] [PubMed]
- Vasicek, E.M.; Berkow, E.L.; Bruno, V.M.; Mitchell, A.P.; Wiederhold, N.; Barker, K.S.; Rogers, P.D. Disruption of the transcriptional regulator Cas5 results in enhanced killing of Candida albicans by fluconazole. Antimicrob. Agents Chemother. 2014, 58, 6807–6818. [Google Scholar] [CrossRef] [PubMed]
- Bockmuhl, D.P.; Ernst, J.F. A potential phosphorylation site for an A-type kinase in the Efg1 regulator protein contributes to hyphal morphogenesis of Candida albicans. Genetics 2001, 157, 1523–1530. [Google Scholar] [CrossRef]
- Nantel, A.; Dignard, D.; Bachewich, C.; Harcus, D.; Marcil, A.; Bouin, A.-P.; Sensen, C.W.; Hogues, H.; Hoog, M.V.H.; Gordon, P.; et al. Transcription profiling of Candida albicans cells undergoing the yeast-to-hyphal transition. Mol. Biol. Cell 2002, 13, 3452–3465. [Google Scholar] [CrossRef]
- Zordan, R.E.; Galgoczy, D.J.; Johnson, A.D. Epigenetic properties of white-opaque switching in Candida albicans are based on a self-sustaining transcriptional feedback loop. Proc. Natl. Acad. Sci. USA 2006, 103, 12807–12812. [Google Scholar] [CrossRef]
- Noffz, C.S.; Liedschulte, V.; Lengeler, K.; Ernst, J.F. Functional mapping of the Candida albicans Efg1 regulator. Eukaryot. Cell 2008, 7, 881–893. [Google Scholar] [CrossRef] [PubMed]
- Stichternoth, C.; Ernst, J.F. Hypoxic Adaptation by Efg1 Regulates biofilm formation by Candida albicans. Appl. Environ. Microbiol. 2009, 75, 3663–3672. [Google Scholar] [CrossRef]
- Langford, M.L.; Hargarten, J.C.; Patefield, K.D.; Marta, E.; Blankenship, J.; Fanning, S.; Nickerson, K.W.; Atkin, A.L. Candida albicans Czf1 and Efg1 coordinate the response to farnesol during quorum sensing, white-opaque thermal dimorphism, and cell death. Eukaryot. Cell 2013, 12, 1281–1292. [Google Scholar] [CrossRef]
- Mottola, A.; Ramírez-Zavala, B.; Hünniger, K.; Kurzai, O.; Morschhäuser, J. The zinc cluster transcription factor Czf1 regulates cell wall architecture and integrity in Candida albicans. Mol. Microbiol. 2021, 116, 483–497. [Google Scholar] [CrossRef]
- Dodou, E.; Treisman, R. The Saccharomyces cerevisiae MADS-box transcription factor Rlm1 is a target for the Mpk1 mitogen-activated protein kinase pathway. Mol. Cell. Biol. 1997, 17, 1848–1859. [Google Scholar] [CrossRef]
- Jung, U.S.; Levin, D. Genome-wide analysis of gene expression regulated by the yeast cell wall integrity signalling pathway. Mol. Microbiol. 1999, 34, 1049–1057. [Google Scholar] [CrossRef]
- Rosenwald, A.G.; Arora, G.; Ferrandino, R.; Gerace, E.L.; Mohammednetej, M.; Nosair, W.; Rattila, S.; Subic, A.Z.; Rolfes, R. Identification of genes in Candida glabrata conferring altered responses to caspofungin, a cell wall synthesis inhibitor. G3 Genes Genomes Genet. 2016, 6, 2893–2907. [Google Scholar] [CrossRef]
- Saito, H.; Posas, F. Response to Hyperosmotic Stress. Genetics 2012, 192, 289–318. [Google Scholar] [CrossRef] [PubMed]
- Román, E.; Correia, I.; Prieto, A.D.; Alonso, R.; Pla, J. The HOG MAPK pathway in Candida albicans: More than an osmosensing pathway. Int. Microbiol. 2019, 23, 23–29. [Google Scholar] [CrossRef] [PubMed]
- Marotta, D.H.; Nantel, A.; Sukala, L.; Teubl, J.R.; Rauceo, J.M. Genome-wide transcriptional profiling and enrichment mapping reveal divergent and conserved roles of Sko1 in the Candida albicans osmotic stress response. Genomics 2013, 102, 363–371. [Google Scholar] [CrossRef] [PubMed]
