MFS1, a Pleiotropic Transporter in Dermatophytes That Plays a Key Role in Their Intrinsic Resistance to Chloramphenicol and Fluconazole
Abstract
:1. Introduction
2. Materials and Methods
2.1. Strains and Growth Media
2.2. Chemicals
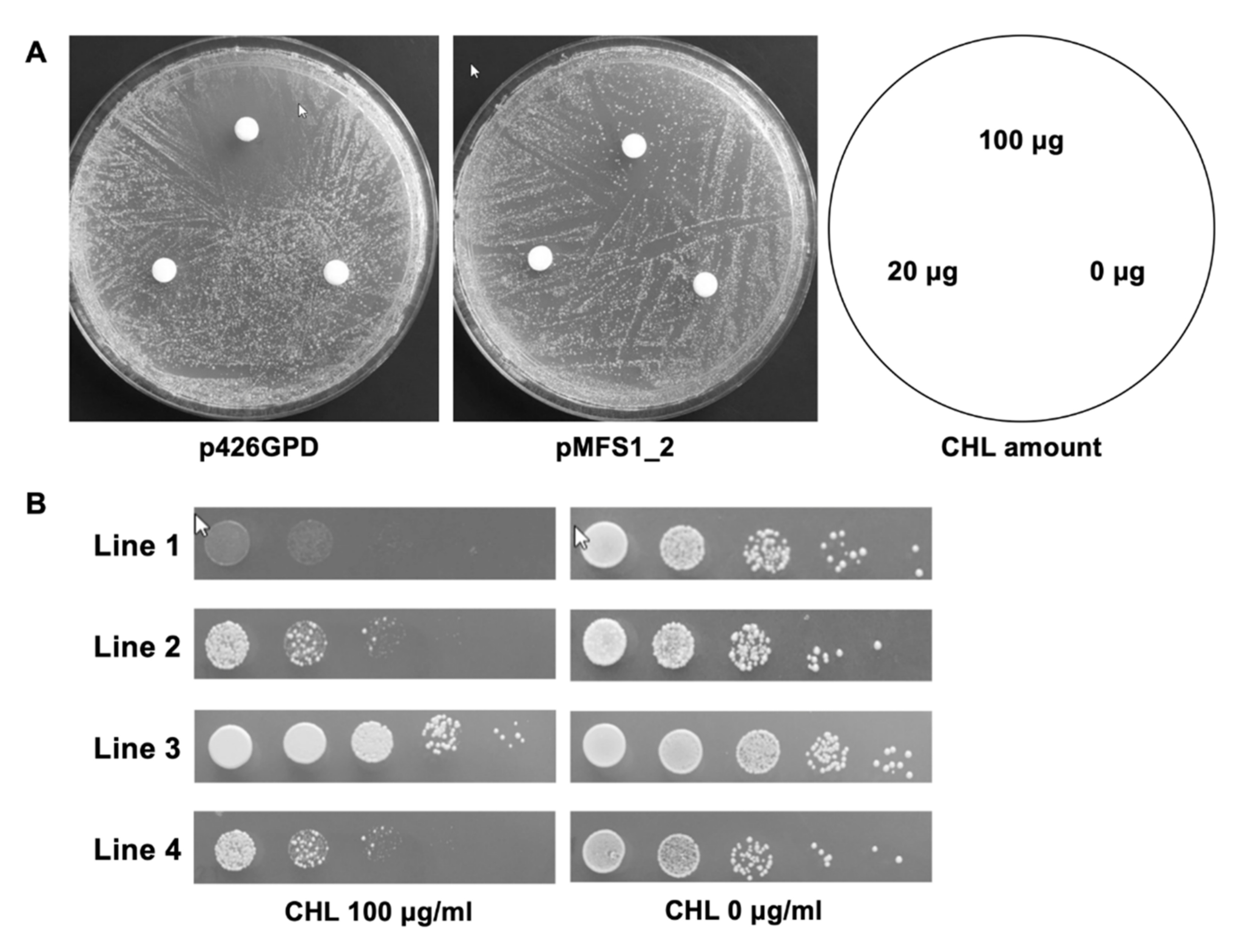
2.3. TruMFS1 Heterologous Expression in S. cerevisiae and CHL Susceptibility Assays
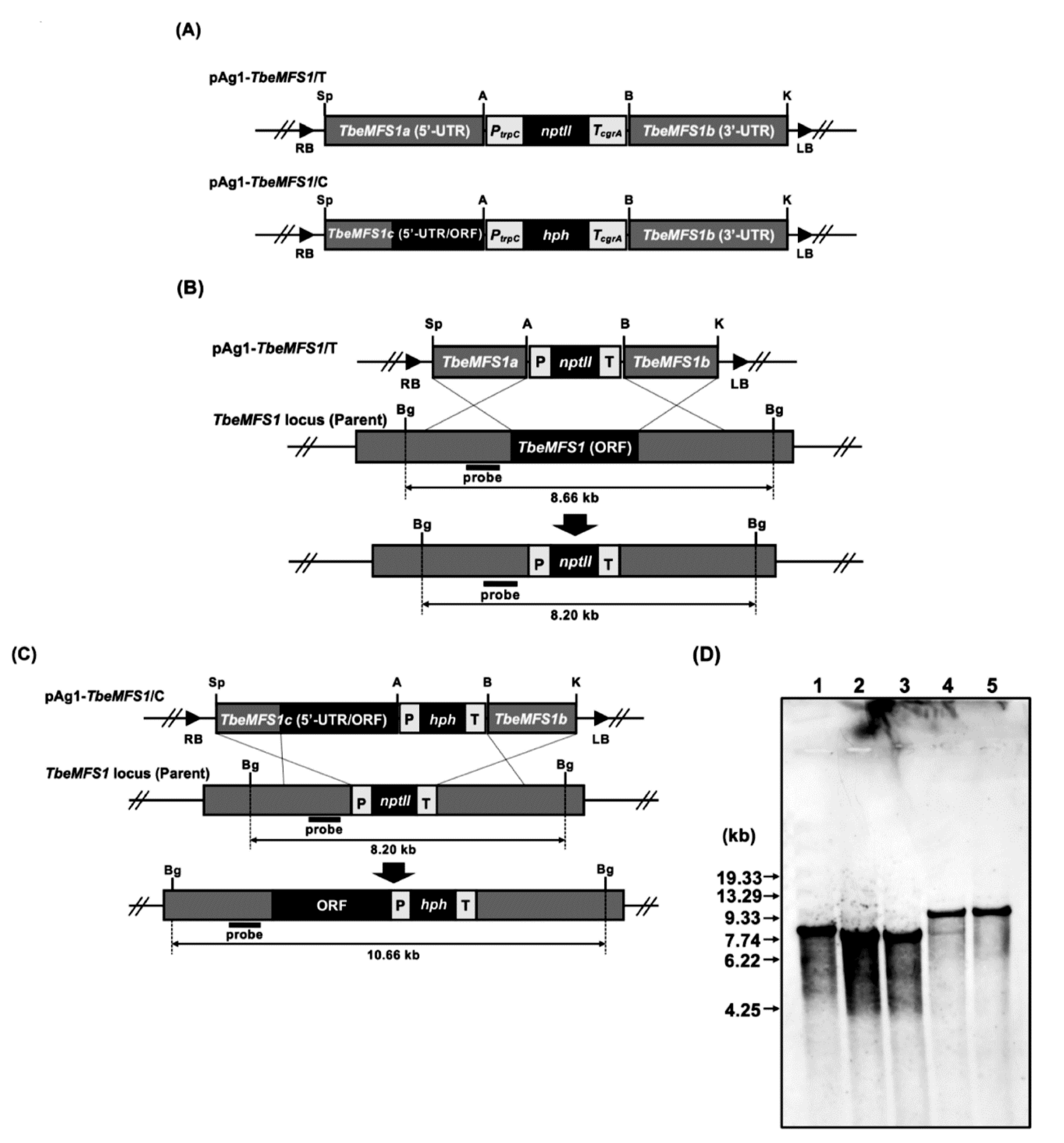
2.4. Construction of Gene Replacement Vectors for Targeted-Gene Disruption
2.5. Fungal Genetic Transformation
2.6. Screening of the Desired Transformants
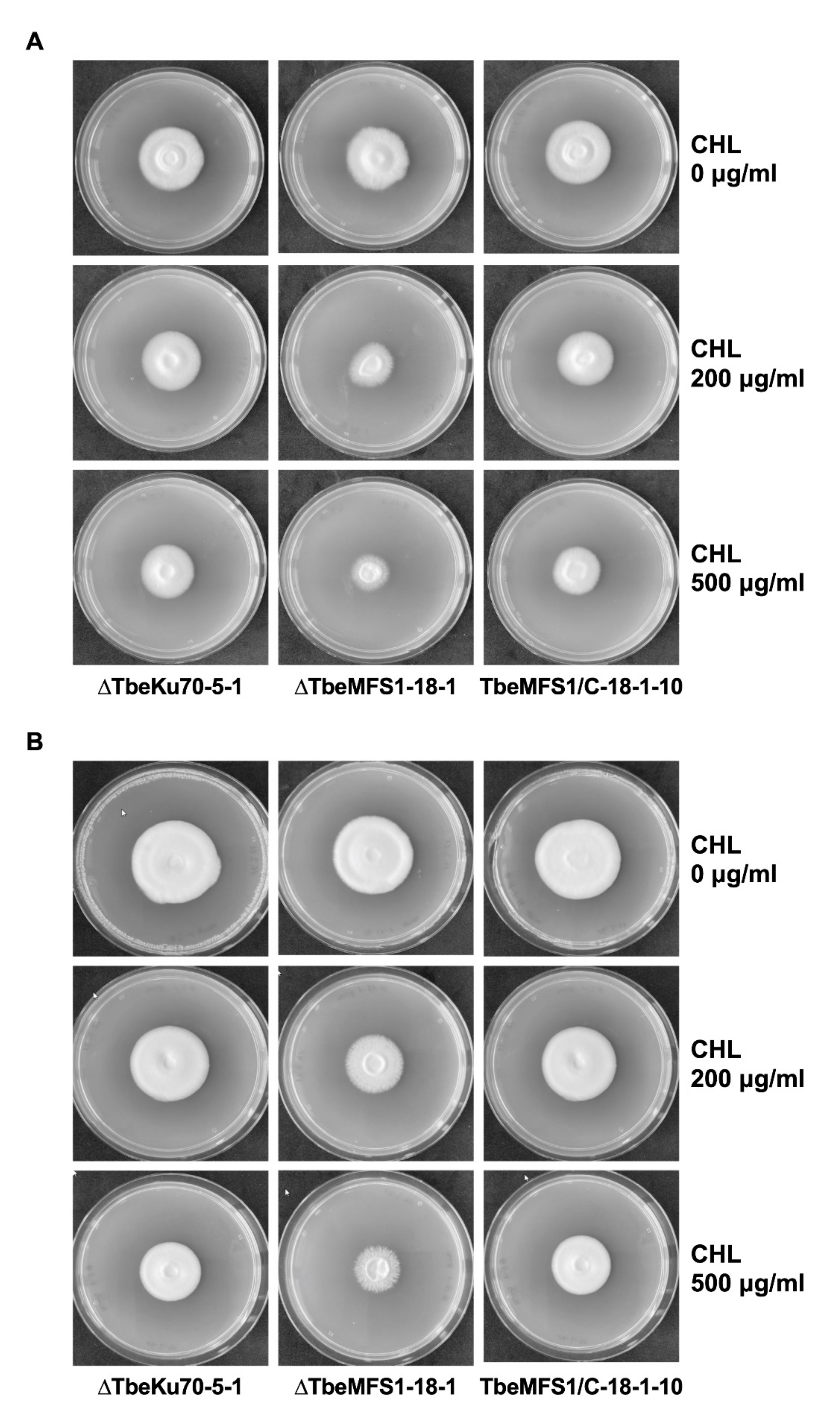
2.7. Drug Susceptibility Testing of T. benhamiae TbeMFS1-Lacking Mutants
2.8. Search for Additional T. rubrum Potential CYH-Resistance Genes by Heterologous Expression in S. cerevisiae
2.9. Nucleotide Sequence Accession Numbers
3. Results
3.1. MFS1 Characterization in T. rubrum and T. benhamiae
3.2. TruMFS1 Operates as a CHL Efflux Pump in S. cerevisiae
3.3. MFS1 Plays a Major Role in Tolerance of T. benhamiae to CHL but Not to CYH
3.4. MFS1 Is Involved in Resistance to Azole Compounds but Not in CYH Resistance in T. benhamiae
3.5. Search for Additional T. rubrum Potential CYH-Resistance Genes in S. cerevisiae
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Kwon-Chung, K.J.; Bennett, J.E. Medical Mycology; Lea and Febiger: Philadelphia, PA, USA, 1992. [Google Scholar]
- Weitzman, I.; Summerbell, R.C. The dermatophytes. Clin. Microbiol. Rev. 1995, 8, 240–259. [Google Scholar] [CrossRef] [PubMed]
- Baran, R.; Hay, R.; Haneke, E.; Tosti, A. Onychomycosis, the Current Approach to Diagnosis and Therapy, 2nd ed.; Taylor and Francis: Abington, UK, 2006; pp. 1–145. [Google Scholar]
- Barnhill, A.E.; Brewer, M.T.; Carlson, S.A. Adverse effects of antimicrobials via predictable or idiosyncratic inhibition of host mitochondrial components. Antimicrob. Agents Chemother. 2012, 56, 4046–4051. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Joseph, M.R.; Al-Hakami, A.M.; Assiry, M.M.; Jamil, A.S.; Assiry, A.M.; Shaker, M.A.; Hamid, M.E. In vitro anti-yeast activity of chloramphenicol: A preliminary report. J. Mycol. Med. 2015, 25, 17–22. [Google Scholar] [CrossRef]
