Cultivable Yeast Microbiota from the Marine Fish Species Genypterus chilensis and Seriolella violacea
Abstract
:1. Introduction
2. Materials and Methods
2.1. Fish, Intestinal Collection and Ethics Statement
2.2. Isolation of Yeasts from Intestinal Samples by Culture Method
2.3. Yeast DNA Extraction and Identification
2.4. Bioinformatic Workflow
2.5. Enzymatic Characterization of Yeasts
3. Results
3.1. Identification and Phylogeny of Isolated Yeasts
3.2. Enzymatic Characterization of Yeasts
4. Discussions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Banerjee, S.; Ghosh, K. Enumeration of Gut Associated Extracellular Enzyme-Producing Yeasts in Some Freshwater Fishes. J. Appl. Ichthyol. 2014, 30, 986–993. [Google Scholar] [CrossRef]
- Mandal, S.; Ghosh, K. Isolation of Tannase-Producing Microbiota from the Gastrointestinal Tracts of Some Freshwater Fish. J. Appl. Ichthyol. 2013, 29, 145–153. [Google Scholar] [CrossRef]
- Gatesoupe, F.J. Live Yeasts in the Gut: Natural Occurrence, Dietary Introduction, and Their Effects on Fish Health and Development. Aquaculture 2007, 267, 20–30. [Google Scholar] [CrossRef] [Green Version]
- Gilbert, J.A.; Blaser, M.J.; Caporaso, J.G.; Jansson, J.K.; Lynch, S.V.; Knight, R. Current Understanding of the Human Microbiome. Nat. Med. 2018, 24, 392–400. [Google Scholar] [CrossRef]
- Rawls, J.F.; Samuel, B.S.; Gordon, J.I. Gnotobiotic Zebrafish Reveal Evolutionarily Conserved Responses to the Gut Microbiota. Proc. Natl. Acad. Sci. USA 2004, 101, 4596–4601. [Google Scholar] [CrossRef] [Green Version]
- Levican, A.; Fisher, J.C.; McLellan, S.L.; Avendaño-Herrera, R. Microbial Communities Associated with Farmed Genypterus Chilensis: Detection in Water Prior to Bacterial Outbreaks Using Culturing High-Throughput Sequencing. Animals 2020, 10, 1055. [Google Scholar] [CrossRef]
- Hill, C.; Guarner, F.; Reid, G.; Gibson, G.R.; Merenstein, D.J.; Pot, B.; Morelli, L.; Canani, R.B.; Flint, H.J.; Salminen, S.; et al. Expert Consensus Document: The International Scientific Association for Probiotics and Prebiotics Consensus Statement on the Scope and Appropriate Use of the Term Probiotic. Nat. Rev. Gastroenterol. Hepatol. 2014, 11, 506–514. [Google Scholar] [CrossRef] [Green Version]
- Tovar, D.; Zambonino, J.; Cahu, C.; Gatesoupe, F.J.; Vázquez-Juárez, R.; Lésel, R. Effect of Live Yeast Incorporation in Compound Diet on Digestive Enzyme Activity in Sea Bass (Dicentrarchus Labrax) Larvae. Aquaculture 2002, 204, 113–123. [Google Scholar] [CrossRef]
- Caruffo, M.; Navarrete, N.C.; Salgado, O.A.; Faúndez, N.B.; Gajardo, M.C.; Feijóo, C.G.; Reyes-Jara, A.; García, K.; Navarrete, P. Protective Yeasts Control V. Anguillarum Pathogenicity and Modulate the Innate Immune Response of Challenged Zebrafish (Danio Rerio) Larvae. Front. Cell. Infect. Microbiol. 2016, 6. [Google Scholar] [CrossRef] [Green Version]
- Reyes-Becerril, M.; Salinas, I.; Cuesta, A.; Meseguer, J.; Tovar-Ramirez, D.; Ascencio-Valle, F.; Esteban, M.Á. Oral Delivery of Live Yeast Debaryomyces Hansenii Modulates the Main Innate Immune Parameters and the Expression of Immune-Relevant Genes in the Gilthead Seabream (Sparus Aurata L.). Fish Shellfish Immunol. 2008, 9, 731–739. [Google Scholar] [CrossRef]
