Characterisation of Aspergillus fumigatus Endocytic Trafficking within Airway Epithelial Cells Using High-Resolution Automated Quantitative Confocal Microscopy
Abstract
1. Introduction
2. Materials and Methods
2.1. Fungal Strains
2.2. Cell Lines
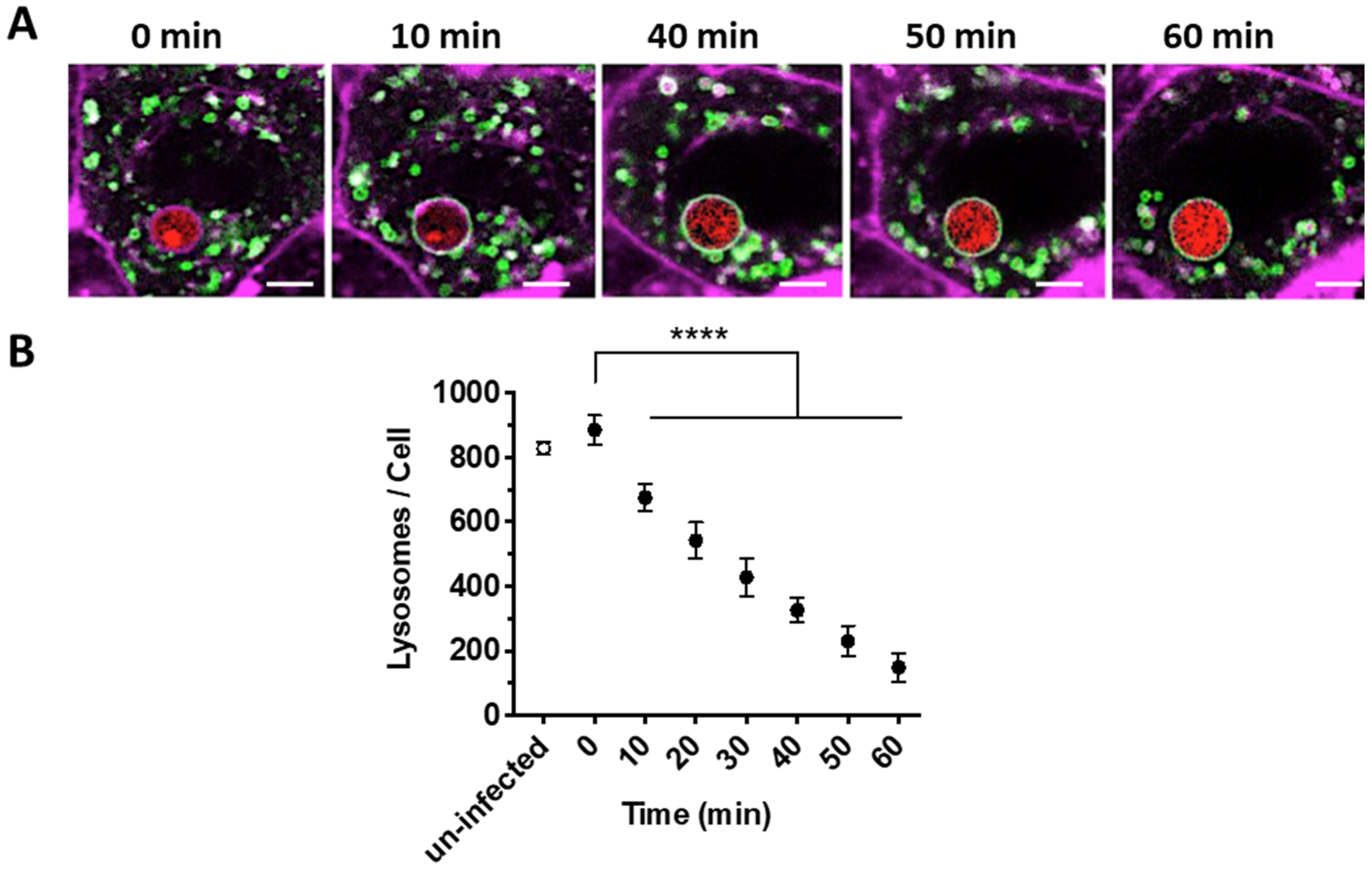
2.3. Quantification of the Number of Lysosomes during A. fumigatus Infection of Lung Epithelial Cells
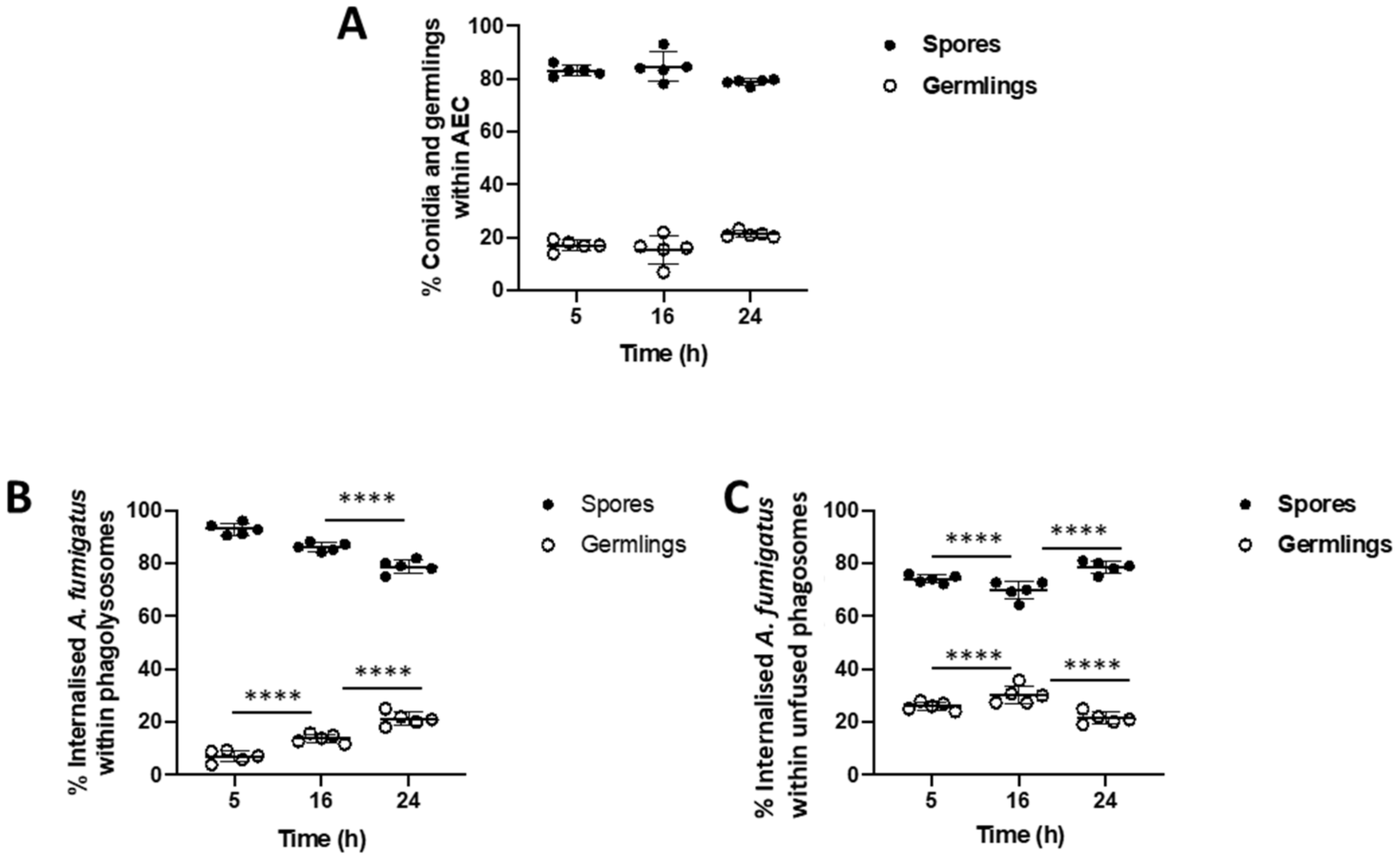
2.4. Quantification of the Rate of Lysosome—Phagosome Fusion during A. fumigatus Infection of Epithelial Cells
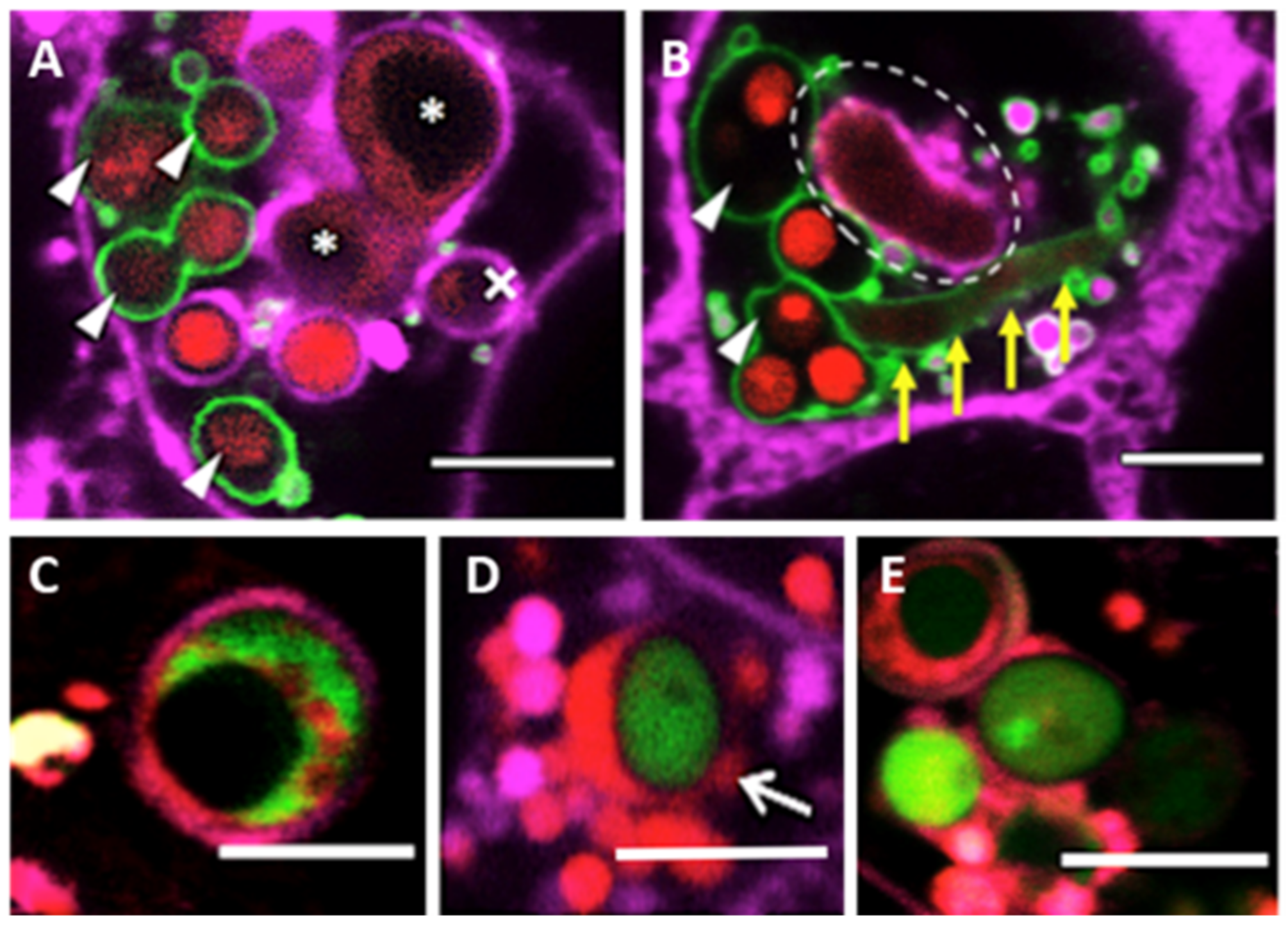
2.5. Determining A. fumigatus Viability within Phagolysosomes by Confocal Microscopy
2.6. Assessing the Impact of A. fumigatus Cell Wall Components on Phagosome-Lysosome Fusion Rates in the Lung Epithelium
2.7. Determining Synchronization of Phagosome Acidification and Phagosome Maturation upon A. fumigatus Infection of the Lung Epithelium
2.8. Fungal Killing Assays
2.9. siRNA Treatment
2.10. Image Acquisition and Data Analyses
2.11. Statistical Analyses
3. Results
3.1. Rate of Lysosome Recruitment to the Phagosome Determines A. fumigatus Fate within the Lung Alveolar Epithelial Cells
3.2. Bronchial Epithelial Cell Phagolysosomes Demonstrate Increased Fungal-Killing, Compared to Alveolar Cells
3.3. Phagolysosome Maturation Relies on A. fumigatus Cell Wall Integrity
3.4. Validation of High Content Live-Cell Imaging on vATPase Silenced Lung Epithelial Cells
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Bongomin, F.; Gago, S.; Oladele, R.O.; Denning, D.W. Global and multi-national prevalence of fungal diseases—Estimate precision. J. Fungi 2017, 3, 57. [Google Scholar] [CrossRef]
- Brown, G.D.; Denning, D.W.; Gow, N.A.R.; Levitz, S.M.; Netea, M.G.; White, T.C. Hidden killers: Human fungal infections. Sci. Transl. Med. 2012, 4, 165rv13. [Google Scholar] [CrossRef]
- Latge, J.P.; Chamilos, G. Aspergillus fumigatus and Aspergillosis in 2019. Clin. Microbiol. Rev. 2019, 33, e00140-18. [Google Scholar] [CrossRef]
- Van De Veerdonk, F.L.; Gresnigt, M.S.; Romani, L.; Netea, M.G.; Latgé, J.P. Aspergillus fumigatus morphology and dynamic host interactions. Nat. Rev. Microbiol. 2017, 15, 661–674. [Google Scholar] [CrossRef]
- Heinekamp, T.; Schmidt, H.; Lapp, K.; Pähtz, V.; Shopova, I.; Köster-Eiserfunke, N.; Krüger, T.; Kniemeyer, O.; Brakhage, A.A. Interference of Aspergillus fumigatus with the immune response. Semin. Immunopathol. 2015, 37, 141–152. [Google Scholar] [CrossRef] [PubMed]
- Margalit, A.; Kavanagh, K. The innate immune response to Aspergillus fumigatus at the alveolar surface. FEMS Microbiol. Rev. 2015, 39, 670–687. [Google Scholar] [CrossRef]
- Herbrecht, R.; Bories, P.; Moulin, J.C.; Ledoux, M.P.; Letscher-Bru, V. Risk stratification for invasive aspergillosis in immunocompromised patients. Ann. N. Y. Acad. Sci. 2012, 1272, 23–30. [Google Scholar] [CrossRef] [PubMed]
