Analysis of Microbiota and Mycobiota in Fungal Ball Rhinosinusitis: Specific Interaction between Aspergillus fumigatus and Haemophilus influenza?
Abstract
:1. Introduction
2. Materials and Methods
2.1. Patients and Clinical Specimens
2.2. Methods
2.2.1. Direct and Histopathologic Examination and Microbiological Cultures
2.2.2. DNA Extraction Protocol for High Throughput Sequencing
2.2.3. Library Preparation and Sequencing
2.2.4. Taxonomic Assignment, Diversity
2.2.5. Statistical Analysis
3. Results
3.1. Patient Characteristics
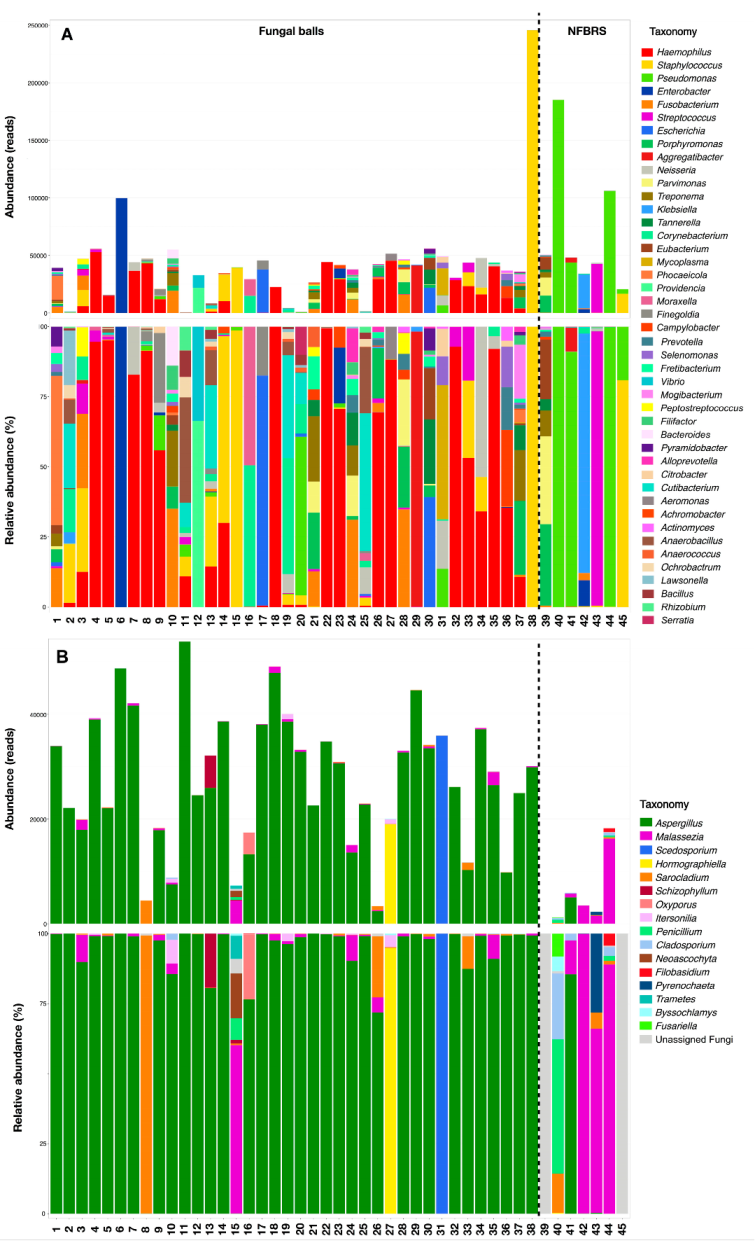
3.2. 16S, ITS1 and ITS2 Targeted Amplicon Sequencing Results
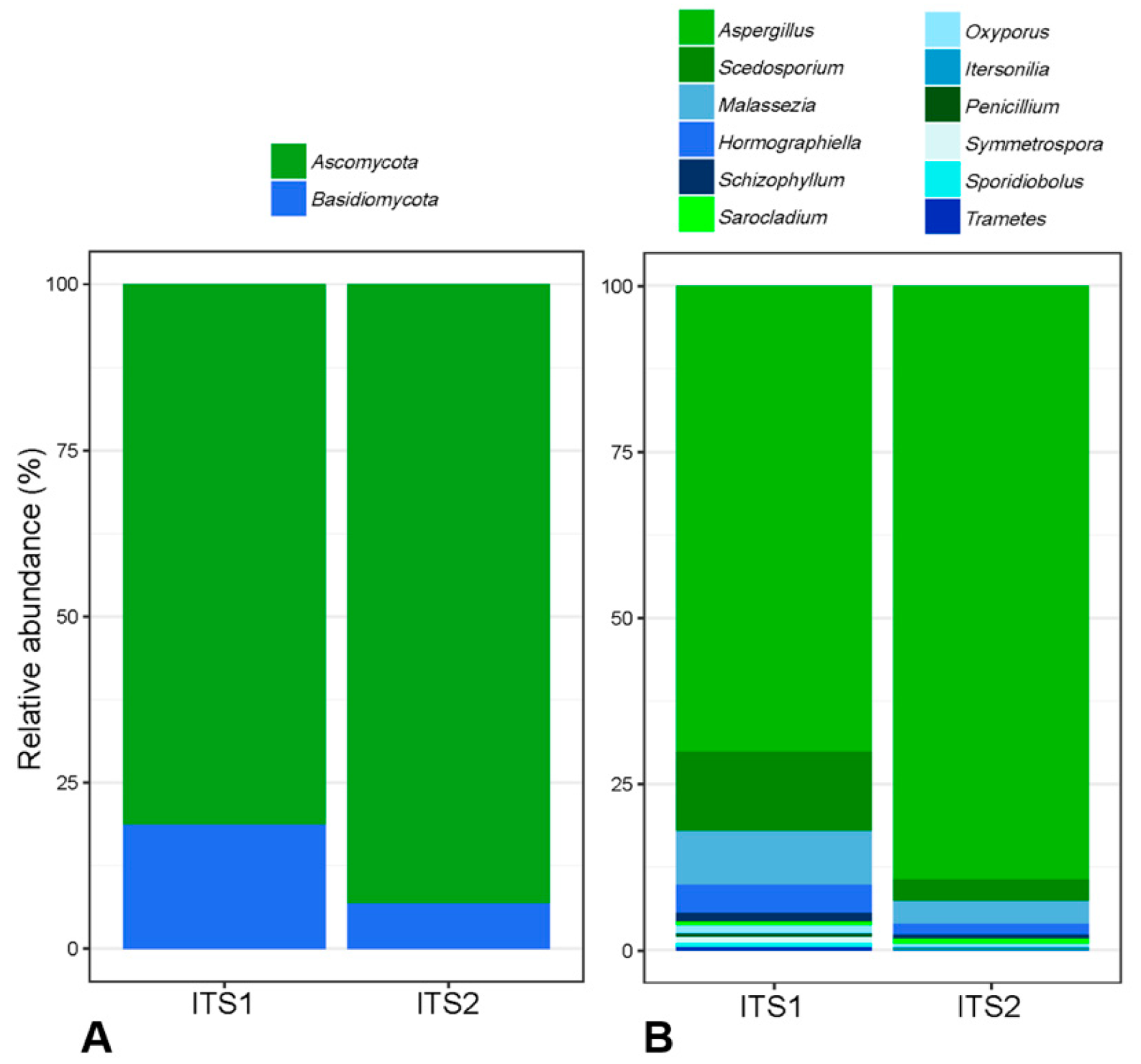
3.3. Comparison of ITS1 and ITS2 Regions for Mycobiota Analysis
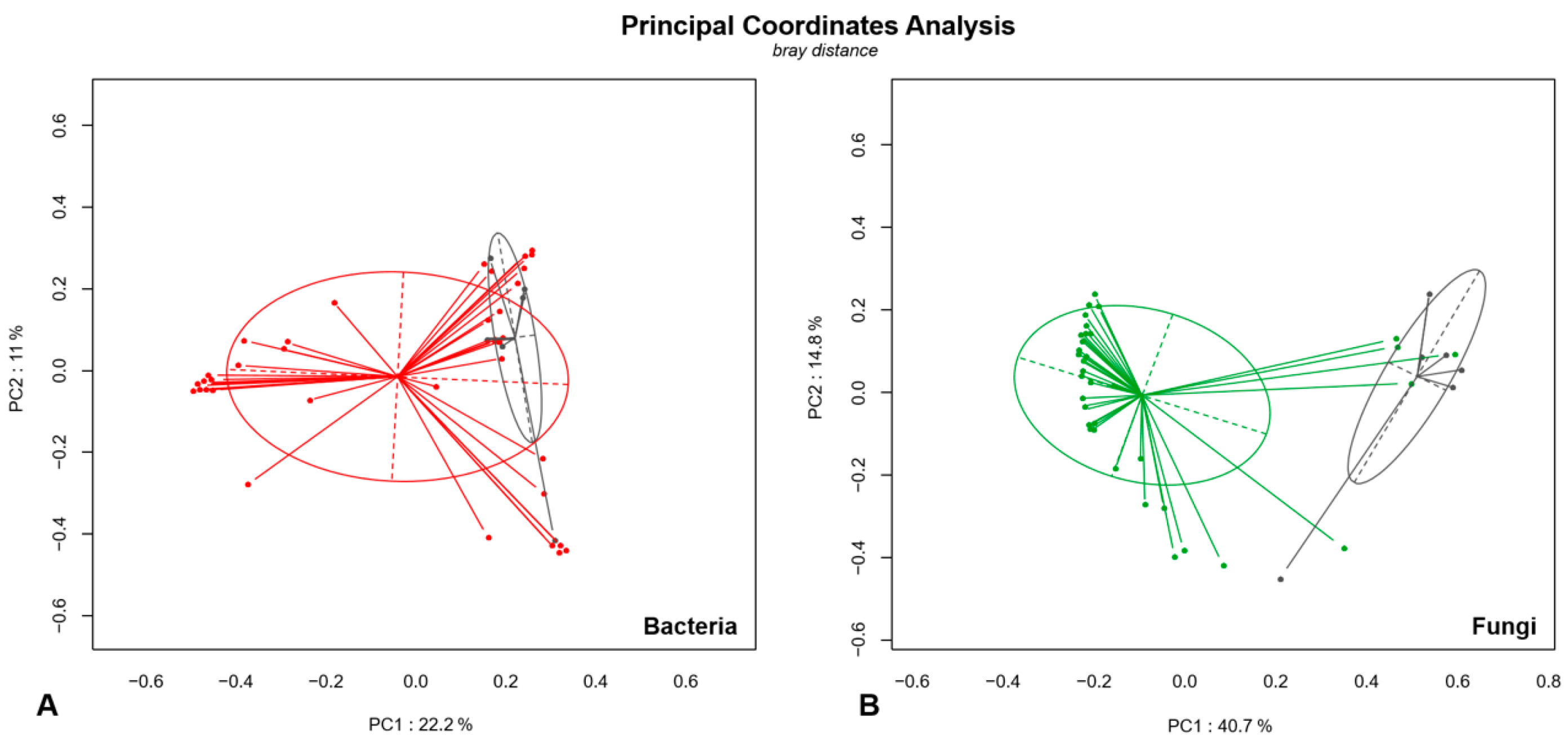
3.4. Analysis of Microbial Diversity in Fungal Ball (FB) and Non-Fungal Ball Chronic Rhinosinusitis (NFBRS)
4. Discussion
4.1. Targeting ITS1 or ITS2 to Study Fungal Diversity
4.2. Fungal Diversity in FBRS
