Oxidative Stress Causes Vacuolar Fragmentation in the Human Fungal Pathogen Cryptococcus neoformans
Abstract
:1. Introduction
2. Materials and Methods
2.1. Strains and Growth Media
2.2. Construction of Strains Expressing the Sod1-GFP or the Sod2-GFP Fusion Protein
2.3. Construction of the fab1 Knock-Out Mutant
2.4. Confocal Fluorescence Microscopy
2.5. Flow Cytometric Analysis
2.6. Western Blot Analysis
2.7. Superoxide Dismutase Activity Assay
3. Results
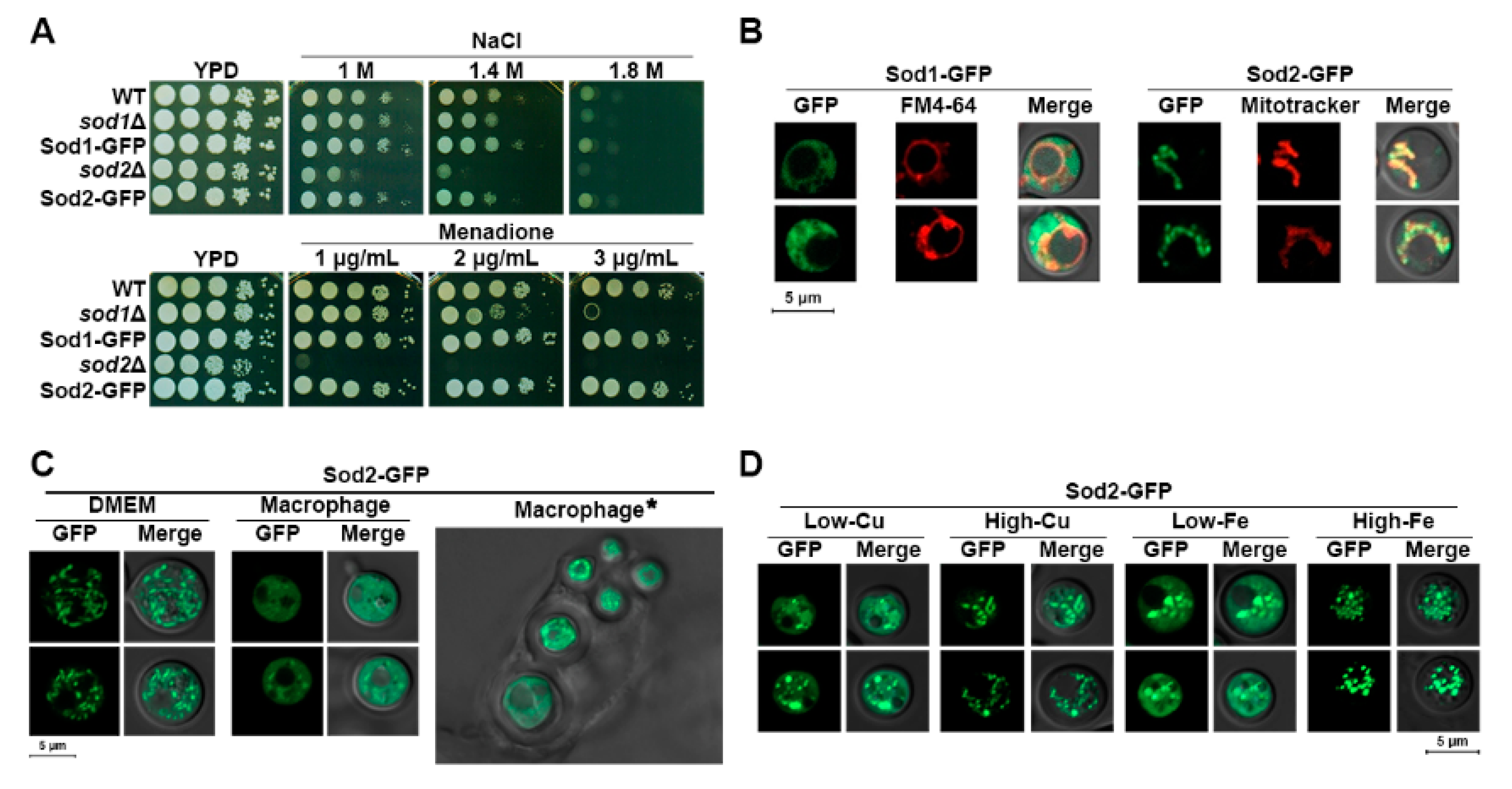
3.1. Localization of Sod2 Is Influenced by Iron and Copper, and Iron Levels Impact the Expression and Activity of Sod2
3.2. Deletion of SOD Influences Expression of Iron Regulators as Well as Vacuole Morphology
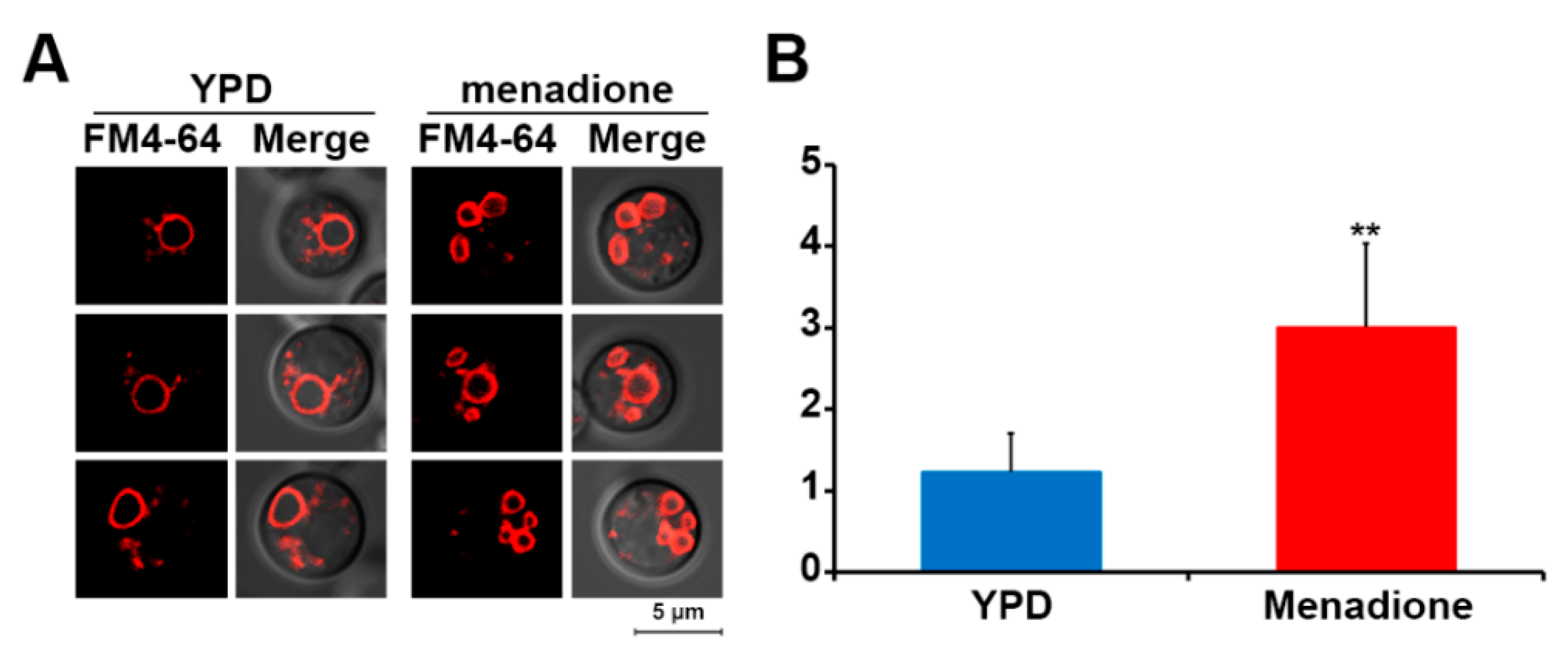
3.3. Oxidative Stress Induces Vacuole Fragmentation
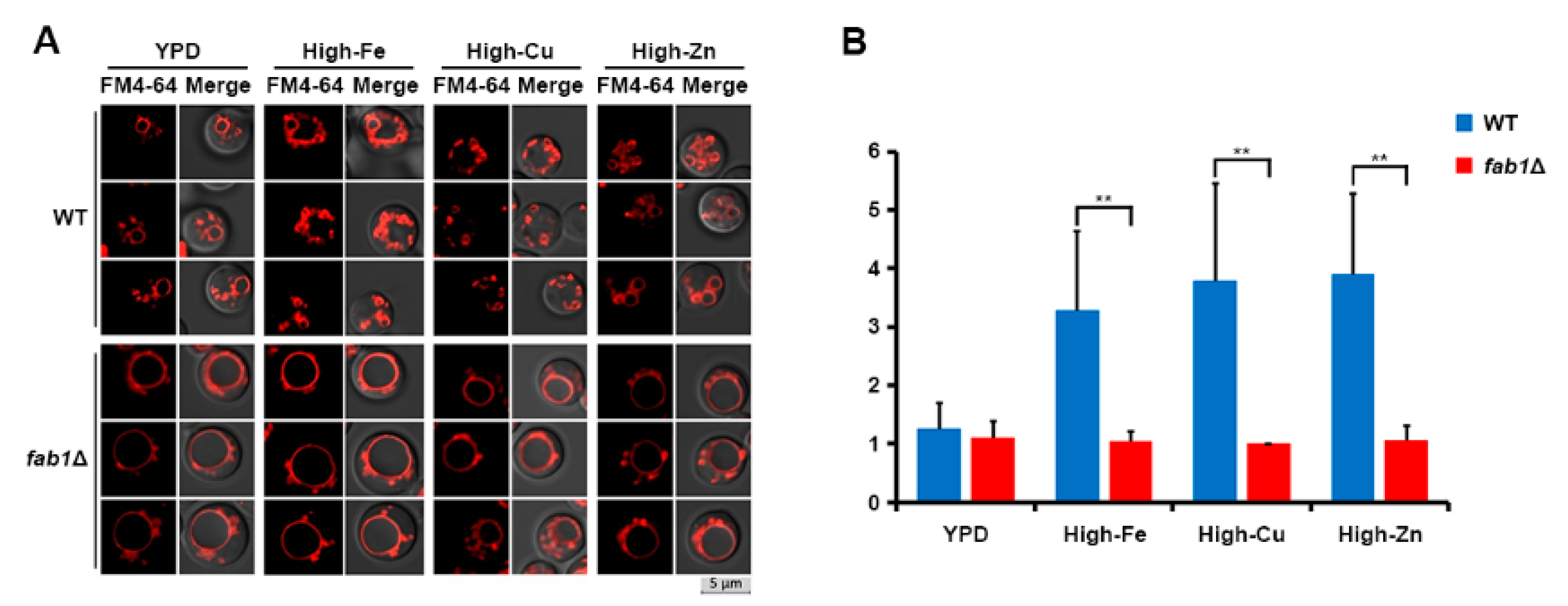
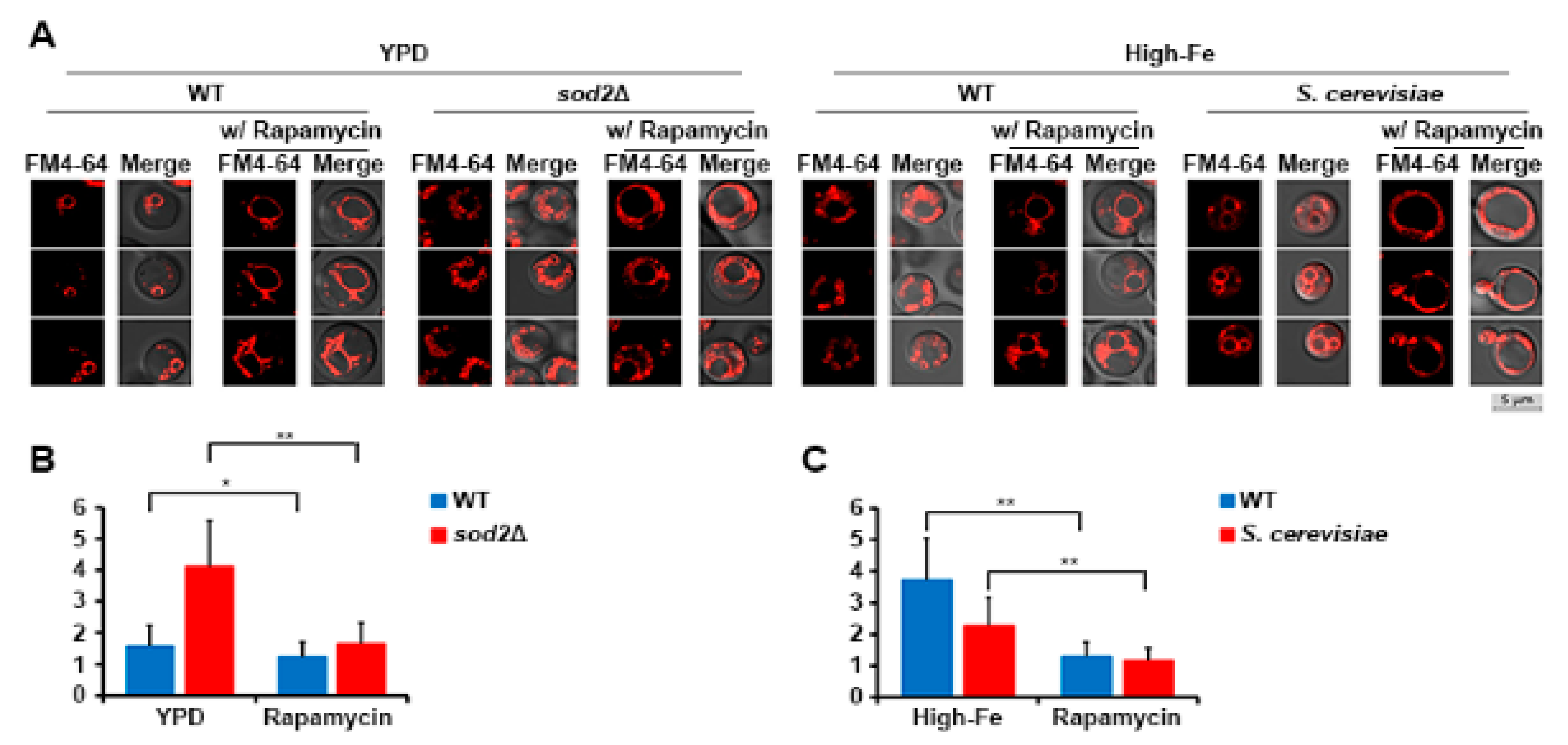
3.4. Vacuole Fragmentation Is Also Influenced by Fab1 and TORC1
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Aufschnaiter, A.; Buttner, S. The vacuolar shapes of ageing: From function to morphology. Biochim. Biophys. Acta Mol. Cell Res. 2019, 1866, 957–970. [Google Scholar] [CrossRef] [PubMed]
- Bonangelino, C.J.; Nau, J.J.; Duex, J.E.; Brinkman, M.; Wurmser, A.E.; Gary, J.D.; Emr, S.D.; Weisman, L.S. Osmotic stress-induced increase of phosphatidylinositol 3,5-bisphosphate requires Vac14p, an activator of the lipid kinase Fab1p. J. Cell Biol. 2002, 156, 1015–1028. [Google Scholar] [CrossRef] [Green Version]
- Gebre, S.; Connor, R.; Xia, Y.; Jawed, S.; Bush, J.M.; Bard, M.; Elsalloukh, H.; Tang, F. Osh6 overexpression extends the lifespan of yeast by increasing vacuole fusion. Cell Cycle 2012, 11, 2176–2188. [Google Scholar] [CrossRef] [Green Version]