- Alonso-Monge, R.; Román, E.; Arana, D.M.; Prieto, A.D.; Urrialde, V.; Nombela, C.; Pla, J. The Sko1 protein represses the yeast-to-hypha transition and regulates the oxidative stress response in Candida albicans. Fungal Genet. Biol. 2010, 47, 587–601. [Google Scholar] [CrossRef] [PubMed]
- Shivarathri, R.; Jenull, S.; Stoiber, A.; Chauhan, M.; Mazumdar, R.; Singh, A.; Nogueira, F.; Kuchler, K.; Chowdhary, A.; Chauhan, N. The two-component response regulator Ssk1 and the mitogen-activated protein Kinase Hog1 control antifungal drug resistance and cell wall architecture of Candida auris. mSphere 2020, 5, e00973-20. [Google Scholar] [CrossRef]
- Day, A.M.; Smith, D.A.; Ikeh, M.A.C.; Haider, M.; Herrero-De-Dios, C.M.; Brown, A.J.; Morgan, B.A.; Erwig, L.P.; Maccallum, D.M.; Quinn, J. Blocking two-component signalling enhances Candida albicans virulence and reveals adaptive mechanisms that counteract sustained SAPK activation. PLoS Pathog. 2017, 13, e1006131. [Google Scholar] [CrossRef] [PubMed]
- Mavrianos, J.; Desai, C.; Chauhan, N. Two-component histidine phosphotransfer protein Ypd1 Is not essential for viability in Candida albicans. Eukaryot. Cell 2014, 13, 452–460. [Google Scholar] [CrossRef]
- Herrero, C.; Alonso-Monge, R.; Pla, J. The lack of upstream elements of the Cek1 and Hog1 mediated pathways leads to a synthetic lethal phenotype upon osmotic stress in Candida albicans. Fungal Genet. Biol. 2014, 69, 31–42. [Google Scholar] [CrossRef] [PubMed]
- Román, E.; Cottier, F.; Ernst, J.F.; Pla, J. Msb2 signaling mucin controls activation of Cek1 mitogen-activated protein kinase in Candida albicans. Eukaryot. Cell 2009, 8, 1235–1249. [Google Scholar] [CrossRef]
- Román, E.; Nombela, C.; Pla, J. The Sho1 adaptor protein links oxidative stress to morphogenesis and cell wall biosynthesis in the fungal pathogen Candida albicans. Mol. Cell. Biol. 2005, 25, 10611–10627. [Google Scholar] [CrossRef]
- Ramírez-Zavala, B.; Weyler, M.; Gildor, T.; Schmauch, C.; Kornitzer, D.; Arkowitz, R.; Morschhäuser, J. Activation of the Cph1-dependent MAP kinase signaling pathway induces white-opaque switching in Candida albicans. PLoS Pathog. 2013, 9, e1003696. [Google Scholar] [CrossRef]
- Sahni, N.; Yi, S.; Daniels, K.J.; Huang, G.; Srikantha, T.; Soll, D.R. Tec1 mediates the pheromone response of the white phenotype of Candida albicans: Insights into the evolution of new signal transduction pathways. PLoS Biol. 2010, 8, e1000363. [Google Scholar] [CrossRef]
- Huang, H.; Harcus, D.; Whiteway, M. Transcript profiling of a MAP kinase pathway in C. albicans. Microbiol. Res. 2008, 163, 380–393. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Van Wijlick, L.; Swidergall, M.; Brandt, P.; Ernst, J.F. Candida albicans responds to glycostructure damage by Ace2-mediated feedback regulation of Cek1 signaling. Mol. Microbiol. 2016, 102, 827–849. [Google Scholar] [CrossRef] [PubMed]
- Walker, L.A.; Lee, K.K.; Munro, C.A.; Gow, N.A. Caspofungin treatment of Aspergillus fumigatus results in ChsG-dependent upregulation of chitin synthesis and the formation of chitin-rich microcolonies. Antimicrob. Agents Chemother. 2015, 59, 5932–5941. [Google Scholar] [CrossRef]
- Lee, K.K.; MacCallum, D.M.; Jacobsen, M.D.; Walker, L.A.; Odds, F.C.; Gow, N.A.R.; Munro, C.A. Elevated cell wall chitin in Candida albicans confers echinocandin resistance In Vivo. Antimicrob. Agents Chemother. 2012, 56, 208–217. [Google Scholar] [CrossRef]
- Walker, L.A.; Gow, N.A.R.; Munro, C.A. Elevated chitin content reduces the susceptibility of Candida species to caspofungin. Antimicrob. Agents Chemother. 2013, 57, 146–154. [Google Scholar] [CrossRef]