- Salkin, I.F.; Hurd, N. Quantitative evaluation of the antifungal properties of cycloheximide. Antimicrob. Agents Chemother. 1972, 1, 177–184. [Google Scholar] [CrossRef] [Green Version]
- Whiffen, A.J. The production, assay, and antibiotic activity of actidione, an antiobiotic from Streptomyces griseus. J. Bacteriol. 1948, 56, 283–291. [Google Scholar] [CrossRef] [Green Version]
- Whiffen, A.J. The activity in vitro of cycloheximide (Actidione) against fungi pathogenic to plants. Mycologia 1950, 42, 253. [Google Scholar] [CrossRef]
- Monod, M.; Feuermann, M.; Salamin, K.; Fratti, M.; Makino, M.; Alshahni, M.M.; Makimura, K.; Yamada, T. Trichophyton rubrum azole resistance mediated by a new ABC transporter, TruMDR3. Antimicrob. Agents Chemother. 2019, 63, e00863-19. [Google Scholar] [CrossRef] [Green Version]
- Cervelatti, E.P.; Fachin, A.L.; Ferreira-Nozawa, M.S.; Martinez-Rossi, N.M. Molecular cloning and characterization of a novel ABC transporter gene in the human pathogen Trichophyton rubrum. Med. Mycol. 2006, 44, 141–147. [Google Scholar] [CrossRef] [Green Version]
- Fachin, A.L.; Ferreira-Nozawa, M.S.; Maccheroni, W.; Martinez-Rossi, N.M. Role of the ABC transporter TruMDR2 in terbinafine, 4-nitroquinoline N-oxide and ethidium bromide susceptibility in Trichophyton rubrum. J. Med. Microbiol. 2006, 55, 1093–1099. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Monod, M.; Feuermann, M.; Yamada, T. Terbinafine and itraconazole resistance in dermatophytes. In Dermatophytes and Dermatophytosis, 1st ed.; Bouchara, J.P., Nenoff, P., Gupta, A.K., Chaturvedi, V., Eds.; Springer: Cham, Switzerland, 2021; pp. 415–429. [Google Scholar]
- Fumeaux, J.; Mock, M.; Ninet, B.; Jan, I.; Bontems, O.; Lechenne, B.; Lew, D.; Panizzon, R.G.; Jousson, O.; Monod, M. First report of Arthroderma benhamiae in Switzerland. Dermatology 2004, 208, 244–250. [Google Scholar] [CrossRef] [PubMed]