- Yamamoto, A.; DeWald, D.B.; Boronenkov, I.V.; Anderson, R.A.; Emr, S.D.; Koshland, D. Novel PI(4)P 5-kinase homologue, Fab1p, essential for normal vacuole function and morphology in yeast. Mol. Biol. Cell 1995, 6, 525–539. [Google Scholar] [CrossRef] [Green Version]
- Michaillat, L.; Baars, T.L.; Mayer, A. Cell-free reconstitution of vacuole membrane fragmentation reveals regulation of vacuole size and number by TORC1. Mol. Biol. Cell 2012, 23, 881–895. [Google Scholar] [CrossRef] [PubMed]
- Corson, L.B.; Folmer, J.; Strain, J.J.; Culotta, V.C.; Cleveland, D.W. Oxidative stress and iron are implicated in fragmenting vacuoles of Saccharomyces cerevisiae lacking Cu, Zn-superoxide dismutase. J. Biol. Chem. 1999, 274, 27590–27596. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tesfa-Selase, F.; Hay, R.J. Superoxide dismutase of Cryptococcus neoformans: Purification and characterization. J. Med. Vet. Mycol 1995, 33, 253–259. [Google Scholar] [CrossRef]
- Cox, G.M.; Harrison, T.S.; McDade, H.C.; Taborda, C.P.; Heinrich, G.; Casadevall, A.; Perfect, J.R. Superoxide dismutase influences the virulence of Cryptococcus neoformans by affecting growth within macrophages. Infect. Immun. 2003, 71, 173–180. [Google Scholar] [CrossRef] [Green Version]
- Giles, S.S.; Batinic-Haberle, I.; Perfect, J.R.; Cox, G.M. Cryptococcus neoformans mitochondrial superoxide dismutase: An essential link between antioxidant function and high-temperature growth. Eukaryot. Cell 2005, 4, 46–54. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Narasipura, S.D.; Ault, J.G.; Behr, M.J.; Chaturvedi, V.; Chaturvedi, S. Characterization of Cu, Zn superoxide dismutase (SOD1) gene knock-out mutant of Cryptococcus neoformans var. gattii: Role in biology and virulence. Mol. Microbiol. 2003, 47, 1681–1694. [Google Scholar] [CrossRef]