- Cabib, E.; Blanco, N.; Grau, C.; Rodriguez-Pena, J.M.; Arroyo, J. Crh1p and Crh2p are required for the cross-linking of chitin to beta(1–6)glucan in the Saccharomyces cerevisiae cell wall. Mol. Microbiol. 2007, 63, 921–935. [Google Scholar] [CrossRef]
- Cabib, E. Two novel techniques for determination of polysaccharide cross-links show that Crh1p and Crh2p attach chitin to both beta(1–6)- and beta(1–3)glucan in the Saccharomyces cerevisiae cell wall. Eukaryot. Cell. 2009, 8, 1626–1636. [Google Scholar] [CrossRef]
- Terashima, H.; Yabuki, N.; Arisawa, M.; Hamada, K.; Kitada, K. Up-regulation of genes encoding glycosylphosphatidylinositol (GPI)-attached proteins in response to cell wall damage caused by disruption of FKS1 in Saccharomyces cerevisiae. Mol. Genet. Genom. 2000, 264, 64–74. [Google Scholar] [CrossRef] [PubMed]
- Yoshimoto, H.; Saltsman, K.; Gasch, A.P.; Li, H.X.; Ogawa, N.; Botstein, D.; Brown, P.O.; Cyert, M.S. Genome-wide analysis of gene expression regulated by the calcineurin/Crz1p signaling pathway in Saccharomyces cerevisiae. J. Biol. Chem. 2002, 277, 31079–31088. [Google Scholar] [CrossRef] [PubMed]
- Xu, H.; Fang, T.; Omran, R.P.; Whiteway, M.; Jiang, L. RNA sequencing reveals an additional Crz1-binding motif in promoters of its target genes in the human fungal pathogen Candida albicans. Cell Commun. Signal. 2020, 18, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Sanglard, D.; Ischer, F.; Marchetti, O.; Entenza, J.; Bille, J. Calcineurin A of Candida albicans: Involvement in antifungal tolerance, cell morphogenesis and virulence. Mol. Microbiol. 2003, 48, 959–976. [Google Scholar] [CrossRef]
- Blankenship, J.R.; Wormley, F.L.; Boyce, M.K.; Schell, W.A.; Filler, S.G.; Perfect, J.R.; Heitman, J. Calcineurin is essential for Candida albicans survival in serum and virulence. Eukaryot. Cell. 2003, 2, 422–430. [Google Scholar] [CrossRef] [PubMed]
- Cruz, M.C.; Goldstein, A.L.; Blankenship, J.R.; Del Poeta, M.; Davis, D.; Cardenas, M.E.; Perfect, J.R.; McCusker, J.H.; Heitman, J. Calcineurin is essential for survival during membrane stress in Candida albicans. EMBO J. 2002, 21, 546–559. [Google Scholar] [CrossRef]
- Jiang, L.; Alber, J.; Wang, J.; Du, W.; Yang, X.; Li, X.; Sanglard, D.; Geyer, J. The Candida albicans plasma membrane protein Rch1p, a member of the vertebrate SLC10 carrier family, is a novel regulator of cytosolic Ca2+ homoeostasis. Biochem. J. 2012, 444, 497–502. [Google Scholar] [CrossRef] [PubMed]
- Mille, C.; Janbon, G.; Delplace, F.; Ibata-Ombetta, S.; Gaillardin, C.; Strecker, G.; Jouault, T.; Trinel, P.-A.; Poulain, D. Inactivation of CaMIT1 inhibits Candida albicans phospholipomannan beta-mannosylation, reduces virulence, and alters cell wall protein beta-mannosylation. J. Biol. Chem. 2004, 279, 47952–47960. [Google Scholar] [CrossRef]
- Ichikawa, Y.; Bruno, V.M.; Woolford, C.A.; Kim, H.; Do, E.; Brewer, G.; Mitchell, A.P. Environmentally contingent control of Candida albicans cell wall integrity by transcriptional regulator Cup9. Genetics 2021, 218, iyab075. [Google Scholar] [CrossRef]
- Garcia-Rubio, R.; Hernandez, R.Y.; Clear, A.; Healey, K.R.; Shor, E.; Perlin, D.S. Critical assessment of cell wall integrity factors contributing to in vivo echinocandin tolerance and resistance in Candida glabrata. Front. Microbiol. 2021, 12, 702779. [Google Scholar] [CrossRef]