- Lokesh, J.; Fernandes, J.M.O.; Korsnes, K.; Bergh, O.; Brinchmann, M.F.; Kiron, V. Transcriptional Regulation of Cytokines in the Intestine of Atlantic Cod Fed Yeast Derived Mannan Oligosaccharide or β-Glucan and Challenged with Vibrio Anguillarum. Fish Shellfish Immunol. 2012, 33, 626–631. [Google Scholar] [CrossRef] [PubMed]
- Navarrete, P.; Tovar-Ramrez, D. Use of Yeasts as Probiotics in Fish Aquaculture. Sustain. Aquac. Tech. 2014. [Google Scholar] [CrossRef] [Green Version]
- Raggi, P.; Lopez, P.; Diaz, A.; Carrasco, D.; Silva, A.; Velez, A.; Opazo, R.; Magne, F.; Navarrete, P.A. Debaryomyces Hansenii and Rhodotorula Mucilaginosa Comprised the Yeast Core Gut Microbiota of Wild and Reared Carnivorous Salmonids, Croaker and Yellowtail. Environ. Microbiol. 2014, 16, 2791–2803. [Google Scholar] [CrossRef]
- White, T.J.; Bruns, T.; Lee, S.; Taylor, J. Amplification and Direct Sequencing of Fungal Ribosomal R.N.A: Genes for Phylogenetics. In PCR Protocols: A Guide to Methods and Applications; Innis, M.A., Gelfand, D.H., Sninsky, J.J., White, T.J., Eds.; Academic Press: San Diego, CA, USA, 1990; pp. 315–322. [Google Scholar]
- O’Donnell, K.; Reynolds, D.R.; Taylor, J.W. Fusarium and Its Near Relatives, International. In Proceedings of the Fungal Holomorph: Mitotic, Meiotic and Pleomorphic Speciation in Fungal Systematics, International Symposium, Portland, OR, USA, 4–7 August 1993; CAB. International: Wallingford, UK, 1993; pp. 225–236. [Google Scholar]
- Kearse, M.; Moir, R.; Wilson, A.; Stones-Havas, S.; Cheung, M.; Sturrock, S.; Buxton, S.; Cooper, A.; Markowitz, S.; Duran, C.; et al. Geneious Basic: An Integrated and Extendable Desktop Software Platform for the Organization and Analysis of Sequence Data. Bioinformatics 2012, 28, 1647–1649. [Google Scholar] [CrossRef] [PubMed]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic Local Alignment Search Tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Hofstetter, V.; Buyck, B.; Eyssartier, G.; Schnee, S.; Gindro, K. The Unbearable Lightness of Sequenced-Based Identification. Fungal Divers. 2019, 96, 243–284. [Google Scholar] [CrossRef] [Green Version]
- Nilsson, R.H.; Anslan, S.; Bahram, M.; Wurzbacher, C.; Baldrian, P.; Tedersoo, L. Mycobiome Diversity: High-Throughput Sequencing and Identification of Fungi. Nat. Rev. Microbiol. 2019, 17, 95–109. [Google Scholar] [CrossRef] [PubMed]
- Raja, H.A.; Miller, A.N.; Pearce, C.J.; Oberlies, N.H. Fungal Identification Using Molecular Tools: A Primer for the Natural Products Research Community. J. Nat. Prod. 2017, 80, 756–770. [Google Scholar] [CrossRef] [PubMed]
- Sievers, F.; Wilm, A.; Dineen, D.; Gibson, T.J.; Karplus, K.; Li, W.; Lopez, R.; McWilliam, H.; Remmert, M.; Söding, J.; et al. Fast, Scalable Generation of High-quality Protein Multiple Sequence Alignments Using Clustal Omega. Mol. Syst. Biol. 2011, 7, 539. [Google Scholar] [CrossRef]
- R Core Team. R: A Language and Environment for Statistical Computing. 2020. Available online: https://cran.r-project.org (accessed on 25 March 2021).