- Narasipura, S.D.; Chaturvedi, V.; Chaturvedi, S. Characterization of Cryptococcus neoformans variety gattii SOD2 reveals distinct roles of the two superoxide dismutases in fungal biology and virulence. Mol. Microbiol. 2005, 55, 1782–1800. [Google Scholar] [CrossRef] [PubMed]
- Smith, A.D.; Garcia-Santamarina, S.; Ralle, M.; Loiselle, D.R.; Haystead, T.A.; Thiele, D.J. Transcription factor-driven alternative localization of Cryptococcus neoformans superoxide dismutase. J. Biol. Chem. 2021, 100391. [Google Scholar] [CrossRef] [PubMed]
- Bairwa, G.; Sanchez-Leon, E.; Do, E.; Jung, W.H.; Kronstad, J.W. A Cytoplasmic Heme Sensor Illuminates the Impacts of Mitochondrial and Vacuolar Functions and Oxidative Stress on Heme-Iron Homeostasis in Cryptococcus neoformans. mBio 2020, 11. [Google Scholar] [CrossRef] [PubMed]
- Jung, W.H.; Hu, G.; Kuo, W.; Kronstad, J.W. Role of ferroxidases in iron uptake and virulence of Cryptococcus neoformans. Eukaryot. Cell 2009, 8, 1511–1520. [Google Scholar] [CrossRef] [Green Version]
- Toffaletti, D.L.; Rude, T.H.; Johnston, S.A.; Durack, D.; Perfect, J.R. Gene transfer in Cryptococcus neoformans by use of biolistic delivery of DNA. J. Bacteriol. 1993, 175, 1405–1411. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kauffman, M.E.; Kauffman, M.K.; Traore, K.; Zhu, H.; Trush, M.A.; Jia, Z.; Li, Y.R. MitoSOX-Based Flow Cytometry for Detecting Mitochondrial ROS. React. Oxyg. Species (Apex) 2016, 2, 361–370. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bradford, M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef]
- Mutoh, N.; Nakagawa, C.W.; Yamada, K. Characterization of Cu, Zn-superoxide dismutase-deficient mutant of fission yeast Schizosaccharomyces pombe. Curr. Genet. 2002, 41, 82–88. [Google Scholar] [CrossRef]
