Identification of a Novel Biosynthetic Gene Cluster in Aspergillus niger Using Comparative Genomics
Abstract
1. Introduction
2. Materials and Methods
3. Results
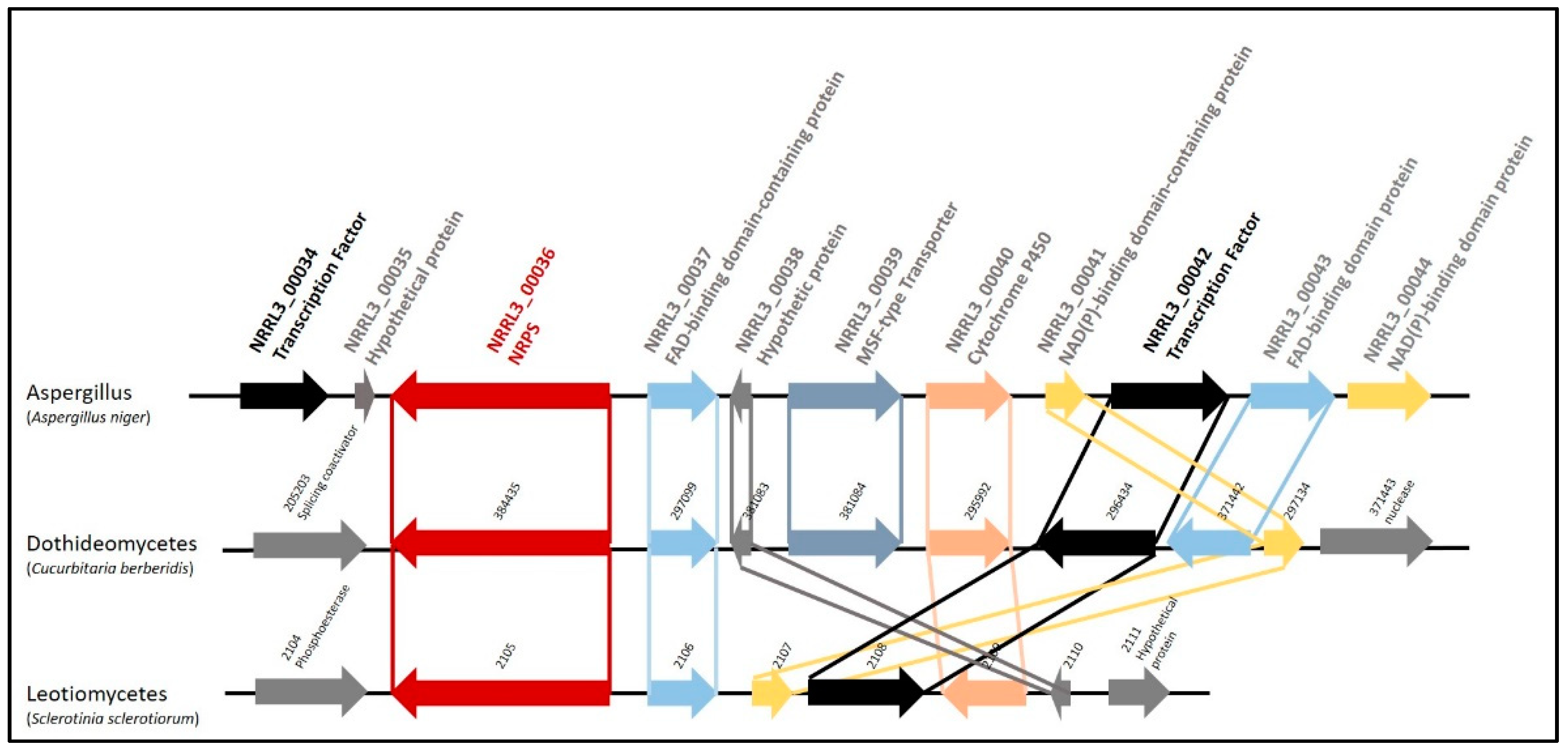
3.1. Biosynthetic Gene Cluster Structure and Phylogeny
3.2. Overexpression of the In-Cluster Transcription Factor Gene Results in Impaired Growth and Overproduction of New Compounds
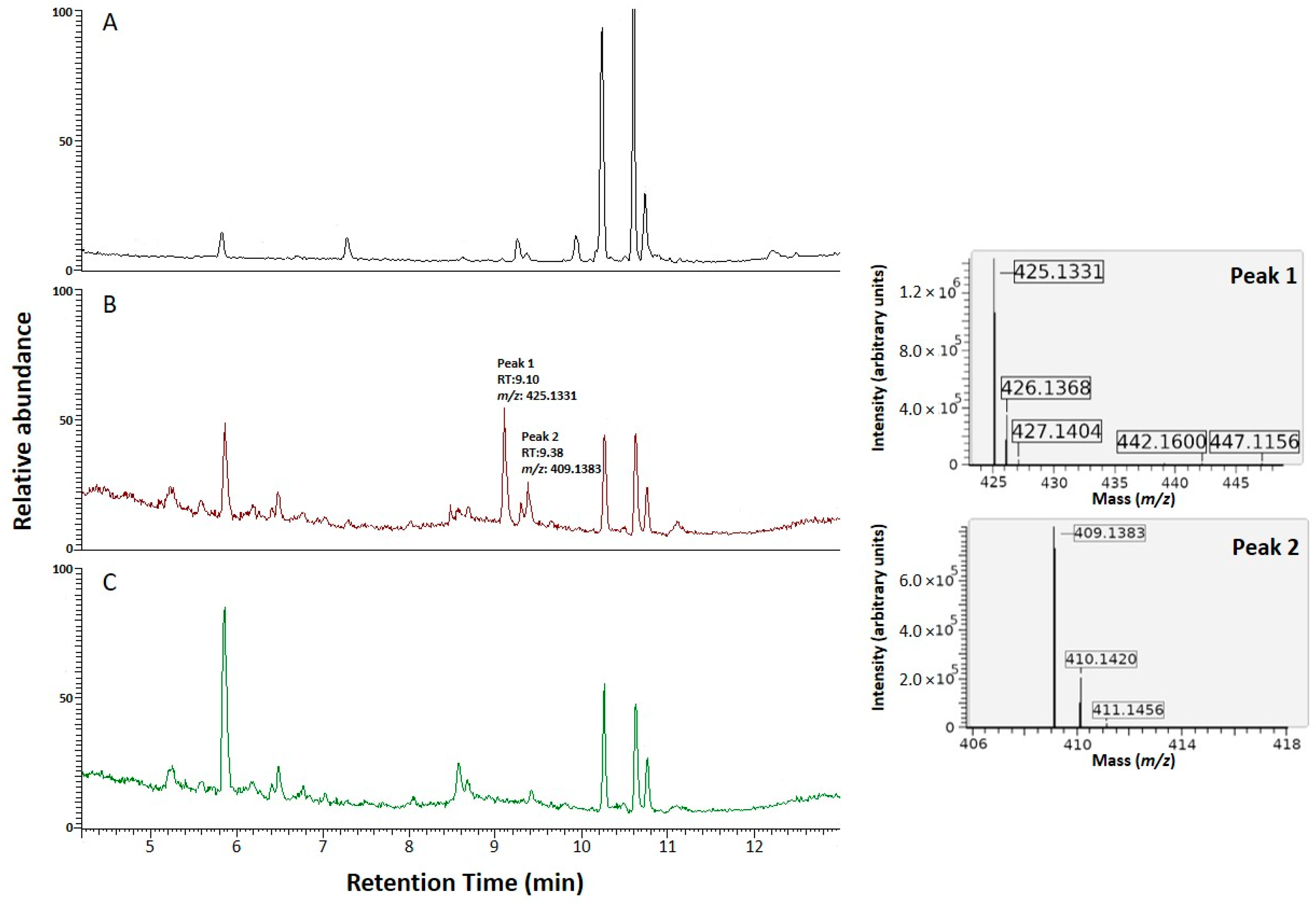
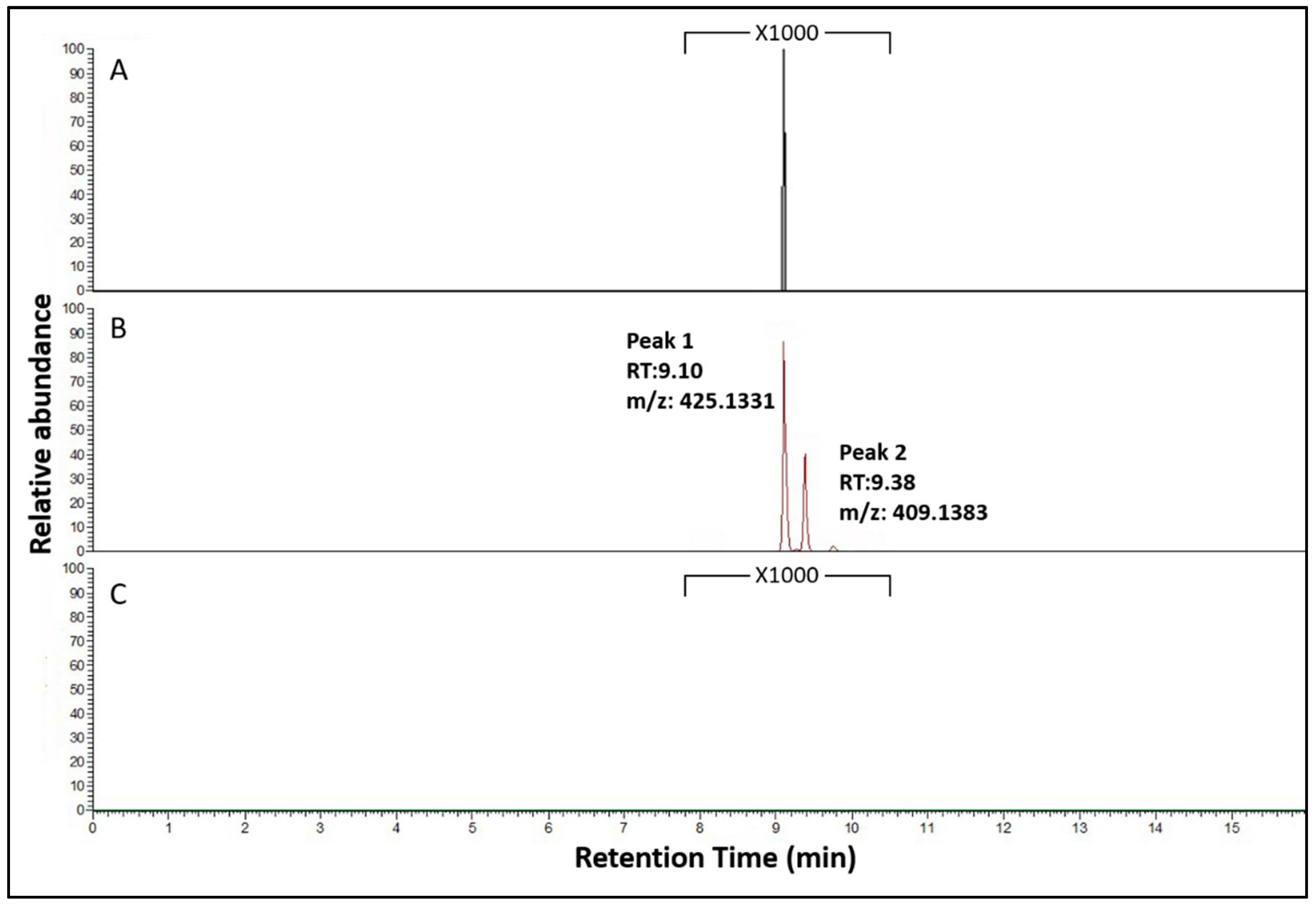
3.3. Functional Characterization of the NRPS NRRL3_00036
3.4. Antimicrobial Assays
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Kwon, M.J.; Jørgensen, T.R.; Nitsche, B.M.; Arentshorst, M.; Park, J.; Ram, A.F.J.; Meyer, V. The transcriptomic fingerprint of glucoamylase over-expression in Aspergillus niger. BMC Genom. 2012, 13, 701. [Google Scholar] [CrossRef]
- Papagianni, M. Advances in citric acid fermentation by Aspergillus niger: Biochemical aspects, membrane transport and modeling. Biotechnol. Adv. 2007, 25, 244–263. [Google Scholar] [CrossRef]