- Smith, N.L.; Denning, D.W. Underlying conditions in chronic pulmonary aspergillosis including simple aspergilloma. Eur. Respir. J. 2011, 37, 865–872. [Google Scholar] [CrossRef] [PubMed]
- Denning, D.W.; Cadranel, J.; Beigelman-Aubry, C.; Ader, F.; Chakrabarti, A.; Blot, S.; Ullmann, A.J.; Dimopoulos, G.; Lange, C. Chronic pulmonary aspergillosis: Rationale and clinical guidelines for diagnosis and management. Eur. Respir. J. 2016, 47, 45–68. [Google Scholar] [CrossRef]
- Vanderbeke, L.; Spriet, I.; Breynaert, C.; Rijnders, B.J.A.; Verweij, P.E.; Wauters, J. Invasive pulmonary aspergillosis complicating severe influenza: Epidemiology, diagnosis and treatment. Curr. Opin. Infect. Dis. 2018, 31, 471–480. [Google Scholar] [CrossRef] [PubMed]
- Schauwvlieghe, A.F.A.D.; Rijnders, B.J.A.; Philips, N.; Verwijs, R.; Vanderbeke, L.; Van Tienen, C.; Lagrou, K.; Verweij, P.E.; Van de Veerdonk, F.L.; Gommers, D.; et al. Invasive aspergillosis in patients admitted to the intensive care unit with severe influenza: A retrospective cohort study. Lancet Respir. Med. 2018, 6, 782–792. [Google Scholar] [CrossRef]
- Brakhage, A.A.; Bruns, S.; Thywissen, A.; Zipfel, P.F.; Behnsen, J. Interaction of phagocytes with filamentous fungi. Curr. Opin. Microbiol. 2010, 13, 409–415. [Google Scholar] [CrossRef] [PubMed]
- Ibrahim-Granet, O.; Philippe, B.; Boleti, H.; Boisvieux-Ulrich, E.; Grenet, D.; Stern, M.; Latgé, J.P. Phagocytosis and intracellular fate of Aspergillus fumigatus conidia in alveolar macrophages. Infect. Immun. 2003, 71, 891–903. [Google Scholar] [CrossRef]
- Philippe, B.; Ibrahim-Granet, O.; Prévost, M.C.; Gougerot-Pocidalo, M.A.; Perez, M.S.; Van der Meeren, A.; Latgé, J.P. Killing of Aspergillus fumigatus by alveolar macrophages is mediated by reactive oxidant intermediates. Infect. Immun. 2003, 71, 3034–3042. [Google Scholar] [CrossRef] [PubMed]
- Steele, C.; Rapaka, R.R.; Metz, A.; Pop, S.M.; Williams, D.L.; Gordon, S.; Kolls, J.K.; Brown, G.D. The beta-glucan receptor dectin-1 recognizes specific morphologies of Aspergillus fumigatus. PLoS Pathog. 2005, 1, e42. [Google Scholar] [CrossRef]
- Amin, S.; Thywissen, A.; Heinekamp, T.; Saluz, H.P.; Brakhage, A.A. Melanin dependent survival of Apergillus fumigatus conidia in lung epithelial cells. Int. J. Med. Microbiol. 2014, 304, 626–636. [Google Scholar] [CrossRef]
- Beisswenger, C.; Hess, C.; Bals, R. Aspergillus fumigatus conidia induce interferon-β signalling in respiratory epithelial cells. Eur. Respir. J. 2012, 39, 411–418. [Google Scholar] [CrossRef]
- Clark, H.R.; Powell, A.B.; Simmons, K.A.; Ayubi, T.; Kale, S.D. Endocytic markers associated with the internalization and processing of Aspergillus fumigatus Conidia by BEAS-2B cells. mSphere 2019, 4, e00663-18. [Google Scholar] [CrossRef] [PubMed]
- Wasylnka, J.A.; Hissen, A.H.T.; Wan, A.N.C.; Moore, M.M. Intracellular and extracellular growth of Aspergillus fumigatus. Med. Mycol. 2005, 43, S27–S30. [Google Scholar] [CrossRef][Green Version]