- Oberegger, H.; Zadra, I.; Schoeser, M.; Haas, H. Iron starvation leads to increased expression of Cu/Zn-superoxide dismutase in Aspergillus. FEBS Lett. 2000, 485, 113–116. [Google Scholar] [CrossRef] [Green Version]
- Schatzman, S.S.; Peterson, R.L.; Teka, M.; He, B.; Cabelli, D.E.; Cormack, B.P.; Culotta, V.C. Copper-only superoxide dismutase enzymes and iron starvation stress in Candida fungal pathogens. J. Biol. Chem. 2020, 295, 570–583. [Google Scholar] [CrossRef]
- Jung, W.H.; Kronstad, J.W. Iron influences the abundance of the iron regulatory protein Cir1 in the fungal pathogen Cryptococcus neoformans. FEBS Lett. 2011, 585, 3342–3347. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Akhter, S.; McDade, H.C.; Gorlach, J.M.; Heinrich, G.; Cox, G.M.; Perfect, J.R. Role of alternative oxidase gene in pathogenesis of Cryptococcus neoformans. Infect. Immun. 2003, 71, 5794–5802. [Google Scholar] [CrossRef] [Green Version]
- Kobayashi, D.; Kondo, K.; Uehara, N.; Otokozawa, S.; Tsuji, N.; Yagihashi, A.; Watanabe, N. Endogenous reactive oxygen species is an important mediator of miconazole antifungal effect. Antimicrob. Agents Chemother. 2002, 46, 3113–3117. [Google Scholar] [CrossRef] [Green Version]
- Kim, J.; Cho, Y.J.; Do, E.; Choi, J.; Hu, G.; Cadieux, B.; Chun, J.; Lee, Y.; Kronstad, J.W.; Jung, W.H. A defect in iron uptake enhances the susceptibility of Cryptococcus neoformans to azole antifungal drugs. Fungal Genet. Biol. 2012, 49, 955–966. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, Z.; Malia, P.C.; Hatakeyama, R.; Nicastro, R.; Hu, Z.; Peli-Gulli, M.P.; Gao, J.; Nishimura, T.; Eskes, E.; Stefan, C.J.; et al. TORC1 Determines Fab1 Lipid Kinase Function at Signaling Endosomes and Vacuoles. Curr. Biol. 2021, 31, 297–309.e298. [Google Scholar] [CrossRef] [PubMed]
- Jomova, K.; Valko, M. Advances in metal-induced oxidative stress and human disease. Toxicology 2011, 283, 65–87. [Google Scholar] [CrossRef]
