Impact of Membrane Lipids on UapA and AzgA Transporter Subcellular Localization and Activity in Aspergillus nidulans
Abstract
1. Introduction
2. Materials and Methods
2.1. Media, Strains and Transformation
2.2. Nucleic Acid Manipulations and Plasmid Constructions
2.3. Protein Extraction and Western Blots
2.4. Kinetic Analysis
2.5. Epifluorescence Microscopy
3. Results and Discussion
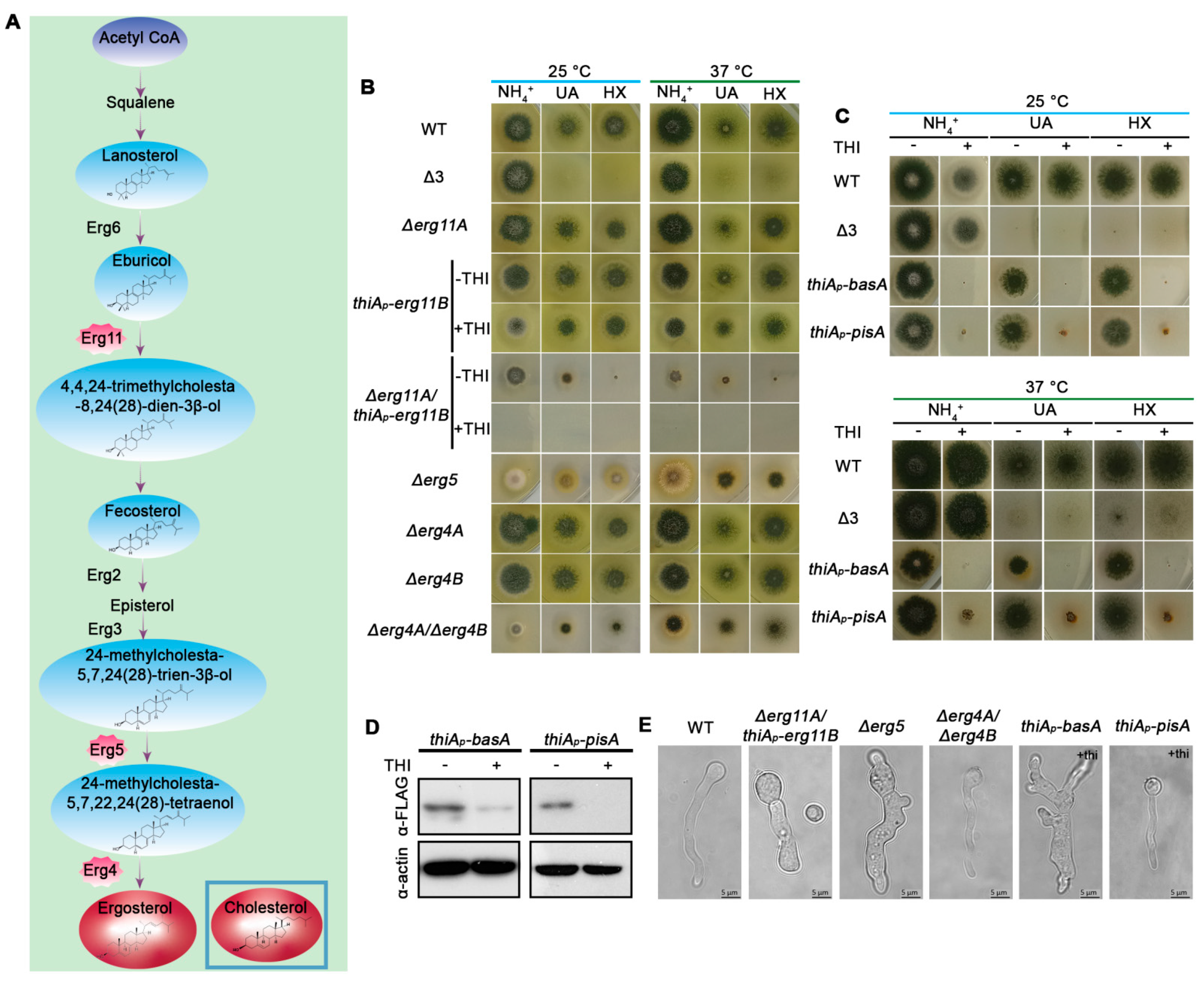
3.1. Biosynthesis of Phosphatidylinositol (PI), Sphingolipids or Ergosterol Is Essential for A. nidulans Growth
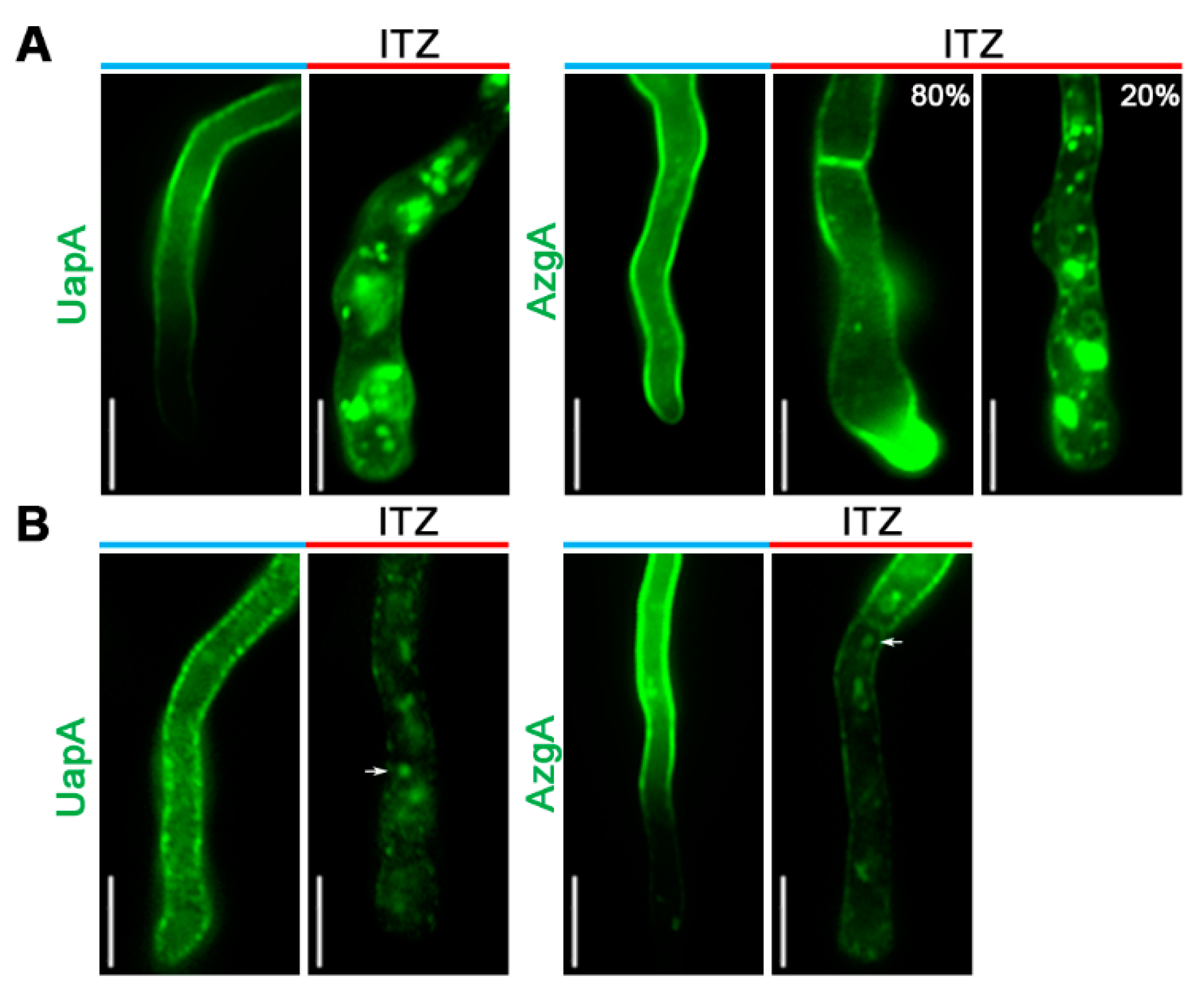
3.2. Subcellular Localization of UapA and AzgA Purine Transporters Is Differentially Affected in Specific Lipid Biosynthesis Mutants
3.3. Itraconazole Leads to Turnover of De Novo Made and PM-Localized UapA and AzgA Turnover, Mimicking the Effect of erg11 Knockdown
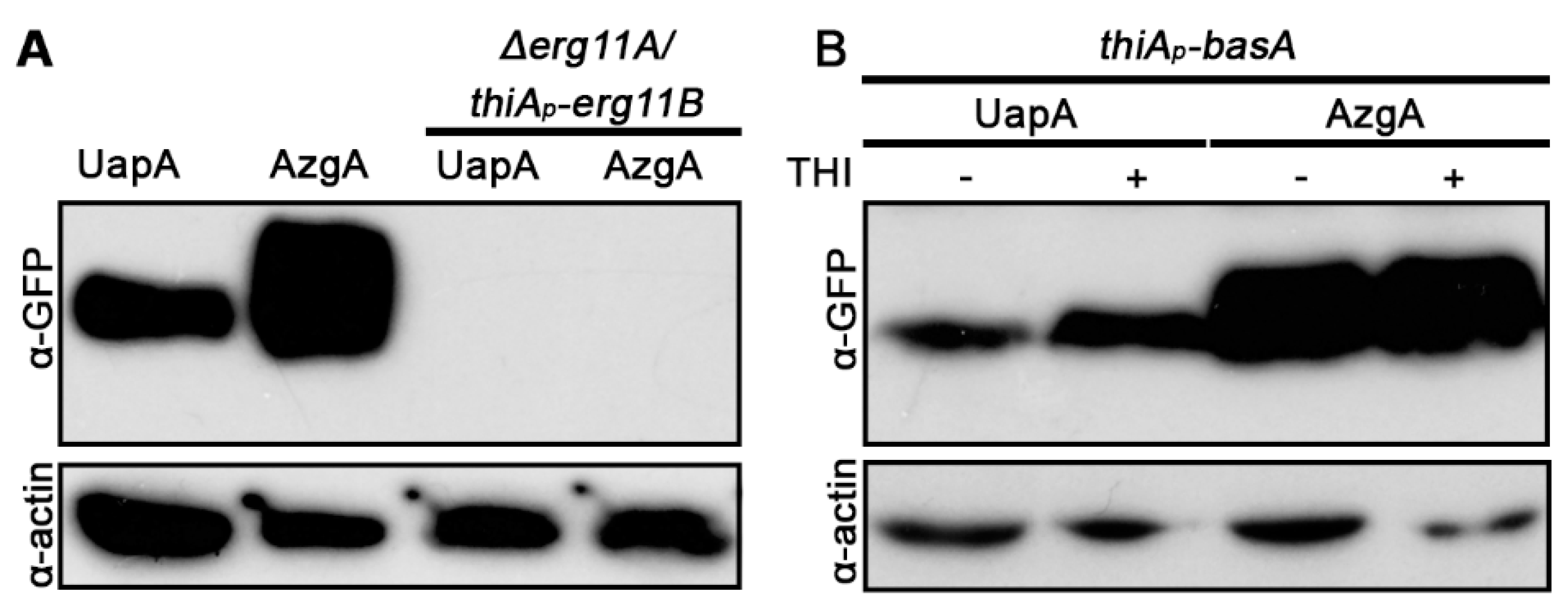
3.4. UapA and AzgA Steady State Levels Are Dramatically Reduced in erg11 Mutants, but Not When basA Is Repressed
3.5. UapA and AzgA Transport Activities Are Not Affected by Repression of Ergosterol, Sphingolipid or PI Biosynthesis
3.6. Turnover of UapA in Response to Ergosterol Depletion Occurs by Multiple Mechanisms Operating during Trafficking and after PM Translocation
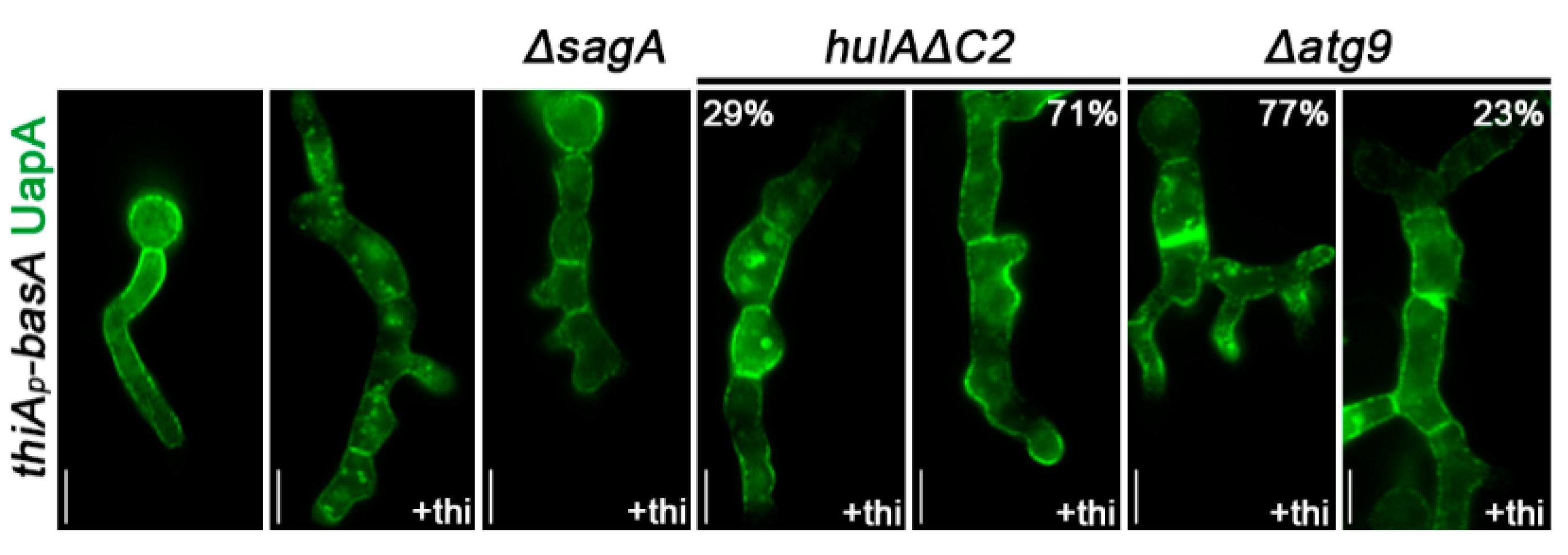
3.7. BasA-Dependent Turnover of UapA Occurs by HulA-Dependent Endocytosis and Selective Autophagy
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Aviram, N.; Schuldiner, M. Targeting and translocation of proteins to the endoplasmic reticulum at a glance. J. Cell Sci. 2017, 130, 4079–4085. [Google Scholar] [CrossRef]
- Zanetti, G.; Pahuja, K.B.; Studer, S.; Shim, S.; Schekman, R. COPII and the regulation of protein sorting in mammals. Nat. Cell Biol. 2012, 14, 20–28. [Google Scholar] [CrossRef]
- D’Arcangelo, J.G.; Stahmer, K.R.; Miller, E.A. Vesicle-mediated export from the ER: COPII coat function and regulation. Biochim. Biophys. Acta Mol. Cell Res. 2013, 1833, 2464–2472. [Google Scholar] [CrossRef]
- Borgese, N. Getting membrane proteins on and off the shuttle bus between the endoplasmic reticulum and the Golgi complex. J. Cell Sci. 2016, 129, 1537–1545. [Google Scholar] [CrossRef] [PubMed]