- Chai, L.Y.A.; Netea, M.G.; Sugui, J.; Vonk, A.G.; van de Sande, W.W.J.; Warris, A.; Kwon-Chung, K.J.; Jan Kullberg, B. Aspergillus fumigatus conidial melanin modulates host cytokine response. Immunobiology 2010, 215, 915–920. [Google Scholar] [CrossRef]
- Botterel, F.; Gross, K.; Ibrahim-Granet, O.; Khoufache, K.; Escabasse, V.; Coste, A.; Cordonnier, C.; Escudier, E.; Bretagne, S. Phagocytosis of Aspergillus fumigatus conidia by primary nasal epithelial cells in vitro. BMC Microbiol. 2008, 8, 97. [Google Scholar] [CrossRef]
- Gauthier, T.; Wang, X.; Dos Santos, J.; Fysikopoulos, A.; Tadrist, S.; Canlet, C.; Artigot, M.P.; Loiseau, N.; Oswald, I.P.; Puel, O. Trypacidin, a spore-borne toxin from Aspergillus fumigatus, is cytotoxic to lung cells. PLoS ONE 2012, 7, e29906. [Google Scholar] [CrossRef] [PubMed]
- Wiesner, D.L.; Merkhofer, R.M.; Ober, C.; Kujoth, G.C.; Niu, M.; Keller, N.P.; Gern, J.E.; Brockman-Schneider, R.A.; Evans, M.D.; Jackson, D.J.; et al. Club cell TRPV4 serves as a damage sensor driving lung allergic inflammation. Cell Host Microbe 2020, 27, 614–628.e616. [Google Scholar] [CrossRef] [PubMed]
- Hernández-Santos, N.; Wiesner, D.L.; Fites, J.S.; McDermott, A.J.; Warner, T.; Wüthrich, M.; Klein, B.S. Lung epithelial cells coordinate innate lymphocytes and immunity against pulmonary fungal infection. Cell Host Microbe 2018, 23, 511–522.e515. [Google Scholar] [CrossRef]
- Jhingran, A.; Kasahara, S.; Shepardson, K.M.; Junecko, B.A.F.; Heung, L.J.; Kumasaka, D.K.; Knoblaugh, S.E.; Lin, X.; Kazmierczak, B.I.; Reinhart, T.A.; et al. Compartment-specific and sequential role of MyD88 and CARD9 in chemokine induction and innate defense during respiratory fungal infection. PLoS Pathog. 2015, 11, e1004589. [Google Scholar] [CrossRef] [PubMed]
- Filler, S.G.; Sheppard, D.C. Fungal invasion of normally non-phagocytic host cells. PLoS Pathog. 2006, 2, e129. [Google Scholar] [CrossRef]
- Wasylnka, J.A.; Moore, M.M. Uptake of Aspergillus fumigatus conidia by phagocytic and nonphagocytic cells in vitro: Quantitation using strains expressing green fluorescent protein. Infect. Immun. 2002, 70, 3156–3163. [Google Scholar] [CrossRef]
- Bertuzzi, M.; Schrettl, M.; Alcazar-Fuoli, L.; Cairns, T.C.; Muñoz, A.; Walker, L.A.; Herbst, S.; Safari, M.; Cheverton, A.M.; Chen, D.; et al. The pH-Responsive PacC transcription factor of Aspergillus fumigatus governs epithelial eentry and tissue invasion during pulmonary aspergillosis. PLoS Pathog. 2014, 10, e1004943. [Google Scholar] [CrossRef]
- Han, X.; Yu, R.; Zhen, D.; Tao, S.; Schmidt, M.; Han, L. β-1,3-Glucan-induced host phospholipase D activation is involved in Aspergillus fumigatus internalization into type II human pneumocyte A549 cells. PLoS ONE 2011, 6, e21468. [Google Scholar] [CrossRef]