- Yamada, T.; Makimura, K.; Satoh, K.; Umeda, Y.; Ishihara, Y.; Abe, S. Agrobacterium tumefaciens-mediated transformation of the dermatophyte, Trichophyton mentagrophytes: An efficient tool for gene transfer. Med. Mycol. 2009, 47, 485–494. [Google Scholar] [CrossRef] [Green Version]
- Sherman, F.; Wakem, P. Mapping yeast genes. Methods Enzymol. 1991, 194, 38–57. [Google Scholar] [PubMed]
- Mumberg, D.; Müller, R.; Funk, M. Yeast vectors for the controlled expression of heterologous proteins in different backgrounds. Gene 1995, 156, 119–122. [Google Scholar] [CrossRef]
- Zhang, A.; Lu, P.; Dahl-Roshak, A.M.; Paress, P.S.; Kennedy, S.; Tkacz, J.S.; An, Z. Efficient disruption of a polyketide synthase gene (pks1) required for melanin synthesis through Agrobacterium-mediated transformation of Glarea lozoyensis. Mol. Genet. Genom. 2003, 268, 645–655. [Google Scholar] [CrossRef] [PubMed]
- Girardin, H.; Latge, J.P. DNA extraction and quantification. In Molecular Biology of Pathogenic Fungi, 2nd ed.; Maresca, B., Kobayashi, G.S., Eds.; Telos Press: New York, NY, USA, 1994; pp. 5–9. [Google Scholar]
- Clinical and Laboratory Standards Institute. Reference Method for Broth Dilution Antifungal Susceptibility Testing of Filamentous Fungi, 2nd ed.; Approved Standard; CLSI document M38-A2; Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2008. [Google Scholar]
- Togni, G.; Sanglard, D.; Quadroni, M.; Foundling, S.I.; Monod, M. Acid proteinase secreted by Candida tropicalis: Functional analysis of preproregion cleavages in C. tropicalis and Saccharomyces cerevisiae. Microbiology 1996, 142, 493–503. [Google Scholar]
- Martinez, D.A.; Oliver, B.G.; Graeser, Y.; Goldberg, J.M.; Li, W.; Martinez-Rossi, N.M.; Monod, M.; Shelest, E.; Barton, R.C.; Birch, E.; et al. Comparative genome analysis of Trichophyton rubrum and related dermatophytes reveals candidate genes involved in infection. mBio 2012, 3, E259. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Reddy, V.S.; Shlykov, M.A.; Castillo, R.; Sun, E.I.; Saier, M.H., Jr. The major facilitator superfamily (MFS) revisited. FEBS J. 2012, 279, 2022–2035. [Google Scholar] [CrossRef]