4.3. Hypothesis to Explain the Poor Positive Culture Rate of FBRS Samples
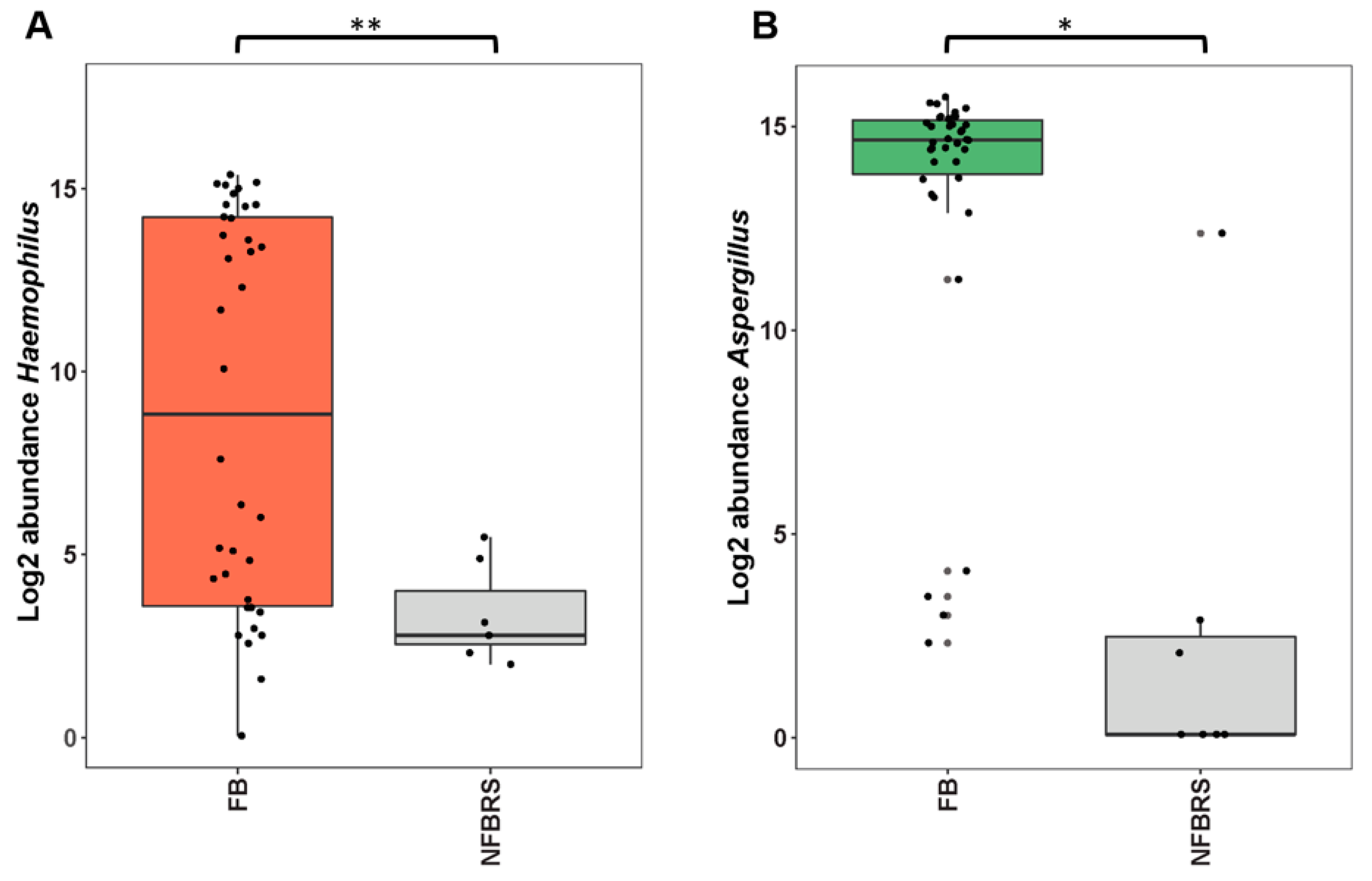
4.4. Could FBRS Result from Aspergillus–Haemophilus Microbial Interaction?
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Pleis, J.R.; Lucas, J.W.; Ward, B.W. Summary Health Statistics for U.S. Adults: National Health Interview Survey; Vital and Health statistics. Series 10; Centers for Disease Control and Prevention: Atlanta, GA, USA, 2009; pp. 1–161.