- Oosthuizen, J.L.; Gomez, P.; Ruan, J.; Hackett, T.L.; Moore, M.M.; Knight, D.A.; Tebbutt, S.J. Dual organism transcriptomics of airway epithelial cells interacting with conidia of Aspergillus fumigatus. PLoS ONE 2011, 6, e0020527. [Google Scholar] [CrossRef] [PubMed]
- Kogan, T.V.; Jadoun, J.; Mittelman, L.; Hirschberg, K.; Osherov, N. Involvement of secreted Aspergillus fumigatus proteases in disruption of the actin fiber cytoskeleton and loss of focal adhesion sites in infected A549 lung pneumocytes. J. Infect. Dis. 2004, 189, 1965–1973. [Google Scholar] [CrossRef] [PubMed]
- Bao, Z.; Han, X.; Chen, F.; Jia, X.; Zhao, J.; Zhang, C.; Yong, C.; Tian, S.; Zhou, X.; Han, L. Evidence for the involvement of cofilin in Aspergillus fumigatus internalization into type II alveolar epithelial cells. BMC Microbiol. 2015, 15, 161. [Google Scholar] [CrossRef]
- Richard, N.; Marti, L.; Varrot, A.; Guillot, L.; Guitard, J.; Hennequin, C.; Imberty, A.; Corvol, H.; Chignard, M.; Balloy, V. Human bronchial epithelial cells inhibit Aspergillus fumigatus germination of extracellular conidia via FleA recognition. Sci. Rep. 2018, 8, 15699. [Google Scholar] [CrossRef]
- Uribe-Quero, E.; Rosales, C. Control of phagocytosis by microbial pathogens. Front. Immunol. 2017, 8, 1368. [Google Scholar] [CrossRef] [PubMed]
- Seidel, C.; Moreno-Velásquez, S.D.; Ben-Ghazzi, N.; Gago, S.; Read, N.D.; Bowyer, P. Phagolysosomal survival enables non-lytic hyphal escape and ramification through lung epithelium during Aspergillus fumigatus infection. Front. Microbiol. 2020, 11, 1955. [Google Scholar] [CrossRef] [PubMed]
- De Leon-Rodriguez, C.M.; Fu, M.S.; Çorbali, M.O.; Cordero, R.J.B.; Casadevall, A. The capsule of Cryptococcus neoformans modulates phagosomal pH through its acid-base properties. mSphere 2018, 3, e00437-18. [Google Scholar] [CrossRef]
- Westman, J.; Walpole, G.F.W.; Kasper, L.; Xue, B.Y.; Elshafee, O.; Hube, B.; Grinstein, S. Lysosome fusion maintains phagosome integrity during fungal infection. Cell Host Microbe 2020, 28, 798–812.e796. [Google Scholar] [CrossRef]
- Bayry, J.; Beaussart, A.; Dufrêne, Y.F.; Sharma, M.; Bansal, K.; Kniemeyer, O.; Aimanianda, V.; Brakhage, A.A.; Kaveri, S.V.; Kwon-Chung, K.J.; et al. Surface structure characterization of Aspergillus fumigatus conidia mutated in the melanin synthesis pathway and their human cellular immune response. Infect. Immun. 2014, 82, 3141–3153. [Google Scholar] [CrossRef] [PubMed]
- Jahn, B.; Langfelder, K.; Schneider, U.; Schindel, C.; Brakhage, A.A. PKSP-dependent reduction of phagolysosome fusion and intracellular kill of Aspergillus fumigatus conidia by human monocyte-derived macrophages. Cell. Microbiol. 2002, 4, 793–803. [Google Scholar] [CrossRef]
- Langfelder, K.; Streibel, M.; Jahn, B.; Haase, G.; Brakhage, A.A. Biosynthesis of fungal melanins and their importance for human pathogenic fungi. Fungal Genet. Biol. 2003, 38, 143–158. [Google Scholar] [CrossRef]