- Gee, H.Y.; Kim, J.; Lee, M.G. Unconventional secretion of transmembrane proteins. Semin. Cell Dev. Biol. 2018, 83. [Google Scholar] [CrossRef] [PubMed]
- Dimou, S.; Diallinas, G. Life and death of fungal transporters under the challenge of polarity. Int. J. Mol. Sci. 2020, 21, 5376. [Google Scholar] [CrossRef] [PubMed]
- Diallinas, G.; Martzoukou, O. Transporter membrane traffic and function: Lessons from a mould. FEBS J. 2019, 286. [Google Scholar] [CrossRef] [PubMed]
- Payet, L.-A.; Pineau, L.; Snyder, E.C.R.; Colas, J.; Moussa, A.; Vannier, B.; Bigay, J.; Clarhaut, J.; Becq, F.; Berjeaud, J.-M.; et al. Saturated fatty acids alter the late secretory pathway by modulating membrane properties. Traffic 2013, 14, 1228–1241. [Google Scholar] [CrossRef]
- Koshy, C.; Ziegler, C. Structural insights into functional lipid-protein interactions in secondary transporters. Biochim. Biophys. Acta Gen. Subj. 2015, 1850, 476–487. [Google Scholar] [CrossRef]
- Martens, C.; Shekhar, M.; Borysik, A.J.; Lau, A.M.; Reading, E.; Tajkhorshid, E.; Booth, P.J.; Politis, A. Direct protein-lipid interactions shape the conformational landscape of secondary transporters. Nat. Commun. 2018, 9, 4151. [Google Scholar] [CrossRef]
- Kourkoulou, A.; Grevias, P.; Lambrinidis, G.; Pyle, E.; Dionysopoulou, M.; Politis, A.; Mikros, E.; Byrne, B.; Diallinas, G. Specific residues in a purine transporter are critical for dimerization, ER Exit, and function. Genetics 2019, 213, 1357–1372. [Google Scholar] [CrossRef] [PubMed]
- Stieger, B.; Steiger, J.; Locher, K.P. Membrane lipids and transporter function. Biochim. Biophys. Acta Mol. Basis Dis. 2021, 1867, 166079. [Google Scholar] [CrossRef]
- Fratti, R.A. Editorial: Effects of membrane lipids on protein function. Front. Cell Dev. Biol. 2021, 9. [Google Scholar] [CrossRef] [PubMed]
- Harayama, T.; Riezman, H. Understanding the diversity of membrane lipid composition. Nat. Rev. Mol. Cell Biol. 2018, 19, 281–296. [Google Scholar] [CrossRef] [PubMed]
- Blom, T.; Somerharju, P.; Ikonen, E. Synthesis and biosynthetic trafficking of membrane lipids. Cold Spring Harb. Perspect. Biol. 2011, 3, a004713. [Google Scholar] [CrossRef] [PubMed]