- Pao, S.S.; Paulsen, I.T.; Saier, M.H., Jr. Major facilitator superfamily. Microbiol. Mol. Biol. Rev. 1998, 62, 1–34. [Google Scholar] [CrossRef] [Green Version]
- Walmsley, A.R.; Barrett, M.P.; Bringaud, F.; Gould, G.W. Sugar transporters from bacteria, parasites and mammals: Structure-activity relationships. Trends Biochem. Sci. 1998, 23, 476–481. [Google Scholar] [CrossRef]
- Burmester, A.; Shelest, E.; Gloeckner, G.; Heddergott, C.; Schindler, S.; Staib, P.; Heidel, A.; Felder, M.; Petzold, A.; Szafranski, K.; et al. Comparative and functional genomics provide insights into the pathogenicity of dermatophytic fungi. Genome Biol. 2011, 12, R7.1–R7.16. [Google Scholar] [CrossRef] [Green Version]
- Yamada, Y.; Maeda, M.; Alshahni, M.M.; Monod, M.; Staib, P.; Yamada, T. Flippase (FLP) recombinase-mediated marker recycling in the dermatophyte Arthroderma vanbreuseghemii. Microbiology 2014, 160, 2122–2135. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Harmsen, D.; Schwinn, A.; Weig, M.; Broecker, E.B.; Heesemann, J. Phylogeny and dating of some pathogenic keratinophilic fungi using small subunit ribosomal RNA. J. Med. Vet. Mycol. 1995, 33, 299–303. [Google Scholar] [CrossRef]
- Chin, B.; Knight, S. Growth of Trichophyton mentagrophytes and Trichophyton rubrum in increased carbon dioxide tensions. J. Gen. Microbiol. 1957, 16, 642–646. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Laurent, A.; Monod, M. Production of Trichophyton rubrum microspores in large quantities and its application to evaluate amorolfine/azole compound interactions in vitro. Mycoses 2017, 60, 581–586. [Google Scholar] [CrossRef] [PubMed]
- Gupta, A.K.; Foley, K.A.; Versteeg, S.G. New antifungal agents and new formulations against dermatophytes. Mycopathologia 2017, 182, 127–141. [Google Scholar] [CrossRef] [PubMed]
- Hay, R.J. Tinea capitis: Current status. Mycopathologia 2017, 182, 87–93. [Google Scholar] [CrossRef] [Green Version]
- Asz-Sigall, D.; Tosti, A.; Arenas, R. Tinea unguium: Diagnosis and treatment in practice. Mycopathologia 2017, 182, 95–100. [Google Scholar] [CrossRef]