- Charif, D.; Lobry, J.R. SeqinR 1.0–2: A Contributed Package to the R Project for Statistical Computing Devoted to Biological Sequences Retrieval and Analysis; Springer: Berlin/Heidelberg, Germany, 2007; pp. 207–232. [Google Scholar]
- Paradis, E.; Schliep, K. Ape 5.0: An Environment for Modern Phylogenetics and Evolutionary Analyses in R. Bioinformatics 2019, 35, 526–528. [Google Scholar] [CrossRef]
- Kimura, M. A Simple Method for Estimating Evolutionary Rates of Base Substitutions through Comparative Studies of Nucleotide Sequences. J. Mol. Evol. 1980, 16, 111–120. [Google Scholar] [CrossRef]
- Saitou, N.; Nei, M. The Neighbor-Joining Method: A New Method for Reconstructing Phylogenetic Trees. Mol. Biol. Evol. 1987, 4, 406–425. [Google Scholar] [CrossRef]
- Felsenstein, J. Confidence Limits on Phylogenies: An Approach Using the Bootstrap. Evolution 1985, 39, 783. [Google Scholar] [CrossRef] [PubMed]
- Yu, G. Using Ggtree to Visualize Data on Tree-Like Structures. Curr. Protoc. Bioinform. 2020, 69. [Google Scholar] [CrossRef]
- Yu, G.; Smith, D.K.; Zhu, H.; Guan, Y.; Lam, T.T.Y. Ggtree: An R Package for Visualization and Annotation of Phylogenetic Trees with Their Covariates and Other Associated Data. Methods Ecol. Evol. 2017, 8, 28–36. [Google Scholar] [CrossRef]
- Yu, G.; Lam, T.T.-Y.; Zhu, H.; Guan, Y. Two Methods for Mapping and Visualizing Associated Data on Phylogeny Using Ggtree. Mol. Biol. Evol. 2018, 35, 3041–3043. [Google Scholar] [CrossRef] [PubMed]
- Khunnamwong, P.; Lertwattanasakul, N.; Jindamorakot, S.; Limtong, S.; Lachance, M.A. Description of Diutina Gen. Nov Diutina Siamensis, f.a. sp. Nov and Reassignment of Candida Catenulata, Candida Mesorugosa, Candida Neorugosa, Candida Pseudorugosa, Candida Ranongensis, Candida Rugosa and Candida Scorzettiae to the Genus Diutina. Int. J. Syst. Evol. Microbiol. 2015, 65, 4701–4709. [Google Scholar] [CrossRef]