- Lee, A. Lipid-protein interactions in biological membranes: A structural perspective. Biochim. Biophys. Acta Biomembr. 2003, 1612, 1–40. [Google Scholar] [CrossRef]
- Contreras, F.-X.; Ernst, A.M.; Wieland, F.; Brugger, B. Specificity of intramembrane protein-lipid interactions. Cold Spring Harb. Perspect. Biol. 2011, 3, a004705. [Google Scholar] [CrossRef]
- Corin, K.; Bowie, J.U. How bilayer properties influence membrane protein folding. Protein Sci. 2020, 29, 2348–2362. [Google Scholar] [CrossRef] [PubMed]
- Lindblom, G.; Orädd, G. Lipid lateral diffusion and membrane heterogeneity. Biochim. Biophys. Acta Biomembr. 2009, 1788, 234–244. [Google Scholar] [CrossRef]
- Malinsky, J.; Opekarová, M.; Tanner, W. The lateral compartmentation of the yeast plasma membrane. Yeast 2010, 27, 473–478. [Google Scholar] [CrossRef] [PubMed]
- Athanasopoulos, A.; André, B.; Sophianopoulou, V.; Gournas, C. Fungal plasma membrane domains. FEMS Microbiol. Rev. 2019, 43, 642–673. [Google Scholar] [CrossRef]
- Brach, T.; Specht, T.; Kaksonen, M. Reassessment of the role of plasma membrane domains in the regulation of vesicular traffic in yeast. J. Cell Sci. 2011, 124, 328–337. [Google Scholar] [CrossRef] [PubMed]
- Bianchi, F.; van’t Klooster, J.S.; Ruiz, S.J.; Poolman, B. Regulation of amino acid transport in Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 2019, 83, e00024-19. [Google Scholar] [CrossRef]
- Laganowsky, A.; Reading, E.; Allison, T.M.; Ulmschneider, M.B.; Degiacomi, M.T.; Baldwin, A.J.; Robinson, C.V. Membrane proteins bind lipids selectively to modulate their structure and function. Nature 2014, 510, 172–175. [Google Scholar] [CrossRef]
- Corradi, V.; Sejdiu, B.I.; Mesa-Galloso, H.; Abdizadeh, H.; Noskov, S.Y.; Marrink, S.J.; Tieleman, D.P. Emerging diversity in lipid-protein interactions. Chem. Rev. 2019, 119, 5775–5848. [Google Scholar] [CrossRef]
- Bolla, J.R.; Agasid, M.T.; Mehmood, S.; Robinson, C.V. Membrane protein–lipid interactions probed using mass spectrometry. Annu. Rev. Biochem. 2019, 88, 85–111. [Google Scholar] [CrossRef]