- Fokkens, W.J.; Lund, V.J.; Hopkins, C.; Hellings, P.W.; Kern, R.; Reitsma, S.; Toppila-Salmi, S.; Bernal-Sprekelsen, M.; Mullol, J.; Alobid, I.; et al. European Position Paper on Rhinosinusitis and Nasal Polyps. Rhinology 2020, 58, 1–464. [Google Scholar] [CrossRef]
- Poddighe, D.; Brambilla, I.; Licari, A.; Marseglia, G.L. Pediatric rhinosinusitis and asthma. Respir. Med. 2018, 141, 94–99. [Google Scholar] [CrossRef]
- Seybt, M.W.; McMains, K.C.; Kountakis, S.E. The prevalence and effect of asthma on adults with chronic rhinosinusitis. Ear Nose Throat J. 2007, 86, 409–411. [Google Scholar] [CrossRef] [Green Version]
- Turner, J.H.; Soudry, E.; Nayak, J.V.; Hwang, P.H. Survival outcomes in acute invasive fungal sinusitis: A systematic review and quantitative synthesis of published evidence. Laryngoscope 2013, 123, 1112–1118. [Google Scholar] [CrossRef]
- Mensi, M.; Piccioni, M.; Marsili, F.; Nicolai, P.; Sapelli, P.L.; Latronico, N. Risk of maxillary fungus ball in patients with endodontic treatment on maxillary teeth: A case-control study. Oral Surg. Oral Med. Oral Pathol. Oral Radiol. 2007, 103, 433–436. [Google Scholar] [CrossRef]
- Raz, E.; Win, W.; Hagiwara, M.; Lui, Y.W.; Cohen, B.; Fatterpekar, G.M. Fungal Sinusitis. Neuroimaging Clin. N. Am. 2015, 25, 569–576. [Google Scholar] [CrossRef]
- Cabaret, O.; Toussain, G.; Abermil, N.; Alsamad, I.A.; Botterel, F.; Costa, J.-M.; Papon, J.-F.; Bretagne, S. Degradation of fungal DNA in formalin-fixed paraffin-embedded sinus fungal balls hampers reliable sequence-based identification of fungi. Med. Mycol. 2011, 49, 329–332. [Google Scholar] [CrossRef] [Green Version]
- Pagella, F.; Matti, E.; Bernardi, F.D.; Semino, L.; Cavanna, C.; Marone, P.; Farina, C.; Castelnuovo, P. Paranasal sinus fungus ball: Diagnosis and management. Mycoses 2007, 50, 451–456. [Google Scholar] [CrossRef] [PubMed]
- Morio, F.; Dannaoui, E.; Chouaki, T.; Cateau, E.; Malard, O.; Bonfils, P.; Page, C.; Dufour, X.; Cottrel, C.; Erwan, T.; et al. PCR-based detection of Aspergillus fumigatus and absence of azole resistance due to TR34/L98H in a french multicenter cohort of 137 patients with fungal rhinosinusitis. Mycoses 2018, 61, 30–34. [Google Scholar] [CrossRef] [PubMed]
- Cui, L.; Morris, A.; Ghedin, E. The human mycobiome in health and disease. Genome Med. BioMed. Central 2013, 5, 63. [Google Scholar] [CrossRef] [Green Version]
- Kumpitsch, C.; Koskinen, K.; Schöpf, V.; Moissl-Eichinger, C. The microbiome of the upper respiratory tract in health and disease. BMC Biol. 2019, 17, 87. [Google Scholar] [CrossRef] [Green Version]
- Tiew, P.Y.; Mac Aogain, M.; Ali, N.A.B.M.; Thng, K.X.; Goh, K.; Lau, K.J.X.; Chotirmall, S.H. The Mycobiome in Health and Disease: Emerging Concepts, Methodologies and Challenges. Mycopathologia 2020, 185, 207–231. [Google Scholar] [CrossRef]
- Cleland, E.J.; Bassioni, A.; Boase, S.; Dowd, S.; Vreugde, S.; Wormald, P.-J. The fungal microbiome in chronic rhinosinusitis: Richness, diversity, postoperative changes and patient outcomes. Int. Forum. Allergy Rhinology. 2014, 4, 259–265. [Google Scholar] [CrossRef]
- Zhao, Y.C.; Bassiouni, A.; Tanjararak, K.; Vreugde, S.; Wormald, P.-J.; Psaltis, A.J. Role of fungi in chronic rhinosinusitis through ITS sequencing. Laryngoscope 2017, 128, 16–22. [Google Scholar] [CrossRef] [PubMed]
- Jung, W.H.; Croll, D.; Cho, J.H.; Kim, Y.R.; Lee, Y.W. Analysis of the nasal vestibule mycobiome in patients with allergic rhinitis. Mycoses 2015, 58, 167–172. [Google Scholar] [CrossRef] [PubMed]
- Healy, D.Y.; Leid, J.G.; Sanderson, A.R.; Hunsaker, D.H. Biofilms with fungi in chronic rhinosinusitis. Otolaryngol. Head Neck Surg. 2008, 138, 641–647. [Google Scholar] [CrossRef]