- Gil-Serna, J.; García-Díaz, M.; Vázquez, C.; González-Jaén, M.T.; Patiño, B. Significance of Aspergillus niger aggregate species as contaminants of food products in Spain regarding their occurrence and their ability to produce mycotoxins. Food Microbiol. 2019, 82, 240–248. [Google Scholar] [CrossRef] [PubMed]
- Liu, D.; Li, X.-M.; Meng, L.; Li, C.-S.; Gao, S.-S.; Shang, Z.; Proksch, P.; Huang, C.-G.; Wang, B.-G. Nigerapyrones A–H, α-Pyrone Derivatives from the Marine Mangrove-Derived Endophytic Fungus Aspergillus niger MA-132. J. Nat. Prod. 2011, 74, 1787–1791. [Google Scholar] [CrossRef] [PubMed]
- Nielsen, K.F.; Mogensen, J.M.; Johansen, M.; Larsen, T.O.; Frisvad, J.C. Review of secondary metabolites and mycotoxins from the Aspergillus niger group. Anal. Bioanal. Chem. 2009, 395, 1225–1242. [Google Scholar] [CrossRef]
- Benoit, I.; van den Esker, M.H.; Patyshakuliyeva, A.; Mattern, D.J.; Blei, F.; Zhou, M.; Dijksterhuis, J.; Brakhage, A.A.; Kuipers, O.P.; de Vries, R.P.; et al. Bacillus subtilis attachment to Aspergillus niger hyphae results in mutually altered metabolism. Environ. Microbiol. 2015, 17, 2099–2113. [Google Scholar] [CrossRef]