- Lauwers, E.; Grossmann, G.; André, B. Evidence for coupled biogenesis of yeast Gap1 permease and sphingolipids: Essential role in transport activity and normal control by ubiquitination. Mol. Biol. Cell 2007, 18, 3068–3080. [Google Scholar] [CrossRef][Green Version]
- Wang, Q.; Chang, A. Sphingoid base synthesis is required for oligomerization and cell surface stability of the yeast plasma membrane ATPase, Pma1. Proc. Natl. Acad. Sci. USA 2002, 99, 12853–12858. [Google Scholar] [CrossRef] [PubMed]
- Lee, M.C.S.; Hamamoto, S.; Schekman, R. Ceramide biosynthesis is required for the formation of the oligomeric H+-ATPase Pma1p in the yeast endoplasmic reticulum. J. Biol. Chem. 2002, 277, 22395–22401. [Google Scholar] [CrossRef] [PubMed]
- Toulmay, A.; Schneiter, R. Lipid-dependent surface transport of the proton pumping ATPase: A model to study plasma membrane biogenesis in yeast. Biochimie 2007, 89, 249–254. [Google Scholar] [CrossRef]
- Gaigg, B.; Timischl, B.; Corbino, L.; Schneiter, R. Synthesis of sphingolipids with very long chain fatty acids but not ergosterol is required for routing of newly synthesized plasma membrane ATPase to the cell surface of yeast. J. Biol. Chem. 2005, 280, 22515–22522. [Google Scholar] [CrossRef]
- Umebayashi, K.; Nakano, A. Ergosterol is required for targeting of tryptophan permease to the yeast plasma membrane. J. Cell Biol. 2003, 161, 1117–1131. [Google Scholar] [CrossRef] [PubMed]
- Pineau, L.; Bonifait, L.; Berjeaud, J.-M.; Alimardani-Theuil, P.; Bergès, T.; Ferreira, T. A lipid-mediated quality control process in the golgi apparatus in yeast. Mol. Biol. Cell 2008, 19, 807–821. [Google Scholar] [CrossRef] [PubMed]
- Gournas, C.; Gkionis, S.; Carquin, M.; Twyffels, L.; Tyteca, D.; André, B. Conformation-dependent partitioning of yeast nutrient transporters into starvation-protective membrane domains. Proc. Natl. Acad. Sci. USA 2018, 115, E3145–E3154. [Google Scholar] [CrossRef] [PubMed]
- Diallinas, G. Dissection of transporter function: From genetics to structure. Trends Genet. 2016, 32, 576–590. [Google Scholar] [CrossRef] [PubMed]