- Klindworth, A.; Pruesse, E.; Schweer, T.; Peplies, J.; Quast, C.; Horn, M.; Glöckner, F.O. Evaluation of general 16S ribosomal RNA gene PCR primers for classical and next-generation sequencing-based diversity studies. Nucleic Acids Res. 2013, 41, e1. [Google Scholar] [CrossRef]
- White, T.J.; Bruns, T.; Lee, S.; Taylor, J. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In PCR Protocols a Guide to Methods and Applications; Academic Press: Cambridge, MA, USA, 1990; pp. 315–322. [Google Scholar]
- Angebault, C.; Payen, M.; Woerther, P.-L.; Rodriguez, C.; Botterel, F. Combined bacterial and fungal targeted amplicon sequencing of respiratory samples: Does the DNA extraction method matter? PLoS ONE 2020, 15, e0232215. [Google Scholar] [CrossRef] [PubMed]
- Bokulich, N.A.; Subramanian, S.; Faith, J.J.; Gevers, D.; Gordon, J.I.; Knight, R.; Mills, D.A.; Caporaso, J.G. Quality-filtering vastly improves diversity estimates from Illumina amplicon sequencing. Nat. Methods 2013, 10, 57–59. [Google Scholar] [CrossRef]
- Bolyen, E.; Rideout, J.R.; Dillon, M.R.; Bokulich, N.A.; Abnet, C.C.; Al-Ghalith, G.A.; Alexander, H.; Alm, E.J.; Arumugam, M.; Asnicar, F.; et al. Reproducible, interactive, scalable and extensible microbiome data science using QIIME. Nat. Biotechnol. 2019, 37, 852–857. [Google Scholar] [CrossRef]
- Amir, A.; McDonald, D.; Navas-Molina, J.A.; Kopylova, E.; Morton, J.T.; Xu, Z.; Kightley, E.P.; Thompson, L.R.; Hyde, E.R.; Gonzalez, A.; et al. Deblur Rapidly Resolves Single-Nucleotide Community Sequence Patterns. mSystems 2017, 2, e00191-16. [Google Scholar] [CrossRef] [Green Version]
- Quast, C.; Pruesse, E.; Yilmaz, P.; Gerken, J.; Schweer, T.; Yarza, P.; Peplies, J.; Glöckner, F.O. The SILVA ribosomal RNA gene database project: Improved data processing and web-based tools. Nucleic Acids Res. 2012, 41, D590–D596. [Google Scholar] [CrossRef]
- Irinyi, L.; Serena, C.; Garcia-Hermoso, D.; Arabatzis, M.; Desnos-Ollivier, M.; Vu, D.; Cardinali, G.; Arthur, I.; Normand, A.C.; Giraldo, A.; et al. International Society of Human and Animal Mycology (ISHAM)-ITS reference DNA barcoding database--the quality controlled standard tool for routine identification of human and animal pathogenic fungi. Med. Mycol. 2015, 53, 313–337. [Google Scholar] [CrossRef]
- Rognes, T.; Flouri, T.; Nichols, B.; Quince, C.; Mahé, F. VSEARCH: A versatile open source tool for metagenomics. PeerJ 2016, 4, e2584-22. [Google Scholar] [CrossRef]
- Robert, V.; Vu, D.; Amor, A.B.H.; Van de Wiele, N.; Brouwer, C.; Jabas, B.; Szoke, S.; Dridi, A.; Triki, M.; Daoud, S.B.; et al. MycoBank gearing up for new horizons. IMA Fungus 2013, 4, 371–379. [Google Scholar] [CrossRef] [PubMed]
- DeSantis, T.Z.; Hugenholtz, P.; Larsen, N.; Rojas, M.; Brodie, E.L.; Keller, K.; Huber, T.; Dalevi, D.; Hu, P.; Andersen, G.L. Greengenes, a Chimera-Checked 16S rRNA Gene Database and Workbench Compatible with ARB. Appl. Environ. Microbiol. 2006, 72, 5069–5072. [Google Scholar] [CrossRef] [Green Version]
- Cole, J.R.; Wang, Q.; Fish, J.A.; Chai, B.; McGarrell, D.M.; Sun, Y.; Brown, C.T.; Porras-Alfaro, A.; Kuske, C.R.; Tiedje, J.M. Ribosomal Database Project: Data and tools for high throughput rRNA analysis. Nucleic Acids Res. 2013, 42, D633–D642. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Park, S.-C.; Won, S. Evaluation of 16S rRNA Databases for Taxonomic Assignments Using a Mock Community. Genom. Inf. 2018, 16, e24. [Google Scholar] [CrossRef] [Green Version]
- Yarza, P.; Yilmaz, P.; Pruesse, E.; Glöckner, F.O.; Ludwig, W.; Schleifer, K.-H.; Whitman, W.B.; Euzéby, J.; Amann, R.; Rosselló-Móra, R. Uniting the classification of cultured and uncultured bacteria and archaea using 16S rRNA gene sequences. Nat. Biotechnol. 2014, 12, 635–645. [Google Scholar] [CrossRef]