- Brown, D.H., Jr.; Giusani, A.D.; Chen, X.; Kumamoto, C.A. Filamentous growth of Candida albicans in response to physical environmental cues and its regulation by the unique CZF1 gene. Mol. Microbiol. 1999, 34, 651–662. [Google Scholar] [CrossRef]
- Leach, M.D.; Budge, S.; Walker, L.; Munro, C.; Cowen, L.; Brown, A.J. Hsp90 orchestrates transcriptional regulation by Hsf1 and cell wall remodelling by MAPK signalling during thermal adaptation in a pathogenic yeast. PLoS Pathog. 2012, 8, e1003069. [Google Scholar] [CrossRef] [PubMed]
- Puri, S.; Kumar, R.; Chadha, S.; Tati, S.; Conti, H.R.; Hube, B.; Cullen, P.J.; Edgerton, M. Secreted aspartic protease cleavage of Candida albicans Msb2 activates Cek1 MAPK signaling affecting biofilm formation and oropharyngeal candidiasis. PLoS ONE 2012, 7, e46020. [Google Scholar] [CrossRef]
- Cullen, P.J.; Sabbagh, W.; Graham, E.; Irick, M.M.; Van Olden, E.K.; Neal, C.; Delrow, J.; Bardwell, L.; Sprague, G.F., Jr. A signaling mucin at the head of the Cdc42- and MAPK-dependent filamentous growth pathway in yeast. Genes Dev. 2004, 18, 1695–1708. [Google Scholar] [CrossRef] [PubMed]
- Sorger, P.; Pelham, H.R. Yeast heat shock factor is an essential DNA-binding protein that exhibits temperature-dependent phosphorylation. Cell 1988, 54, 855–864. [Google Scholar] [CrossRef]
- Leach, M.; Tyc, K.M.; Brown, A.J.P.; Klipp, E. Modelling the regulation of thermal adaptation in Candida albicans, a major fungal pathogen of humans. PLoS ONE 2012, 7, e32467. [Google Scholar] [CrossRef] [PubMed]
- Jakobsen, B.K.; Pelham, H.R. Constitutive binding of yeast heat shock factor to DNA in vivo. Mol. Cell Biol. 1988, 8, 5040–5042. [Google Scholar] [CrossRef]
- Sorger, P.; Lewis, M.J.; Pelham, H.R.B. Heat shock factor is regulated differently in yeast and HeLa cells. Nature 1987, 329, 81–84. [Google Scholar] [CrossRef] [PubMed]
- Feder, M.E.; Hofmann, G.E. Heat-shock proteins, molecular chaperones, and the stress response: Evolutionary and ecological physiology. Annu. Rev. Physiol. 1999, 61, 243–282. [Google Scholar] [CrossRef] [PubMed]
- Navarro-García, F.; Monge, R.A.; Rico, H.; Pla, J.; Sentandreu, R.; Nombela, C. A role for the MAP kinase gene MKC1 in cell wall construction and morphological transitions in Candida albicans. Microbiology 1998, 144 Pt 2, 411–424. [Google Scholar] [CrossRef]
- Heilmann, C.J.; Sorgo, A.G.; Mohammadi, S.; Sosinska, G.J.; de Koster, C.; Brul, S.; De Koning, L.J.; Klis, F.M. Surface Stress Induces a Conserved Cell Wall Stress response in the pathogenic fungus Candida albicans. Eukaryot. Cell 2013, 12, 254–264. [Google Scholar] [CrossRef]
- Winkler, A.; Arkind, C.; Mattison, C.P.; Burkholder, A.; Knoche, K.; Ota, I. Heat stress activates the yeast high-osmolarity glycerol mitogen-activated protein kinase pathway, and protein tyrosine phosphatases are essential under heat stress. Eukaryot. Cell 2002, 1, 163–173. [Google Scholar] [CrossRef] [PubMed]
- Prill, S.K.-H.; Klinkert, B.; Timpel, C.; Gale, C.A.; Schröppel, K.; Ernst, J.F. PMT family of Candida albicans: Five protein mannosyltransferase isoforms affect growth, morphogenesis and antifungal resistance. Mol. Microbiol. 2005, 55, 546–560. [Google Scholar] [CrossRef] [PubMed]
- Bates, S.; Hughes, H.B.; Munro, C.; Thomas, W.P.; MacCallum, D.; Bertram, G.; Atrih, A.; Ferguson, M.; Brown, A.J.; Odds, F.C.; et al. Outer chain n-glycans are required for cell wall integrity and virulence of Candida albicans. J. Biol. Chem. 2006, 281, 90–98. [Google Scholar] [CrossRef]