- Alguel, Y.; Amillis, S.; Leung, J.; Lambrinidis, G.; Capaldi, S.; Scull, N.J.; Craven, G.; Iwata, S.; Armstrong, A.; Mikros, E.; et al. Structure of eukaryotic purine/H þ symporter UapA suggests a role for homodimerization in transport activity. Nat. Commun. 2016, 7, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Martzoukou, O.; Karachaliou, M.; Yalelis, V.; Leung, J.; Byrne, B.; Amillis, S.; Diallinas, G. Oligomerization of the UapA purine transporter is critical for ER-exit, plasma membrane localization and turnover. J. Mol. Biol. 2015, 427, 2679–2696. [Google Scholar] [CrossRef] [PubMed]
- Pyle, E.; Kalli, A.C.; Amillis, S.; Hall, Z.; Lau, A.M.; Hanyaloglu, A.C.; Diallinas, G.; Byrne, B.; Politis, A. Structural lipids enable the formation of functional oligomers of the eukaryotic purine symporter UapA. Cell Chem. Biol. 2018, 25, 840–848.e4. [Google Scholar] [CrossRef]
- Krypotou, E.; Lambrinidis, G.; Evangelidis, T.; Mikros, E.; Diallinas, G. Modelling, substrate docking and mutational analysis identify residues essential for function and specificity of the major fungal purine transporter AzgA. Mol. Microbiol. 2014, 93, 129–145. [Google Scholar] [CrossRef] [PubMed]
- Apostolaki, A.; Harispe, L.; Calcagno-Pizarelli, A.M.; Vangelatos, I.; Sophianopoulou, V.; Arst, H.N., Jr.; Peñalva, M.A.; Amillis, S.; Scazzocchio, C. Aspergillus nidulans CkiA is an essential casein kinase I required for delivery of amino acid transporters to the plasma membrane. Mol. Microbiol. 2012, 84, 530–549. [Google Scholar] [CrossRef]
- Koukaki, M.; Giannoutsou, E.; Karagouni, A.; Diallinas, G. A novel improved method for Aspergillus nidulans transformation. J. Microbiol. Methods 2003, 55, 687–695. [Google Scholar] [CrossRef]
- Nayak, T.; Szewczyk, E.; Oakley, C.E.; Osmani, A.; Ukil, L.; Murray, S.L.; Hynes, M.J.; Osmani, S.A.; Oakley, B.R. A versatile and efficient gene-targeting system for aspergillus nidulans. Genetics 2006, 172, 1557–1566. [Google Scholar] [CrossRef] [PubMed]
- Yu, J.-H.; Hamari, Z.; Han, K.-H.; Seo, J.-A.; Reyes-Domínguez, Y.; Scazzocchio, C. Double-joint PCR: A PCR-based molecular tool for gene manipulations in filamentous fungi. Fungal Genet. Biol. 2004, 41, 973–981. [Google Scholar] [CrossRef] [PubMed]