- Volant, S.; Lechat, P.; Woringer, P.; Motreff, L.; Campagne, P.; Malabat, C.; Kennedy, S.; Ghozlane, A. SHAMAN: A user-friendly website for metataxonomic analysis from raw reads to statistical analysis. BMC Bioinform. 2020, 21, 345. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Quereda, J.J.; Dussurget, O.; Nahori, M.-A.; Ghozlane, A.; Volant, S.; Dillies, M.-A.; Regnault, B.; Kennedy, S.; Mondot, S.; Villoing, B.; et al. Bacteriocin from epidemic Listeriastrains alters the host intestinal microbiota to favor infection. Proc. Natl. Acad. Sci. USA 2016, 113, 5706–5711. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Benjamini, Y.; Hochberg, Y. Controlling the false discovery rate: A practical and powerful approach to multiple testing. J. R. Stat. Soc. 1995, 57, 289–300. [Google Scholar] [CrossRef]
- Nilsson, R.H.; Anslan, S.; Bahram, M.; Wurzbacher, C.; Baldrian, P.; Tedersoo, L. Mycobiome diversity: High-throughput sequencing and identification of fungi. Nat. Biotechnol. 2019, 17, 95–109. [Google Scholar] [CrossRef]
- NIH HMP Working Group; Peterson, J.; Garges, S.; Giovanni, M.; McInnes, P.; Wang, L.; Schloss, J.A.; Bonazzi, V.; McEwen, J.E.; Wetterstrand, K.A.; et al. The NIH Human Microbiome Project. Genome Res. 2009, 19, 2317–2323. [Google Scholar]
- Alanio, A.; Bretagne, S. Difficulties with molecular diagnostic tests for mould and yeast infections: Where do we stand? Clin. Microbiol. Infect. 2014, 20, 36–41. [Google Scholar] [CrossRef] [Green Version]
- Schoch, C.L.; Seifert, K.A.; Huhndorf, S.; Robert, V.; Spouge, J.L.; Levesque, C.A.; Chen, W.; Fungal Barcoding Consortium. Fungal Barcoding Consortium Author List Nuclear ribosomal internal transcribed spacer (ITS) region as a universal DNA barcode marker for Fungi. Proc. Natl. Acad. Sci. USA 2012, 109, 6241–6246. [Google Scholar] [CrossRef] [Green Version]
- Bazzicalupo, A.L.; Bálint, M.; Schmitt, I. Comparison of ITS1 and ITS2 rDNA in 454 sequencing of hyperdiverse fungal communities. Fungal Ecol. 2012, 6, 102–109. [Google Scholar] [CrossRef]
- Blaalid, R.; Kumar, S.; Nilsson, R.H.; Abarenkov, K.; Kirk, P.M.; Kauserud, H. ITS1 versus ITS2 as DNA metabarcodes for fungi. Mol. Ecol. Resour. 2013, 13, 218–224. [Google Scholar] [CrossRef] [Green Version]
- Monard, C.; Gantner, S.; Stenlid, J. Utilizing ITS1 and ITS2 to study environmental fungal diversity using pyrosequencing. FEMS Microbiol. Ecol. 2012, 84, 165–175. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, X.-C.; Liu, C.; Huang, L.; Bengtsson-Palme, J.; Chen, H.; Zhang, J.-H.; Cai, D.; Li, J.-Q. ITS1: A DNA barcode better than ITS2 in eukaryotes? Mol. Ecol. Resour. 2014, 15, 573–586. [Google Scholar] [CrossRef]
- Yang, R.-H.; Su, J.-H.; Shang, J.-J.; Wu, Y.-Y.; Li, Y.; Bao, D.-P.; Yao, Y.-J. Evaluation of the ribosomal DNA internal transcribed spacer (ITS), specifically ITS1 and ITS2, for the analysis of fungal diversity by deep sequencing. PLoS ONE 2018, 13, e0206428. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nilsson, R.H.; Kristiansson, E.; Ryberg, M.; Hallenberg, N.; Larsson, K.-H. Intraspecific ITS variability in the kingdom fungi as expressed in the international sequence databases and its implications for molecular species identification. Evol. Bioinform. Online 2008, 4, 193–201. [Google Scholar] [CrossRef] [PubMed]
- Lowe, R.; Shirley, N.; Bleackley, M.; Dolan, S.; Shafee, T. Transcriptomics technologies. PLoS Comput. Biol. 2017, 13, e1005457. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lusk, R.W. Diverse and Widespread Contamination Evident in the Unmapped Depths of High Throughput Sequencing Data. PLoS ONE 2014, 9, e110808. [Google Scholar] [CrossRef] [Green Version]
- Zhang, J.; Li, Y.; Lu, X.; Wang, X.; Zang, H.; Wang, T.; Zhou, B.; Zhang, L. Analysis of fungal ball rhinosinusitis by culturing fungal clumps under endoscopic surgery. Int. J. Clin. Exp. Med. 2015, 8, 5925–5930. [Google Scholar]