- Aimanianda, V.; Bayry, J.; Bozza, S.; Kniemeyer, O.; Perruccio, K.; Elluru, S.R.; Clavaud, C.; Paris, S.; Brakhage, A.A.; Kaveri, S.V.; et al. Surface hydrophobin prevents immune recognition of airborne fungal spores. Nature 2009, 460, 1117–1121. [Google Scholar] [CrossRef]
- Bruns, S.; Kniemeyer, O.; Hasenberg, M.; Aimanianda, V.; Nietzsche, S.; Thywien, A.; Jeron, A.; Latgé, J.P.; Brakhage, A.A.; Gunzer, M. Production of extracellular traps against Aspergillus fumigatus in vitro and in infected lung tissue is dependent on invading neutrophils and influenced by hydrophobin rodA. PLoS Pathog. 2010, 6, e1000873. [Google Scholar] [CrossRef] [PubMed]
- Paris, S.; Debeaupuis, J.P.; Crameri, R.; Carey, M.; Charlès, F.; Prévost, M.C.; Schmitt, C.; Philippe, B.; Latgé, J.P. Conidial hydrophobins of Aspergillus fumigatus. Appl. Environ. Microbiol. 2003, 69, 1581–1588. [Google Scholar] [CrossRef] [PubMed]
- Thywißen, A.; Heinekamp, T.; Dahse, H.M.; Schmaler-Ripcke, J.; Nietzsche, S.; Zipfel, P.F.; Brakhage, A.A. Conidial dihydroxynaphthalene melanin of the human pathogenic fungus Aspergillus fumigatus interferes with the host endocytosis pathway. Front. Microbiol. 2011, 2, 96. [Google Scholar] [CrossRef] [PubMed]
- Gualtieri, M.; Øvrevik, J.; Holme, J.A.; Perrone, M.G.; Bolzacchini, E.; Schwarze, P.E.; Camatini, M. Differences in cytotoxicity versus pro-inflammatory potency of different PM fractions in human epithelial lung cells. Toxicol. Vitr. 2010, 24, 29–39. [Google Scholar] [CrossRef]
- Schulz, C.; Farkas, L.; Wolf, K.; Krätzel, K.; Eissner, G.; Pfeifer, M. Differences in LPS-induced activation of bronchial epithelial cells (BEAS-2B) and type II-like pneumocytes (A-549). Scand. J. Immunol. 2002, 56, 294–302. [Google Scholar] [CrossRef] [PubMed]
- Cozens, A.L.; Yezzi, M.J.; Kunzelmann, K.; Ohrui, T.; Chin, L.; Eng, K.; Finkbeiner, W.E.; Widdicombe, J.H.; Gruenert, D.C. CFTR expression and chloride secretion in polarized immortal human bronchial epithelial cells. Am. J. Respir. Cell Mol. Biol. 1994, 10, 38–47. [Google Scholar] [CrossRef]
- Bleichrodt, R.J.; Foster, P.; Howell, G.; Latgé, J.P.; Read, N.D. Cell wall composition heterogeneity between single cells in Aspergillus fumigatus leads to heterogeneous behavior during antifungal treatment and phagocytosis. mBio 2020, 11, e03015-19. [Google Scholar] [CrossRef]
- Bigot, J.; Guillot, L.; Guitard, J.; Ruffin, M.; Corvol, H.; Balloy, V.; Hennequin, C. Bronchial Epithelial Cells on the Front Line to Fight Lung Infection-Causing Aspergillus fumigatus. Front. Immunol. 2020, 11, 1041. [Google Scholar] [CrossRef]
- Pollmächer, J.; Figge, M.T. Agent-based model of human alveoli predicts chemotactic signaling by epithelial cells during early Aspergillus fumigatus infection. PLoS ONE 2014, 9, e116030. [Google Scholar] [CrossRef]