- Galanopoulou, K.; Scazzocchio, C.; Galinou, M.E.; Liu, W.; Borbolis, F.; Karachaliou, M.; Oestreicher, N.; Hatzinikolaou, D.G.; Diallinas, G.; Amillis, S. Purine utilization proteins in the Eurotiales: Cellular compartmentalization, phylogenetic conservation and divergence. Fungal Genet. Biol. 2014, 69, 96–108. [Google Scholar] [CrossRef] [PubMed]
- Krypotou, E.; Diallinas, G. Transport assays in filamentous fungi: Kinetic characterization of the UapC purine transporter of Aspergillus nidulans. Fungal Genet. Biol. 2014, 63, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Martzoukou, O.; Amillis, S.; Zervakou, A.; Christoforidis, S.; Diallinas, G. The AP-2 complex has a specialized clathrin-independent role in apical endocytosis and polar growth in fungi. eLife 2017, 6, e20083. [Google Scholar] [CrossRef] [PubMed]
- Dimou, S.; Martzoukou, O.; Dionysopoulou, M.; Bouris, V.; Amillis, S.; Diallinas, G. Translocation of nutrient transporters to cell membrane via Golgi bypass in Aspergillus nidulans. EMBO Rep. 2020, 21, e49929. [Google Scholar] [CrossRef] [PubMed]
- Karachaliou, M.; Amillis, S.; Evangelinos, M.; Kokotos, A.C.; Yalelis, V.; Diallinas, G. The arrestin-like protein ArtA is essential for ubiquitination and endocytosis of the UapA transporter in response to both broad-range and specific signals. Mol. Microbiol. 2013, 88, 301–317. [Google Scholar] [CrossRef] [PubMed]
- Martzoukou, O.; Diallinas, G.; Amillis, S. Secretory vesicle polar sorting, endosome recycling and cytoskeleton organization require the AP-1 complex in Aspergillus nidulans. Genetics 2018, 209, 1121–1138. [Google Scholar] [CrossRef]
- Amillis, S.; Cecchetto, G.; Sophianopoulou, V.; Koukaki, M.; Scazzocchio, C.; Diallinas, G. Transcription of purine transporter genes is activated during the isotropic growth phase of Aspergillus nidulans conidia. Mol. Microbiol. 2004, 52, 205–216. [Google Scholar] [CrossRef]
- Nikawa, J.; Kodaki, T.; Yamashita, S. Primary structure and disruption of the phosphatidylinositol synthase gene of Saccharomyces cerevisiae. J. Biol. Chem. 1987, 262, 4876–4881. [Google Scholar] [CrossRef]
- Li, S.; Bao, D.; Yuen, G.; Harris, S.D.; Calvo, A.M. basA regulates cell wall organization and asexual/sexual sporulation ratio in aspergillus nidulans. Genetics 2007, 176, 243–253. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Li, S.; Du, L.; Yuen, G.; Harris, S.D. Distinct ceramide synthases regulate polarized growth in the filamentous fungus Aspergillus nidulans. Mol. Biol. Cell 2006, 17, 1218–1227. [Google Scholar] [CrossRef] [PubMed]