- Vickery, T.W.; Ramakrishnan, V.R.; Suh, J.D. The Role of Staphylococcus aureus in Patients with Chronic Sinusitis and Nasal Polyposis. Curr. Allergy Asthma Rep. 2019, 19, 1–8. [Google Scholar] [CrossRef]
- Poddighe, D.; Vangelista, L. Staphylococcus aureus Infection and Persistence in Chronic Rhinosinusitis: Focus on Leukocidin ED. Toxins 2020, 12, 678. [Google Scholar] [CrossRef]
- Ramage, G.; Rajendran, R.; Gutierrez-Correa, M.; Jones, B.; Williams, C. Aspergillus biofilms: Clinical and industrial significance. FEMS Microbiol Lett. 2011, 324, 89–97. [Google Scholar] [CrossRef] [Green Version]
- Flemming, H.-C.; Wingender, J.; Szewzyk, U.; Steinberg, P.; Rice, S.A.; Kjelleberg, S. Biofilms: An emergent form of bacterial life. Nat. Biotechnol. 2016, 14, 563–575. [Google Scholar] [CrossRef]
- Hommel, B.; Sturny-Leclere, A.; Volant, S.; Veluppillai, N.; Duchateau, M.; Yu, C.-H.; Hourdel, V.; Varet, H.; Matondo, M.; Perfect, J.R. Cryptococcus neoformans resists to drastic conditions by switching to viable but non-culturable cell phenotype. PLoS Pathogens 2019, 15, e1007945. [Google Scholar]
- Salma, M.; Rousseaux, S.; Sequeira-Le Grand, A.; Divol, B.; Alexandre, H. Characterization of the Viable but Nonculturable (VBNC) State in Saccharomyces cerevisiae. PLoS ONE 2013, 8, e77600. [Google Scholar] [CrossRef] [PubMed]
- Loussert, C.; Schmitt, C.; Prevost, M.-C.; Balloy, V.; Fadel, E.; Philippe, B.; Kauffmann-Lacroix, C.; Latgé, J.P.; Beauvais, A. In vivo biofilm composition of Aspergillus fumigatus. Cell Microbiol. 2010, 12, 405–410. [Google Scholar] [CrossRef] [PubMed]
- Morisse, H.; Heyman, L.; Salaün, M.; Favennec, L.; Picquenot, J.M.; Bohn, P.; Thiberville, L. In vivo molecular microimaging of pulmonary aspergillosis. Med. Mycol. 2013, 51, 352–360. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Melloul, E.; Roisin, L.; Durieux, M.-F.; Woerther, P.-L.; Jenot, D.; Risco, V.; Guillot, J.; Dannaoui, E.; Decousser, J.-W.; Botterel, F. Interactions of Aspergillus fumigatus and Stenotrophomonas maltophilia in an in vitro Mixed Biofilm Model: Does the Strain Matter? Front. Microbiol. 2018, 9, 2850. [Google Scholar] [CrossRef] [Green Version]
- Melloul, E.; Luiggi, S.; Anaïs, L.; Arné, P.; Costa, J.M.; Fihman, V.; Briard, B.; Dannaoui, E.; Guillot, J.; Decousser, J.W.; et al. Characteristics of Aspergillus fumigatus in Association with Stenotrophomonas maltophilia in an In Vitro Model of Mixed Biofilm. PLoS ONE 2016, 11, e0166325-18. [Google Scholar] [CrossRef] [PubMed]
- Roisin, L.; Melloul, E.; Woerther, P.-L.; Royer, G.; Decousser, J.-W.; Guillot, J.; Dannaoui, E.; Botterel, F. Modulated Response of Aspergillus fumigatus and Stenotrophomonas maltophilia to Antimicrobial Agents in Polymicrobial Biofilm. Front. Cell Infect. Microbiol. 2020, 10, CD009249-14. [Google Scholar] [CrossRef]
- Seidler, M.J.; Salvenmoser, S.; Muller, F.M.C. Aspergillus fumigatus Forms Biofilms with Reduced Antifungal Drug Susceptibility on Bronchial Epithelial Cells. Antimicrob. Agents Chemother. 2008, 52, 4130–4136. [Google Scholar] [CrossRef] [Green Version]
- Lee, M.J.; Liu, H.; Barker, B.M.; Snarr, B.D.; Gravelat, F.N.; Al Abdallah, Q.; Gavino, C.; Baistrocchi, S.R.; Ostapska, H.; Xiao, T. The Fungal Exopolysaccharide Galactosaminogalactan Mediates Virulence by Enhancing Resistance to Neutrophil Extracellular Traps. PLoS Pathogens 2015, 11, e1005187. [Google Scholar] [CrossRef]
- Van Eldere, J.; Slack, M.P.; Ladhani, S.; Cripps, A.W. Non-typeable Haemophilus influenzae, an under-recognised pathogen. Lancet Infect. Dis. 2014, 14, 1281–1292. [Google Scholar] [CrossRef] [Green Version]