- Mech, F.; Wilson, D.; Lehnert, T.; Hube, B.; Thilo Figge, M. Epithelial invasion outcompetes hypha development during Candida albicans infection as revealed by an image-based systems biology approach. Cytom. Part A 2014, 85, 126–139. [Google Scholar] [CrossRef]
- Fisch, D.; Yakimovich, A.; Clough, B.; Wright, J.; Bunyan, M.; Howell, M.; Mercer, J.; Frickel, E. Defining host–pathogen interactions employing an artificial intelligence workflow. eLife 2019, 8, e40560. [Google Scholar] [CrossRef]
- Prauße, M.T.E.; Lehnert, T.; Timme, S.; Hünniger, K.; Leonhardt, I.; Kurzai, O.; Figge, M.T. Predictive virtual infection modeling of fungal immune evasion in human whole blood. Front. Immunol. 2018, 9, 560. [Google Scholar] [CrossRef] [PubMed]
- Kraibooj, K.; Schoeler, H.; Svensson, C.M.; Brakhage, A.A.; Figge, M.T. Automated quantification of the phagocytosis of Aspergillus fumigatus conidia by a novel image analysis algorithm. Front. Microbiol. 2015, 6, 549. [Google Scholar] [CrossRef]
- Ferling, I.; Dunn, J.D.; Ferling, A.; Soldati, T.; Hillmann, F.; Goldman, G.H. Conidial melanin of the human-pathogenic fungus Aspergillus fumigatus disrupts cell autonomous defenses in amoebae. mBio 2020, 11, e00862-20. [Google Scholar] [CrossRef]
- Ju, X.; Yan, Y.; Liu, Q.; Li, N.; Sheng, M.; Zhang, L.; Li, X.; Liang, Z.; Huang, F.; Liu, K.; et al. Neuraminidase of Influenza A Virus Binds Lysosome-Associated Membrane Proteins Directly and Induces Lysosome Rupture. J. Virol. 2015, 89, 10347–10358. [Google Scholar] [CrossRef] [PubMed]
- Tsuruta, F.; Okajima, T.; Yano, S.; Chiba, T. Quantification of endosome and lysosome motilities in cultured neurons using fluorescent probes. J. Vis. Exp. 2017, 2017, 55488. [Google Scholar] [CrossRef]
- Mrakovic, A.; Kay, J.G.; Furuya, W.; Brumell, J.H.; Botelho, R.J. Rab7 and Arl8 GTPases are Necessary for Lysosome Tubulation in Macrophages. Traffic 2012, 13, 1667–1679. [Google Scholar] [CrossRef]
- Pu, J.; Guardia, C.M.; Keren-Kaplan, T.; Bonifacino, J.S. Mechanisms and functions of lysosome positioning. J. Cell Sci. 2016, 129, 4329–4339. [Google Scholar] [CrossRef]
- Ayala, P.; Lin, L.; Hopper, S.; Fukuda, M.; So, M. Infection of epithelial cells by pathogenic Neisseriae reduces the levels of multiple lysosomal constituents. Infect. Immun. 1998, 66, 5001–5007. [Google Scholar] [CrossRef] [PubMed]
- Lewis, L.E.; Bain, J.M.; Lowes, C.; Gillespie, C.; Rudkin, F.M.; Gow, N.A.R.; Erwig, L.P. Stage specific assessment of Candida albicans phagocytosis by macrophages identifies cell wall composition and morphogenesis as key determinants. PLoS Pathog. 2012, 8, e1002578. [Google Scholar] [CrossRef]
- Prashar, A.; Bhatia, S.; Gigliozzi, D.; Martin, T.; Duncan, C.; Guyard, C.; Terebiznik, M.R. Filamentous morphology of bacteria delays the timing of phagosome morphogenesis in macrophages. J. Cell Biol. 2013, 203, 1081–1097. [Google Scholar] [CrossRef]
- Brakhage, A.A.; Liebmann, B. Aspergillus fumigatus conidial pigment and cAMP signal transduction: Significance for virulence. Med. Mycol. 2005, 43, S75–S82. [Google Scholar] [CrossRef]