- Fonseca, A.; Scorzetti, G.; Fell, J.W. Diversity in the Yeast Cryptococcus Albidus and Related Species as Revealed by Ribosomal DNA Sequence Analysis. Can. J. Microbiol. 1999, 46, 7–27. [Google Scholar] [CrossRef]
- Liu, X.Z.; Wang, Q.M.; Göker, M.; Groenewald, M.; Kachalkin, A.V.; Lumbsch, H.T.; Millanes, A.M.; Wedin, M.; Yurkov, A.M.; Boekhout, T.; et al. Towards an Integrated Phylogenetic Classification of the Tremellomycetes. Stud. Mycol. 2015, 81, 85–147. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ciafardini, G.; Zullo, B.A.; Antonielli, L.; Corte, L.; Roscini, L.; Cardinali, G. Yamadazyma Terventina sp. nov., a Yeast Species of the Yamadazyma Clade from Italian Olive Oils. Int. J. Syst. Evol. Microbiol. 2013, 63, 372–376. [Google Scholar] [CrossRef]
- Nilsson, R.H.; Ryberg, M.; Kristiansson, E.; Abarenkov, K.; Larsson, K.-H.; Koljalg, U. Taxonomic Reliability of DNA Sequences in Public Sequence Databases: A Fungal Perspective. PLoS ONE 2006, 1, e59. [Google Scholar] [CrossRef]
- Bridge, P.D.; Roberts, P.J.; Spooner, B.M.; Panchal, G. On the Unreliability of Published D.N.A. Sequences. New Phytol. 2003, 160, 43–48. [Google Scholar] [CrossRef] [Green Version]
- Liu, H.; Guo, X.; Gooneratne, R.; Lai, R.; Zeng, C.; Zhan, F.; Wang, W. The Gut Microbiome and Degradation Enzyme Activity of Wild Freshwater Fishes Influenced by Their Trophic Levels. Sci. Rep. 2016, 6, 24340. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fernandez-Reiriz, M.J.; Labarta, U.; Ferreiro, M.J. Effects of Commercial Enrichment Diets on the Nutritional Value of the Rotifer (Brachionus Plicatilis). Aquaculture 1993, 112, 195–206. [Google Scholar] [CrossRef] [Green Version]
- Hamre, K. Nutrient Profiles of Rotifers (Brachionus Sp.) and Rotifer Diets from Four Different Marine Fish Hatcheries. Aquaculture 2016, 450, 136–142. [Google Scholar] [CrossRef]
- Kolkovski, S. Digestive Enzymes in Fish Larvae and Juveniles—Implications and Applications to Formulated Diets. Aquaculture 2001, 200, 181–201. [Google Scholar] [CrossRef]