- Wagner Mackenzie, B.; Chang, K.; Zoing, M.; Jain, R.; Hoggard, M.; Biswas, K.; Douglas, R.G.; Taylor, M.W. Longitudinal study of the bacterial and fungal microbiota in the human sinuses reveals seasonal and annual changes in diversity. Sci. Rep. 2019, 9, 17416. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bailey, K.L.; LeVan, T.D.; Yanov, D.A.; Pavlik, J.A.; DeVasure, J.M.; Sisson, J.H.; Wyatt, T.A. Non-typeable Haemophilus influenzae decreases cilia beating via protein kinase C epsilon. Respir. Res. 2012, 13, 49. [Google Scholar] [CrossRef] [Green Version]
- Zhang, J.; He, S.; Li, Y.; Lv, M.; Wei, H.; Qu, B.; Zheng, Y.; Hu, C. Distinguishing the dominant species of pathogen in ethmoidal sinusitis by sequencing DNA dataset analysis. Exp. Ther. Med. 2018, 4207–4212. [Google Scholar] [CrossRef] [PubMed]
- Boase, S.; Jervis-Bardy, J.; Cleland, E.; Pant, H.; Tan, L.; Wormald, P.-J. Bacterial-induced epithelial damage promotes fungal biofilm formation in a sheep model of sinusitis. Int. Forum. Allergy Rhinol. 2013, 3, 341–348. [Google Scholar] [CrossRef]
- Wajima, T.; Anzai, Y.; Yamada, T.; Ikoshi, H.; Noguchi, N. Oldenlandia diffusa Extract Inhibits Biofilm Formation by Haemophilus influenzae Clinical Isolates. PLoS ONE 2016, 11, e0167335. [Google Scholar] [CrossRef]
| FB | NFBRS | p | |
|---|---|---|---|
| Age (+/− SD) (years) | 60 (±13) | 55 (±14) | 0.45 |
| Sex ratio H/F | 0.58 | 0.75 | 0.99 |
| Typeofsinusitisn (%) | 0.18 | ||
| Maxillary | 24 (63.2) | 3 (42.9) | |
| Frontal | 1 (2.6) | 1 (14.3) | |
| Ethmoidal | 1 (2.6) | 1 (14.3) | |
| Sphenoidal | 7 (18.4) | 0 (0) | |
| Pansinusitis | 5 (13.2) | 2 (28.6) | |
| Immunocompromised status | 2 (5.1) | 1 (14.3) | 0.41 |
| Mycology | |||
| Positive direct examination | 37 (97.3) | 0 (0) | <0.001 |
| Positive culture | 12 (31.6) | 0 (0) | 0.16 |
| Bacteriology | |||
| Culture performed | 14 (36.8) | 7 (100) | <0.001 |
| Histology | |||
| Performed | 26 (68.4) | 5 (71.4) | - |
| Hyphae observed | 20 (52.6) | 0 (0) | - |
| ITS1 | ITS2 | p | ||
|---|---|---|---|---|
| Before trimming | Minimum reads/sample | 47,418 | 26,384 | |
| Maximum reads/sample | 76,196 | 67,095 | ||
| Mean reads/sample | 129,894 | 131,564 | NS | |
| After trimming | Minimum reads/sample | 14 | 2 | |
| Maximum reads/sample | 35,815 | 60,335 | ||
| Mean reads/sample | 5967 | 25,374 | <0.001 | |
| >5000 reads/FB sample | 26.3% (10/38) | 97.4% (37/38) | ||
| OTUs produced | 113 | 157 | NS | |
| Mean OTUs/sample | 6 | 9 | ||
| Taxa (genus level) | 48 | 57 | --- |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dellière, S.; Dannaoui, E.; Fieux, M.; Bonfils, P.; Gricourt, G.; Demontant, V.; Podglajen, I.; Woerther, P.-L.; Angebault, C.; Botterel, F. Analysis of Microbiota and Mycobiota in Fungal Ball Rhinosinusitis: Specific Interaction between Aspergillus fumigatus and Haemophilus influenza? J. Fungi 2021, 7, 550. https://doi.org/10.3390/jof7070550
Dellière S, Dannaoui E, Fieux M, Bonfils P, Gricourt G, Demontant V, Podglajen I, Woerther P-L, Angebault C, Botterel F. Analysis of Microbiota and Mycobiota in Fungal Ball Rhinosinusitis: Specific Interaction between Aspergillus fumigatus and Haemophilus influenza? Journal of Fungi. 2021; 7(7):550. https://doi.org/10.3390/jof7070550
Chicago/Turabian StyleDellière, Sarah, Eric Dannaoui, Maxime Fieux, Pierre Bonfils, Guillaume Gricourt, Vanessa Demontant, Isabelle Podglajen, Paul-Louis Woerther, Cécile Angebault, and Françoise Botterel. 2021. "Analysis of Microbiota and Mycobiota in Fungal Ball Rhinosinusitis: Specific Interaction between Aspergillus fumigatus and Haemophilus influenza?" Journal of Fungi 7, no. 7: 550. https://doi.org/10.3390/jof7070550
APA StyleDellière, S., Dannaoui, E., Fieux, M., Bonfils, P., Gricourt, G., Demontant, V., Podglajen, I., Woerther, P.-L., Angebault, C., & Botterel, F. (2021). Analysis of Microbiota and Mycobiota in Fungal Ball Rhinosinusitis: Specific Interaction between Aspergillus fumigatus and Haemophilus influenza? Journal of Fungi, 7(7), 550. https://doi.org/10.3390/jof7070550