- Okawa, Y.; Goto, K. Antigenicity of cell wall mannans of Candida albicans and Candida stellatoidea cultured at high temperatures in BACTEC medium. Biol. Pharm. Bull. 2006, 29, 1723–1727. [Google Scholar] [CrossRef] [PubMed]
- Terashima, H.; Hamada, K.; Kitada, K. The localization change of Ybr078w/Ecm33, a yeast GPI-associated protein, from the plasma membrane to the cell wall, affecting the cellular function. FEMS Microbiol. Lett. 2003, 218, 175–180. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Gil-Bona, A.; Reales-Calderon, J.A.; Giraldo, C.M.P.; Martinez, R.; Monteoliva, L.; Gil, C. The cell wall protein Ecm33 of Candida albicans is involved in chronological life span, morphogenesis, cell wall regeneration, stress tolerance, and host–cell interaction. Front. Microbiol. 2016, 7, 64. [Google Scholar] [CrossRef] [PubMed]
- Douglas, C.M.; Foor, F.; Marrinan, J.A.; Morin, N.; Nielsen, J.B.; Dahl, A.M.; Mazur, P.; Baginsky, W.; Li, W.; El-Sherbeini, M. The Saccharomyces cerevisiae FKS1 (ETG1) gene encodes an integral membrane protein which is a subunit of 1,3-beta-D-glucan synthase. Proc. Natl. Acad. Sci. USA 1994, 91, 12907–12911. [Google Scholar] [CrossRef]
- Qadota, H.; Python, C.P.; Inoue, S.B.; Arisawa, M.; Anraku, Y.; Zheng, Y.; Watanabe, T.; Levin, D.E.; Ohya, Y. Identification of yeast Rho1p GTPase as a regulatory subunit of 1,3-beta-glucan Synthase. Science 1996, 272, 279–281. [Google Scholar] [CrossRef]
- Denning, D.W. Echinocandin antifungal drugs. Lancet 2003, 362, 1142–1151. [Google Scholar] [CrossRef]
- Reinoso-Martín, C.; Schüller, C.; Schuetzer-Muehlbauer, M.; Kuchler, K. The yeast protein kinase C cell integrity pathway mediates tolerance to the antifungal drug caspofungin through activation of Slt2p mitogen-activated protein kinase signaling. Eukaryot. Cell 2003, 2, 1200–1210. [Google Scholar] [CrossRef] [PubMed]
- Tams, R.N.; Wagner, A.S.; Jackson, J.W.; Gann, E.R.; Sparer, T.E.; Reynolds, T.B. Pathways that synthesize phosphatidylethanolamine impact Candida albicans hyphal length and cell wall composition through transcriptional and posttranscriptional mechanisms. Infect. Immun. 2020, 88. [Google Scholar] [CrossRef]
- Walker, L.A.; Lenardon, M.D.; Preechasuth, K.; Munro, C.; Gow, N.A.R. Cell wall stress induces alternative fungal cytokinesis and septation strategies. J. Cell Sci. 2013, 126 Pt 12, 2668–2677. [Google Scholar] [CrossRef]
- Blanco, N.; Sanz, A.B.; Peña, J.M.R.; Nombela, C.; Farkas, V.; Hurtado-Guerrero, R.; Arroyo, J. Structural and functional analysis of yeast Crh1 and Crh2 transglycosylases. FEBS J. 2014, 282, 715–731. [Google Scholar] [CrossRef]
- Pardo, M.; Monteoliva, L.; Vázquez, P.; Martinez, R.; Molero, G.; Nombela, C.; Gil, C. PST1 and ECM33 encode two yeast cell surface GPI proteins important for cell wall integrity. Microbiology 2004, 150 Pt 12, 4157–4170. [Google Scholar] [CrossRef]
- Peña, J.M.R.; Cid, V.J.; Arroyo, J.; Nombela, C. A novel family of cell wall-related proteins regulated differently during the yeast life cycle. Mol. Cell. Biol. 2000, 20, 3245–3255. [Google Scholar] [CrossRef]
- Spreghini, E.; Davis, D.A.; Subaran, R.; Kim, M.; Mitchell, A.P. Roles of Candida albicans Dfg5p and Dcw1p Cell Surface Proteins in Growth and Hypha Formation. Eukaryot. Cell 2003, 2, 746–755. [Google Scholar] [CrossRef] [PubMed]
- Gelis, S.; de Groot, P.W.; Castillo, L.; Moragues, M.-D.; Sentandreu, R.; Gómez, M.-M.; Valentín, E. Pga13 in Candida albicans is localized in the cell wall and influences cell surface properties, morphogenesis and virulence. Fungal Genet. Biol. 2012, 49, 322–331. [Google Scholar] [CrossRef] [PubMed]