- Waché, Y.; Auffray, F.; Gatesoupe, F.J.; Zambonino, J.; Gayet, V.; Labbé, L.; Quentel, C. Cross Effects of the Strain of Dietary Saccharomyces Cerevisiae and Rearing Conditions on the Onset of Intestinal Microbiota and Digestive Enzymes in Rainbow Trout, Onchorhynchus Mykiss, Fry. Aquaculture 2006, 258, 470–478. [Google Scholar] [CrossRef] [Green Version]
- Tovar-Ramírez, D.; Zambonino Infante, J.; Cahu, C.; Gatesoupe, F.J.; Vázquez-Juárez, R. Influence of Dietary Live Yeast on European Sea Bass (Dicentrarchus Labrax) Larval Development. Aquaculture 2004, 234, 415–427. [Google Scholar] [CrossRef] [Green Version]
- Buts, J.-P.; De Keyser, N.; Stilmant, C.; Sokal, E.; Marandi, S. Saccharomyces Boulardii Enhances N-Terminal Peptide Hydrolysis in Suckling Rat Small Intestine by Endoluminal Release of a Zinc-Binding Metalloprotease. Pediatric Res. 2002, 51, 528–534. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Burbank, D.R.; Shah, D.H.; LaPatra, S.E.; Fornshell, G.; Cain, K.D. Enhanced Resistance to Coldwater Disease Following Feeding of Probiotic Bacterial Strains to Rainbow Trout (Oncorhynchus Mykiss). Aquaculture 2011, 321, 185–190. [Google Scholar] [CrossRef]
- Nandi, A.; Dan, S.K.; Banerjee, G.; Ghosh, P.; Ghosh, K.; Ringø, E.; Ray, A.K. Probiotic Potential of Autochthonous Bacteria Isolated from the Gastrointestinal Tract of Four Freshwater Teleosts. Probiotics Antimicrob. Proteins 2017, 9, 12–21. [Google Scholar] [CrossRef] [PubMed]
- Bardone, E.; Bravi, M.; Keshavarz, T.; Rodriguez-Mateus, Z.; Agualimpia, B.; Zafra, G. Isolation and Molecular Characterization of Microorganisms with Potential for the Degradation of Oil and Grease from Palm Oil Refinery Wastes. Chem. Eng. Trans. 2016, 49, 517–522. [Google Scholar]
- Ide-Pérez, M.R.; Fernández-López, M.G.; Sánchez-Reyes, A.; Leija, A.; Batista-García, R.A.; Folch-Mallol, J.L.; Sánchez-Carbente, M.d.R. Aromatic Hydrocarbon Removal by Novel Extremotolerant Exophiala and Rhodotorula spp. from an Oil Polluted Site in Mexico. J. Fungi 2020, 6, 135. [Google Scholar] [CrossRef]