- Gupta, A.K.; Kohli, Y.; Batra, R. In vitro activities of posaconazole, ravuconazole, terbinafine, itraconazole and fluconazole against dermatophyte, yeast and non-dermatophyte species. Med. Mycol. 2005, 43, 179–185. [Google Scholar] [CrossRef] [Green Version]
- Ghannoum, M.A.; Arthington-Skaggs, B.; Chaturvedi, V.; Espinel-Ingroff, A.; Pfaller, M.A.; Rennie, R.; Rinaldi, M.G.; Walsh, T.J. Interlaboratory study of quality control isolates for a broth microdilution method (modified CLSI M38-A) for testing susceptibilities of dermatophytes to antifungals. J. Clin. Microbiol. 2006, 44, 4353–4356. [Google Scholar] [CrossRef] [Green Version]
- Salehi, Z.; Fatahi, N.; Taran, M.; Izadi, A.; Badali, H.; Hashemi, S.J.; Rezaie, S.; Daie Ghazvini, R.; Ghaffari, M.; Aala, F.; et al. Comparison of in vitro antifungal activity of novel triazoles with available antifungal agents against dermatophyte species caused tinea pedis. J. Mycol. Med. 2020, 30, 100935. [Google Scholar] [CrossRef]
- Yu, J.; Chang, P.K.; Ehrlich, K.C.; Cary, J.W.; Bhatnagar, D.; Cleveland, T.E.; Payne, G.A.; Linz, J.E.; Woloshuk, C.P.; Bennett, J.W. Clustered pathway genes in aflatoxin biosynthesis. Appl. Environ. Microbiol. 2004, 70, 1253–1262. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chang, P.K.; Yu, J.; Yu, J.H. AflT, a MFS transporter-encoding gene located in the aflatoxin gene cluster, does not have a significant role in aflatoxin secretion. Fungal Genet. Biol. 2004, 41, 911–920. [Google Scholar] [CrossRef] [PubMed]
- Lin, H.C.; Yu, P.L.; Chen, L.H.; Tsai, H.C.; Chung, K.R. A major facilitator superfamily transporter regulated by the stress-responsive transcription factor Yap1 is required for resistance to fungicides, xenobiotics, and oxidants and full virulence in Alternaria alternata. Front. Microbiol. 2018, 9, 2229. [Google Scholar] [CrossRef]
- Demuyser, L.; van Dijck, P. Can Saccharomyces cerevisiae keep up as a model system in fungal azole susceptibility research? Drug Resist. Updat. 2019, 42, 22–34. [Google Scholar] [CrossRef]