- Osherov, N. Resistance to itraconazole in Aspergillus nidulans and Aspergillus fumigatus is conferred by extra copies of the A. nidulans P-450 14alpha-demethylase gene, pdmA. J. Antimicrob. Chemother. 2001, 48, 75–81. [Google Scholar] [CrossRef] [PubMed]
- Alcazar-Fuoli, L.; Mellado, E.; Garcia-Effron, G.; Lopez, J.F.; Grimalt, J.O.; Cuenca-Estrella, J.M.; Rodriguez-Tudela, J.L. Ergosterol biosynthesis pathway in Aspergillus fumigatus. Steroids 2008, 73, 339–347. [Google Scholar] [CrossRef] [PubMed]
- Alcazar-Fuoli, L.; Mellado, E. Ergosterol biosynthesis in Aspergillus fumigatus: Its relevance as an antifungal target and role in antifungal drug resistance. Front. Microbiol. 2013, 3, 439. [Google Scholar] [CrossRef]
- Abenza, J.F.; Galindo, A.; Pantazopoulou, A.; Gil, C.; de los Ríos, V.; Peñalva, M.A. Aspergillus RabB Rab5 integrates acquisition of degradative identity with the long distance movement of early endosomes. Mol. Biol. Cell 2010, 21, 2756–2769. [Google Scholar] [CrossRef] [PubMed]
- Gournas, C.; Amillis, S.; Vlanti, A.; Diallinas, G. Transport-dependent endocytosis and turnover of a uric acid-xanthine permease. Mol. Microbiol. 2010, 75, 246–260. [Google Scholar] [CrossRef]
- Diallinas, G.; Rafailidou, N.; Kalpaktsi, I.; Komianou, A.C.; Tsouvali, V.; Zantza, I.; Mikros, E.; Skaltsounis, A.L.; Kostakis, I.K. Hydroxytyrosol (HT) analogs act as potent antifungals by direct disruption of the fungal cell membrane. Front. Microbiol. 2018, 9, 2624. [Google Scholar] [CrossRef]
- Evangelinos, M.; Martzoukou, O.; Chorozian, K.; Amillis, S.; Diallinas, G. BsdA Bsd2 -dependent vacuolar turnover of a misfolded version of the UapA transporter along the secretory pathway: Prominent role of selective autophagy. Mol. Microbiol. 2016, 100, 893–911. [Google Scholar] [CrossRef]
- Babst, M. Quality control at the plasma membrane: One mechanism does not fit all. J. Cell Biol. 2014, 205, 11–20. [Google Scholar] [CrossRef] [PubMed]