- Hernalsteens, S.; Maugeri, F. Purification and Characterisation of a Fructosyltransferase from Rhodotorula sp. Appl. Microbiol. Biotechnol. 2008, 79, 589–596. [Google Scholar] [CrossRef] [PubMed]
- Limtong, S.; Kaewwichian, R.; Yongmanitchai, W.; Kawasaki, H. Diversity of Culturable Yeasts in Phylloplane of Sugarcane in Thailand and Their Capability to Produce Indole-3-Acetic Acid. World J. Microbiol. Biotechnol. 2014, 30, 1785–1796. [Google Scholar] [CrossRef] [PubMed]
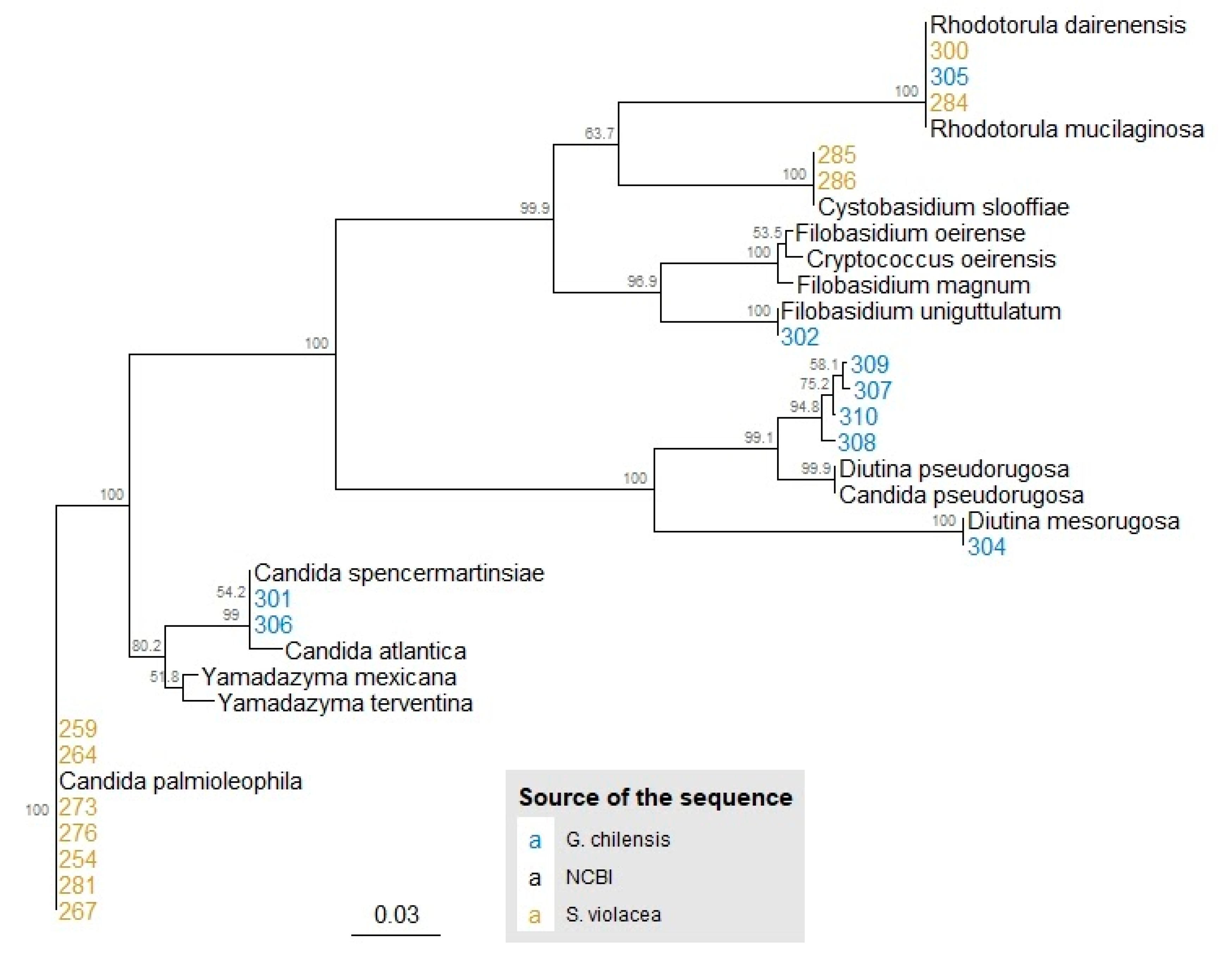
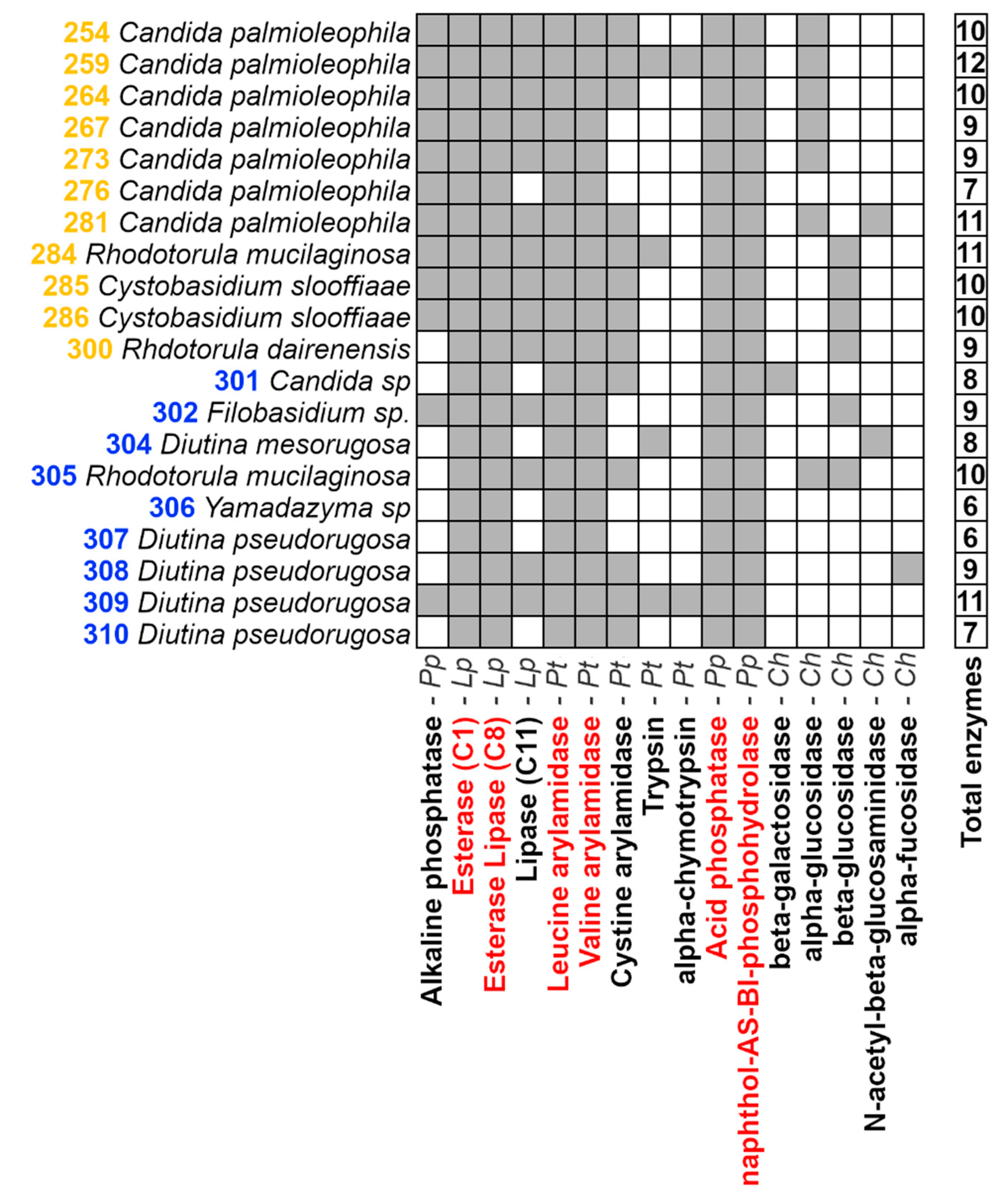
| Fish Species | Fish Specimen | Isolate ID | Length of the Sequence (bp) | Closest Relative (NCBI Access Code of the Best Match) | Identity (%) | Query Cover (%) |
|---|---|---|---|---|---|---|
| S. violacea | 1 | 254 | 960 | Candida palmioleophila (KJ705005.1) a | 99.8 | 100 |
| 1 | 276 | 990 | Candida palmioleophila (KJ705005.1) a | 99.7 | 100 | |
| 1 | 273 | 972 | Candida palmioleophila (KJ705005.1) a | 99.5 | 100 | |
| 1 | 264 | 1029 | Candida palmioleophila (KJ705005.1) a | 99.8 | 100 | |
| 1 | 281 | 925 | Candida palmioleophila (KJ705005.1) a | 99.9 | 100 | |
| 1 | 267 | 1028 | Candida palmioleophila (KJ705005.1) a | 99.3 | 100 | |
| 1 | 259 | 1037 | Candida palmioleophila (KJ705005.1) a | 99.9 | 100 | |
| S. violacea | 2 | 284 | 979 | Rhodotorula mucilaginosa (MN006818.1) b | 99.7 | 99 |
| S. violacea | 3 | 300 | 976 | Rhodotorula dairenensis (AB026010.2) b | 99.6 | 100 |
| S. violacea | 4 | 285 | 1011 | Cystobasidium slooffiae (AB025994.2) b | 99.8 | 99 |
| S. violacea | 5 | 286 | 799 | Cystobasidium slooffiae (AB025994.2) b | 99.6 | 100 |
| G. chilensis | 1 | 301 | 626 | Candida sp. (KP794187.1 and KY101952.1) a | 99.3 99.3 | 94 94 |
| 1 | 302 | 1065 | Filobasidium sp. (KX067801.1) b | 94.2 | 97 | |
| G. chilensis | 2 | 304 | 841 | Diutina mesorugosa (KY464166.1) a | 99.9 | 100 |