- Castillo, L.; Calvo, E.; Martínez, A.I.; Ruiz-Herrera, J.; Valentín, E.; Lopez, J.A.; Sentandreu, R. A study of the Candida albicans cell wall proteome. Proteomics 2008, 8, 3871–3881. [Google Scholar] [CrossRef] [PubMed]
- Childers, D.S.; Avelar, G.M.; Bain, J.M.; Pradhan, A.; Larcombe, D.E.; Netea, M.G.; Erwig, L.P.; Gow, N.A.R.; Brown, A.J.P. Epitope shaving promotes fungal immune evasion. mBio 2020, 11, e00984-20. [Google Scholar] [CrossRef] [PubMed]
- Ballou, E.R.; Avelar, G.M.; Childers, D.S.; Mackie, J.; Bain, J.M.; Wagener, J.; Kastora, S.L.; Panea, M.D.; Hardison, S.E.; Walker, L.A.; et al. Lactate signalling regulates fungal beta-glucan masking and immune evasion. Nat. Microbiol. 2016, 2, 16238. [Google Scholar] [CrossRef] [PubMed]
- Kruppa, M.; Greene, R.R.; Noss, I.; Lowman, D.W.; Williams, D.L. C. albicans increases cell wall mannoprotein, but not mannan, in response to blood, serum and cultivation at physiological temperature. Glycobiology 2011, 21, 1173–1180. [Google Scholar] [CrossRef] [PubMed]
- Sosinska, G.J.; De Koning, L.J.; De Groot, P.; Manders, E.M.M.; Dekker, H.L.; Hellingwerf, K.J.; De Koster, C.G.; Klis, F.M. Mass spectrometric quantification of the adaptations in the wall proteome of Candida albicans in response to ambient pH. Microbiology 2011, 157 Pt 1, 136–146. [Google Scholar] [CrossRef]
| Protein/Family | Features and Functions | Regulation |
|---|---|---|
| GPI modified CWPs Adhesins, invasins Als family (Als1-7, 9) | ||
| N-terminal c. 300-residue Ig domain, bind variety of substrates [27,28]; high (Als1, Als2), intermediate (Als4, Als9), and low (Als5-7) levels of gene expression [27,29,30,31]. Als3 is expressed uniformly all over hyphae [32]. Als1 and Als3 N terminal sequences are used as vaccine antigen [7]. Als1 and Als3 contribute to biofilm formation, and Als3 functions as an invasin, and as a ferritin receptor [27,30,31,33]. | Als proteins are differentially expressed, ALS1, ALS3, and HWP1 are under the positive regulatory control of Bcr1 [34,35,36]. Tup1 (repressor of filamentation] and Ahr1 are required for full expression of ALS3 [37]. | |
| Hwp1, Hwp2, Eap1, Ihd1, and Hyr1 | Hwp1 level is induced by oxygen and iron restriction [38]. N terminal is recognised as substrate for epithelial transglutaminases [39]. Hwp1 facilitates cell to cell interaction important in biofilm development [33]. N terminal 14-mer peptide and recombinant N terminal fragment are used in vaccine and diagnostic development, respectively [40,41]. Hwp2 has sequence identity with Hwp1 and can function in adhesion and invasion; it is also involved in oxidative stress tolerance and protein aggregation [42,43]. Hwp1, Hwp2, Eap1, and Ihd1 contribute to initial cell attachment and adhesion maintenance during biofilm formation [44]. N terminal of Hyr1 has been used in vaccines and diagnostics development [5,45]. | |
| Carbohydrate active enzymes | ||
| 1,3-β-Glucan processing Phr1-3, Pga4, and Pga5 | N terminal glycoside hydrolase (GH) 72 domain; play a role in cell wall construction (β-1,3-glucan modification); incorporated at acidic pH (Phr2) and neutral/alkaline pH (Phr1) [45,46]. Pga4 is transcribed independent of pH, and Phr3 and Pga5 have low expression levels [47]. Pga4 is serum- and host infection-inducible [48]. | PHR1 and PHR2 are differentially regulated by extracellular pH [49]. |