- Kruszewski, M. Labile iron pool: The main determinant of cellular response to oxidative stress. Mutat Res. 2003, 531, 81–92. [Google Scholar] [CrossRef]
- Ho, Y.N.; Hoo, S.Y.; Wang, B.W.; Hsieh, C.T.; Lin, C.C.; Sun, C.H.; Peng, C.C.; Lin, C.; Yang, Y.L. Specific inactivation of an antifungal bacterial siderophore by a fungal plant pathogen. ISME J. 2021. [Google Scholar] [CrossRef] [PubMed]
- Pagani, M.A.; Casamayor, A.; Serrano, R.; Atrian, S.; Arino, J. Disruption of iron homeostasis in Saccharomyces cerevisiae by high zinc levels: A genome-wide study. Mol. Microbiol. 2007, 65, 521–537. [Google Scholar] [CrossRef]
- Zhao, Y.Y.; Cao, C.L.; Liu, Y.L.; Wang, J.; Li, J.; Li, S.Y.; Deng, Y. Identification of the Genetic Requirements for Zinc Tolerance and Toxicity in Saccharomyces cerevisiae. G3 (Bethesda) 2020, 10, 479–488. [Google Scholar] [CrossRef] [Green Version]
- Farres, M.; Pina, B.; Tauler, R. LC-MS based metabolomics and chemometrics study of the toxic effects of copper on Saccharomyces cerevisiae. Metallomics 2016, 8, 790–798. [Google Scholar] [CrossRef]
- Avery, S.V.; Howlett, N.G.; Radice, S. Copper toxicity towards Saccharomyces cerevisiae: Dependence on plasma membrane fatty acid composition. Appl. Environ. Microbiol. 1996, 62, 3960–3966. [Google Scholar] [CrossRef] [Green Version]
- Macomber, L.; Imlay, J.A. The iron-sulfur clusters of dehydratases are primary intracellular targets of copper toxicity. Proc. Natl. Acad. Sci. USA 2009, 106, 8344–8349. [Google Scholar] [CrossRef] [Green Version]
- Kronstad, J.W.; Hu, G.; Jung, W.H. An encapsulation of iron homeostasis and virulence in Cryptococcus neoformans. Trends Microbiol. 2013, 21, 457–465. [Google Scholar] [CrossRef] [Green Version]