| G. chilensis | 3 | 305 | 1018 | Rhodotorula mucilaginosa (MN006818.1) b | 99.8 | 99 |
| G. chilensis | 4 | 306 | 919 | Yamadazyma sp. (JQ247716.1) a | 94.3 | 100 |
| G. chilensis | 5 | 307 | 821 | Candida pseudorugosa (KT336718.1) a Diutina pseudorugosa (MK394157.1) a | 97.7 97.7 | 99 99 |
| 5 | 308 | 828 | Candida pseudorugosa (KT336718.1) a Diutina pseudorugosa (MK394157.1) a | 98.6 98.6 | 96 96 | |
| 5 | 309 | 849 | Candida pseudorugosa (KT336718.1) a Diutina pseudorugosa (MK394157.1) a | 97.0 97.0 | 100 100 | |
| 5 | 310 | 803 | Candida pseudorugosa (KT336718.1) a Diutina pseudorugosa (MK394157.1) a | 98.1 98.1 | 100 100 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Valderrama, B.; Ruiz, J.J.; Gutiérrez, M.S.; Alveal, K.; Caruffo, M.; Oliva, M.; Flores, H.; Silva, A.; Toro, M.; Reyes-Jara, A.; et al. Cultivable Yeast Microbiota from the Marine Fish Species Genypterus chilensis and Seriolella violacea. J. Fungi 2021, 7, 515. https://doi.org/10.3390/jof7070515
Valderrama B, Ruiz JJ, Gutiérrez MS, Alveal K, Caruffo M, Oliva M, Flores H, Silva A, Toro M, Reyes-Jara A, et al. Cultivable Yeast Microbiota from the Marine Fish Species Genypterus chilensis and Seriolella violacea. Journal of Fungi. 2021; 7(7):515. https://doi.org/10.3390/jof7070515
Chicago/Turabian StyleValderrama, Benjamín, José J. Ruiz, María Soledad Gutiérrez, Katherine Alveal, Mario Caruffo, Marcia Oliva, Héctor Flores, Alfonso Silva, Magaly Toro, Angélica Reyes-Jara, and et al. 2021. "Cultivable Yeast Microbiota from the Marine Fish Species Genypterus chilensis and Seriolella violacea" Journal of Fungi 7, no. 7: 515. https://doi.org/10.3390/jof7070515
APA StyleValderrama, B., Ruiz, J. J., Gutiérrez, M. S., Alveal, K., Caruffo, M., Oliva, M., Flores, H., Silva, A., Toro, M., Reyes-Jara, A., & Navarrete, P. (2021). Cultivable Yeast Microbiota from the Marine Fish Species Genypterus chilensis and Seriolella violacea. Journal of Fungi, 7(7), 515. https://doi.org/10.3390/jof7070515