| Chitin-glucan cross-linkersChr family | N terminal GH16 domain; involved in cell wall organization and integrity; cross-linking β-1,3-glucan and chitin; involved in protoplast regeneration [50,51]. Control cell wall elasticity in osmotic resistance [52]. | UTR2 expression is regulated by calcineurin and Crz1 [53]. CRH11 is subject to caspofungin-induced Cas5 regulation [54] |
| Others | ||
| Dfg5 and Dcw1 | Putative glycosyltransferase enzyme activity; involved in the incorporation of GPI anchored proteins into the cell wall [55,56]. Dfg5 and Dcw1 are involved in hyphal morphogenesis and biofilm formation; Dfg5 is required for growth; Dcw1 is required for cell wall integrity response; Dfg5 has synthetic lethality with Dcw1 [56]. | DFG5 has been shown to be regulated by Rlm1 in S. cerevisaie, but not in C. albicans [54]. |
| Pga31-like (Pga29-31) | Enriched in pathogenic fungi [57]. Pga31 has predicted transmembrane domain and with Pga30 they have three conserved cysteine residues (http://www.candidagenome.org/ (accessed on 12 July 2021)). Pga29 and Pga31 are echinocandin induced; Pga29 is required for normal cell surface property [58]. Pga31 is induced during protoplast regeneration [59] and may be involved in cell wall chitin synthesis during remodelling in response to stress [60]. | PGA31 is upregulated by the Pkc pathway [60]. |
Sod4 and Sod5 | Superoxide dismutase; contribute to combating oxidative stress by clearing reactive oxygen species [61]. | Rim101 is required for induction of SOD5 under certain conditions, and Efg1 is required specifically for serum-modulated expression [62]. |
| Sap9 and Sap10 | Yapsin-like proteins are mainly found in the cell membrane (Sap9) and cell wall (Sap10); required for full cell wall integrity [63]. | |
| Pga59 | Cell wall localised [64]; abundant in the cell wall protein coat; mature protein consists of three cysteine residues and cross-links cell wall proteins through disulphide bridges [64]. | |
| Rbt5,Pga10, andPga7 | N terminal CFEM domain; cell membrane (Pga7 and Rbt5) localised; loss of function results in fragile biofilms (Pga10 and Rbt5) [65,66]; function as haeme receptors and involved in haeme-iron utilization [67,68]. Rbt5 levels increase following iron and oxygen restriction [38]. | The proteins have been shown to be expressed during yeast to hyphae switch and thus are regulated by Tup1 [66]. |
Non-GPI modified CWPs | ||
| Pir1 | C terminal conserved four cysteine pattern and seven repeats; predicted to cross-link β-1,3-glucan chains [69]; protein levels increase in hypoxic conditions [38]. | |
| Mp65 | C terminal GH17z domain; present in fibrillar material with putative transglycosylase activity; potential vaccine candidate [70]. |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ibe, C.; Munro, C.A. Fungal Cell Wall Proteins and Signaling Pathways Form a Cytoprotective Network to Combat Stresses. J. Fungi 2021, 7, 739. https://doi.org/10.3390/jof7090739
Ibe C, Munro CA. Fungal Cell Wall Proteins and Signaling Pathways Form a Cytoprotective Network to Combat Stresses. Journal of Fungi. 2021; 7(9):739. https://doi.org/10.3390/jof7090739
Chicago/Turabian StyleIbe, Chibuike, and Carol A. Munro. 2021. "Fungal Cell Wall Proteins and Signaling Pathways Form a Cytoprotective Network to Combat Stresses" Journal of Fungi 7, no. 9: 739. https://doi.org/10.3390/jof7090739
APA StyleIbe, C., & Munro, C. A. (2021). Fungal Cell Wall Proteins and Signaling Pathways Form a Cytoprotective Network to Combat Stresses. Journal of Fungi, 7(9), 739. https://doi.org/10.3390/jof7090739