- Calabrese, D.; Bille, J.; Sanglard, D. A novel multidrug efflux transporter gene of the major facilitator superfamily from Candida albicans (FLU1) conferring resistance to fluconazole. Microbiology 2000, 146, 2743–2754. [Google Scholar] [CrossRef] [Green Version]
- Andrade, A.C.; van Nistelrooy, J.G.; Peery, R.B.; Skatrud, P.L.; de Waard, M.A. The role of ABC transporters from Aspergillus nidulans in protection against cytotoxic agents and in antibiotic production. Mol. Gen. Genet. 2000, 263, 966–977. [Google Scholar] [CrossRef] [PubMed]

| Plasmid | Description a | Source or Reference |
|---|---|---|
| p426GPD | Yeast episomal plasmid (high copy number) carrying the 2 µ ori and containing the glyceraldehyde-3-phosphate dehydrogenase gene (GPD) promoter, flanked by a multiple cloning site and an XhoI-KpnI fragment of the cytochrome-c oxidase gene (CYC1) terminator, and the URA3 marker gene for the transformation of S. cerevisiae. | [16] |
| pMFS1 | pYES2-DEST52 (Invitrogen) containing TruMFS1 cDNA under the control of the GAL1 promoter of S. cerevisiae. | [9] |
| pMFS1-2 | p426GPD containing TruMFS1 cDNA flanked upstream by the GPD promoter and downstream by the CYC1 terminator. | This study |
| pAg1 | Streamlined version of the binary vector pBIN19 containing sequences necessary for replication in E. coli and A. tumefaciens (oriV and trfA), E. coli neomycin phosphotransferase gene (nptII), and the transferable DNA (T-DNA) region, with a multiple cloning site within the T-DNA region | [17] |
| pSP72-PcFLP | pSP72 (Promega) containing the 5′-UTR of Trichophyton mentagrophytes Ku80 (TmKu80) gene (GenBank accession no. AB427108) (1.5 kb), the FLP recombination target (5′-FRT), the promoter sequence of A. nidulans tryptophan C (trpC) gene (PtrpC) (GenBank accession no. X02390), nptII, the terminator sequence of A. fumigatus cgrA gene (TcgrA) (GenBank accession no. EAL84894), the promoter sequence of T. rubrum high affinity copper transporter (ctr4) gene (Pctr4) (TERG_01401), Penicillium chrysogenum-optimized FLP recombinase (Pcflp) gene, the terminator sequence of Cryptococcus neoformans phosphoribosyl anthranilate isomerase (trp1) gene (Ttrp1) (GenBank accession no. M74901), 3′-FRT and the 3′-UTR of TmKu80 gene (1.5 kb). | [9] |