- Heinekamp, T.; Thywißen, A.; Macheleidt, J.; Keller, S.; Valiante, V.; Brakhage, A.A. Aspergillus fumigatus melanins: Interference with the host endocytosis pathway and impact on virulence. Front. Microbiol. 2012, 3, 440. [Google Scholar] [CrossRef]
- Kyrmizi, I.; Ferreira, H.; Carvalho, A.; Figueroa, J.A.L.; Zarmpas, P.; Cunha, C.; Akoumianaki, T.; Stylianou, K.; Deepe, G.S.; Samonis, G.; et al. Calcium sequestration by fungal melanin inhibits calcium-calmodulin signalling to prevent LC3-associated phagocytosis. Nat. Microbiol. 2018, 3, 791–803. [Google Scholar] [CrossRef]
- Stappers, M.H.T.; Clark, A.E.; Aimanianda, V.; Bidula, S.; Reid, D.M.; Asamaphan, P.; Hardison, S.E.; Dambuza, I.M.; Valsecchi, I.; Kerscher, B.; et al. Recognition of DHN-melanin by a C-type lectin receptor is required for immunity to Aspergillus. Nature 2018, 555, 382–386. [Google Scholar] [CrossRef]
- Steinberg, B.E.; Grinstein, S. Assessment of phagosome formation and maturation by fluorescence microscopy. Methods Mol. Biol. Clifton N. J. 2007, 412, 289–300. [Google Scholar] [CrossRef]
- Schmidt, F.; Thywißen, A.; Goldmann, M.; Cunha, C.; Cseresnyés, Z.; Schmidt, H.; Rafiq, M.; Galiani, S.; Gräler, M.H.; Chamilos, G.; et al. Flotillin-dependent membrane microdomains are required for functional phagolysosomes against fungal infections. Cell Rep. 2020, 32, 108017. [Google Scholar] [CrossRef] [PubMed]
- Schmidt, H.; Vlaic, S.; Krüger, T.; Schmidt, F.; Balkenhol, J.; Dandekar, T.; Guthke, R.; Kniemeyer, O.; Heinekamp, T.; Brakhage, A.A. Proteomics of Aspergillus fumigatus conidia-containing phagolysosomes identifies processes governing immune evasion. Mol. Cell Proteom. 2018, 17, 1084–1096. [Google Scholar] [CrossRef]
- Bertuzzi, M.; Hayes, G.E.; Icheoku, U.J.; van Rhijn, N.; Denning, D.W.; Osherov, N.; Bignell, E.M. Anti-Aspergillus activities of the respiratory epithelium in health and disease. J. Fungi 2018, 4, 8. [Google Scholar] [CrossRef] [PubMed]
- Koike, R.; Cueno, M.E.; Nodomi, K.; Tamura, M.; Kamio, N.; Tanaka, H.; Kotani, A.; Imai, K. Heat-killed Fusobacterium nucleatum triggers varying heme-related inflammatory and stress responses depending on primary human respiratory epithelial cell type. Molecules 2020, 25, 3839. [Google Scholar] [CrossRef]
- Mostafa, M.M.; Rider, C.F.; Shah, S.; Traves, S.L.; Gordon, P.M.K.; Miller-Larsson, A.; Leigh, R.; Newton, R. Glucocorticoid-driven transcriptomes in human airway epithelial cells: Commonalities, differences and functional insight from cell lines and primary cells. BMC Med. Genom. 2019, 12, 29. [Google Scholar] [CrossRef]
- Baskoro, H.; Sato, T.; Karasutani, K.; Suzuki, Y.; Mitsui, A.; Arano, N.; Nurwidya, F.; Kato, M.; Takahashi, F.; Kodama, Y.; et al. Regional heterogeneity in response of airway epithelial cells to cigarette smoke. BMC Pulm. Med. 2018, 18, 148. [Google Scholar] [CrossRef] [PubMed]