- Sardana, R.; Zhu, L.; Emr, S.D. Rsp5 Ubiquitin ligase–mediated quality control system clears membrane proteins mistargeted to the vacuole membrane. J. Cell Biol. 2019, 218, 234–250. [Google Scholar] [CrossRef] [PubMed]
- Wilfling, F.; Lee, C.-W.; Erdmann, P.S.; Zheng, Y.; Sherpa, D.; Jentsch, S.; Pfander, B.; Schulman, B.A.; Baumeister, W. A selective autophagy pathway for phase-separated endocytic protein deposits. Mol. Cell 2020, 80, 764–778.e7. [Google Scholar] [CrossRef] [PubMed]


| Xanthine Uptake Kinetics | UapA Localization | Adenine Uptake Kinetics | AzgA Localization | |||
|---|---|---|---|---|---|---|
| Allele | Km (μΜ) | V (%) | Km (μΜ) | V (%) | ||
| wild-type | 5 ± 1 | 100 ± 12 | PM | 3 ± 1 | 100 ± 9 | PM |
| Δerg4A/Β | 4 ± 1 | 68 ± 14 | PM | n.d. | n.d. | PM |
| Δerg5 | 4 ± 2 | 101 ± 12 | PM | n.d. | n.d. | PM |
| Δerg11A | n.d. | 105 ± 10 | n.d. | n.d. | n.d. | n.d. |
| thiAp-erg11B * | n.d. | 95 ± 5 | n.d. | n.d. | n.d. | n.d. |
| Δerg11A/ thiAp-erg11B * | 2 ± 1 | 21 ± 3 | cytoplasmic puncta/foci | 7 ± 2 | 38 ± 9 | PM/cytoplasmic puncta/ foci |
| thiAp-basA * | 7 ± 1 | 57 ± 6 | PM/cytoplasmic puncta/ foci | 2 ± 1 | 99 ± 10 | PM |
| thiAp-pisA * | 2 ± 1 | 94 ± 14 | PM | 2 ± 1 | 104 ± 12 | PM |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dionysopoulou, M.; Diallinas, G. Impact of Membrane Lipids on UapA and AzgA Transporter Subcellular Localization and Activity in Aspergillus nidulans. J. Fungi 2021, 7, 514. https://doi.org/10.3390/jof7070514
Dionysopoulou M, Diallinas G. Impact of Membrane Lipids on UapA and AzgA Transporter Subcellular Localization and Activity in Aspergillus nidulans. Journal of Fungi. 2021; 7(7):514. https://doi.org/10.3390/jof7070514
Chicago/Turabian StyleDionysopoulou, Mariangela, and George Diallinas. 2021. "Impact of Membrane Lipids on UapA and AzgA Transporter Subcellular Localization and Activity in Aspergillus nidulans" Journal of Fungi 7, no. 7: 514. https://doi.org/10.3390/jof7070514
APA StyleDionysopoulou, M., & Diallinas, G. (2021). Impact of Membrane Lipids on UapA and AzgA Transporter Subcellular Localization and Activity in Aspergillus nidulans. Journal of Fungi, 7(7), 514. https://doi.org/10.3390/jof7070514