| pAg1-hph | Cochliobolus heterostrophus promoter 1 (GenBank accession no. M12304), E. coli hygromycin B phosphotransferase gene (hph), the terminator sequence of A. nidulans trpC gene (TtrpC) | [14] |
| pAg1-TbeMFS1/T | TbeMFS1a fragment (the 5′ UTR of TbeMFS1 gene; 2.04 kb) (GenBank accession no. DAA75241), PtrpC, nptII, TcgrA, TbeMFS1b fragment (the 3′ UTR of TbeMFS1 gene; 2.02 kb) | This study |
| pAg1-TbeMFS1/C | TbeMFS1c fragment (the 5′ UTR and ORF of TbeMFS1 gene; 3.50 kb), PtrpC, hph, TcgrA, TbeMFS1b fragment | This study |
| Species and Strains | Antimicrobial Agents (µg/mL) | ||||||
|---|---|---|---|---|---|---|---|
| CHL | TAP | CYH | FLC | ITC | VRC | MCZ | |
| Trichophyton benhamiae | |||||||
| ΔTbeKu70-5-1 | 1280 | >2560 | >4000 | 32 | 0.25 | 0.12 | 2 |
| ΔTbeMFS1-17-1 | 320 | 160 | >4000 | 4 | 0.25 | 0.06 | 0.5 |
| ΔTbeMFS1-18-1 | 320 | 320 | >4000 | 8 | 0.25 | 0.12 | 0.5 |
| TbeMFS1/C-18-1-10 | 1280 | >2560 | >4000 | 32 | 0.25 | 0.12 | 2 |
| TbeMFS1/C-18-1-16 | 1280 | >2560 | >4000 | 32 | 0.25 | 0.12 | 2 |
| T. interdigitale | |||||||
| ATCC MYA-4439 | 160 | >2560 | >4000 | 4 | 0.25 | 0.06 | 0.25 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yamada, T.; Yaguchi, T.; Salamin, K.; Guenova, E.; Feuermann, M.; Monod, M. MFS1, a Pleiotropic Transporter in Dermatophytes That Plays a Key Role in Their Intrinsic Resistance to Chloramphenicol and Fluconazole. J. Fungi 2021, 7, 542. https://doi.org/10.3390/jof7070542
Yamada T, Yaguchi T, Salamin K, Guenova E, Feuermann M, Monod M. MFS1, a Pleiotropic Transporter in Dermatophytes That Plays a Key Role in Their Intrinsic Resistance to Chloramphenicol and Fluconazole. Journal of Fungi. 2021; 7(7):542. https://doi.org/10.3390/jof7070542
Chicago/Turabian StyleYamada, Tsuyoshi, Takashi Yaguchi, Karine Salamin, Emmanuella Guenova, Marc Feuermann, and Michel Monod. 2021. "MFS1, a Pleiotropic Transporter in Dermatophytes That Plays a Key Role in Their Intrinsic Resistance to Chloramphenicol and Fluconazole" Journal of Fungi 7, no. 7: 542. https://doi.org/10.3390/jof7070542