- Callaghan, P.J.; Ferrick, B.; Rybakovsky, E.; Thomas, S.; Mullin, J.M. Epithelial barrier function properties of the 16HBE14o-human bronchial epithelial cell culture model. Biosci. Rep. 2020, 40, BSR20201532. [Google Scholar] [CrossRef] [PubMed]
- Amich, J.; Mokhtari, Z.; Strobel, M.; Vialetto, E.; Sheta, D.; Yu, Y.; Hartweg, J.; Kalleda, N.; Jarick, K.J.; Brede, C.; et al. Three-dimensional light sheet fluorescence microscopy of lungs to dissect local host immune-Aspergillus fumigatus interactions. mBio 2020, 11, e02752-10. [Google Scholar] [CrossRef]
- Fettucciari, K.; Quotadamo, F.; Noce, R.; Palumbo, C.; Modesti, A.; Rosati, E.; Mannucci, R.; Bartoli, A.; Marconi, P. Group B Streptococcus (GBS) disrupts by calpain activation the actin and microtubule cytoskeleton of macrophages. Cell Microbiol. 2011, 13, 859–884. [Google Scholar] [CrossRef] [PubMed]
- Shevchenko, M.A.; Bolkhovitina, E.B.; Ekaterina, A.S.; Sapozhnikov, A.M. Elimination of Aspergillus fumigatus conidia from the airways of mice with allergic airway inflammation. Resp. Res. 2013, 14, 78. [Google Scholar] [CrossRef] [PubMed]
- Schindelin, J.; Arganda-Carreras, I.; Frise, E.; Kaynig, V.; Longair, M.; Pietzsch, T.; Preibisch, S.; Rueden, C.; Saalfeld, S.; Schmid, B.; et al. Fiji: An open-source platform for biological-image analysis. Nat. Methods 2012, 9, 676–682. [Google Scholar] [CrossRef]




Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ben-Ghazzi, N.; Moreno-Velásquez, S.; Seidel, C.; Thomson, D.; Denning, D.W.; Read, N.D.; Bowyer, P.; Gago, S. Characterisation of Aspergillus fumigatus Endocytic Trafficking within Airway Epithelial Cells Using High-Resolution Automated Quantitative Confocal Microscopy. J. Fungi 2021, 7, 454. https://doi.org/10.3390/jof7060454
Ben-Ghazzi N, Moreno-Velásquez S, Seidel C, Thomson D, Denning DW, Read ND, Bowyer P, Gago S. Characterisation of Aspergillus fumigatus Endocytic Trafficking within Airway Epithelial Cells Using High-Resolution Automated Quantitative Confocal Microscopy. Journal of Fungi. 2021; 7(6):454. https://doi.org/10.3390/jof7060454
Chicago/Turabian StyleBen-Ghazzi, Nagwa, Sergio Moreno-Velásquez, Constanze Seidel, Darren Thomson, David W. Denning, Nick D. Read, Paul Bowyer, and Sara Gago. 2021. "Characterisation of Aspergillus fumigatus Endocytic Trafficking within Airway Epithelial Cells Using High-Resolution Automated Quantitative Confocal Microscopy" Journal of Fungi 7, no. 6: 454. https://doi.org/10.3390/jof7060454
APA StyleBen-Ghazzi, N., Moreno-Velásquez, S., Seidel, C., Thomson, D., Denning, D. W., Read, N. D., Bowyer, P., & Gago, S. (2021). Characterisation of Aspergillus fumigatus Endocytic Trafficking within Airway Epithelial Cells Using High-Resolution Automated Quantitative Confocal Microscopy. Journal of Fungi, 7(6), 454. https://doi.org/10.3390/jof7060454