- Lian, T.; Simmer, M.I.; D’Souza, C.A.; Steen, B.R.; Zuyderduyn, S.D.; Jones, S.J.; Marra, M.A.; Kronstad, J.W. Iron-regulated transcription and capsule formation in the fungal pathogen Cryptococcus neoformans. Mol. Microbiol. 2005, 55, 1452–1472. [Google Scholar] [CrossRef] [PubMed]
- Do, E.; Cho, Y.J.; Kim, D.; Kronstad, J.W.; Jung, W.H. A Transcriptional Regulatory Map of Iron Homeostasis Reveals a New Control Circuit for Capsule Formation in Cryptococcus neoformans. Genetics 2020, 215, 1171–1189. [Google Scholar] [CrossRef]
- Ibrahim, W.H.; Habib, H.M.; Kamal, H.; St Clair, D.K.; Chow, C.K. Mitochondrial superoxide mediates labile iron level: Evidence from Mn-SOD-transgenic mice and heterozygous knockout mice and isolated rat liver mitochondria. Free Radic. Biol. Med. 2013, 65, 143–149. [Google Scholar] [CrossRef] [PubMed]
- Ito, H.; Kurokawa, H.; Hirayama, A.; Indo, H.P.; Majima, H.J.; Matsui, H. Cancer cell-specific mitochondrial reactive oxygen species promote non-heme iron uptake and enhance the proliferation of gastric epithelial cancer cell. J. Clin. Biochem. Nutr. 2017, 61, 183–188. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Flint, D.H.; Tuminello, J.F.; Emptage, M.H. The inactivation of Fe-S cluster containing hydro-lyases by superoxide. J. Biol Chem 1993, 268, 22369–22376. [Google Scholar] [CrossRef]
- Murakami, K.; Yoshino, M. Inactivation of aconitase in yeast exposed to oxidative stress. Biochem Mol. Biol Int 1997, 41, 481–486. [Google Scholar] [CrossRef]
- Takatori, S.; Tatematsu, T.; Cheng, J.; Matsumoto, J.; Akano, T.; Fujimoto, T. Phosphatidylinositol 3,5-Bisphosphate-Rich Membrane Domains in Endosomes and Lysosomes. Traffic 2016, 17, 154–167. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Urbanowski, J.L.; Piper, R.C. The iron transporter Fth1p forms a complex with the Fet5 iron oxidase and resides on the vacuolar membrane. J. Biol. Chem. 1999, 274, 38061–38070. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, L.; Chen, O.S.; McVey Ward, D.; Kaplan, J. CCC1 is a transporter that mediates vacuolar iron storage in yeast. J. Biol. Chem. 2001, 276, 29515–29519. [Google Scholar] [CrossRef] [Green Version]
- Stauffer, B.; Powers, T. Target of rapamycin signaling mediates vacuolar fission caused by endoplasmic reticulum stress in Saccharomyces cerevisiae. Mol. Biol. Cell 2015, 26, 4618–4630. [Google Scholar] [CrossRef] [PubMed]
- So, Y.S.; Lee, D.G.; Idnurm, A.; Ianiri, G.; Bahn, Y.S. The TOR Pathway Plays Pleiotropic Roles in Growth and Stress Responses of the Fungal Pathogen Cryptococcus neoformans. Genetics 2019, 212, 1241–1258. [Google Scholar] [CrossRef] [PubMed]



Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kim, D.; Song, M.; Do, E.; Choi, Y.; Kronstad, J.W.; Jung, W.H. Oxidative Stress Causes Vacuolar Fragmentation in the Human Fungal Pathogen Cryptococcus neoformans. J. Fungi 2021, 7, 523. https://doi.org/10.3390/jof7070523
Kim D, Song M, Do E, Choi Y, Kronstad JW, Jung WH. Oxidative Stress Causes Vacuolar Fragmentation in the Human Fungal Pathogen Cryptococcus neoformans. Journal of Fungi. 2021; 7(7):523. https://doi.org/10.3390/jof7070523
Chicago/Turabian StyleKim, Donghyeun, Moonyong Song, Eunsoo Do, Yoojeong Choi, James W. Kronstad, and Won Hee Jung. 2021. "Oxidative Stress Causes Vacuolar Fragmentation in the Human Fungal Pathogen Cryptococcus neoformans" Journal of Fungi 7, no. 7: 523. https://doi.org/10.3390/jof7070523
APA StyleKim, D., Song, M., Do, E., Choi, Y., Kronstad, J. W., & Jung, W. H. (2021). Oxidative Stress Causes Vacuolar Fragmentation in the Human Fungal Pathogen Cryptococcus neoformans. Journal of Fungi, 7(7), 523. https://doi.org/10.3390/jof7070523