- Aguilar-Pontes, M.; Brandl, J.; McDonnell, E.; Strasser, K.; Nguyen, T.; Riley, R.; Mondo, S.; Salamov, A.; Nybo, J.; Vesth, T.; et al. The gold-standard genome of Aspergillus niger NRRL 3 enables a detailed view of the diversity of sugar catabolism in fungi. Stud. Mycol. 2018, 91, 61–78. [Google Scholar] [CrossRef]
- Lin, T.S.; Chen, B.; Chiang, Y.M.; Wang, C.C.C. Recent advances in the genome mining of Aspergillus secondary metabolites (covering 2012–2018). Medchemcomm 2019, 20, 329–334. [Google Scholar]
- Song, L.; Ouedraogo, J.-P.; Kolbusz, M.; Nguyen, T.T.M.; Tsang, A. Efficient genome editing using tRNA promoter-driven CRISPR/Cas9 gRNA in Aspergillus niger. PLoS ONE 2018, 13, e0202868. [Google Scholar] [CrossRef] [PubMed]
- Master, E.R.; Zheng, Y.; Storms, R.; Tsang, A.; Powlowski, J. A xyloglucan-specific family 12 glycosyl hydrolase from As-pergillus niger: Recombinant expression, purification and characterization. Biochem. J. 2008, 411, 161–170. [Google Scholar] [CrossRef] [PubMed]
- ChemSpider. Available online: http://www.chemspider.com/DataSources.aspx (accessed on 7 May 2020).
- Methods for Dilution Antimicrobial Susceptibility Testing of Bacteria That Grow Aerobically, Approved Standard—10th Edition. CLSI Document M07-A10; Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2015.
- JGI MycoCosm Database. Available online: https://mycocosm.jgi.doe.gov/mycocosm/home.S (accessed on 7 January 2021).
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Edgar, R.C. MUSCLE: A multiple sequence alignment method with reduced time and space complexity. BMC Bioinform. 2004, 5, 113. [Google Scholar] [CrossRef]
- Price, M.N.; Dehal, P.S.; Arkin, A.P. FastTree 2—Approximately Maximum-Likelihood Trees for Large Alignments. PLoS ONE 2010, 5, e9490. [Google Scholar] [CrossRef]
- Letunic, I.; Bork, P. Interactive Tree Of Life (iTOL) v5: An online tool for phylogenetic tree display and annotation. Nucleic Acids Res. 2021, 23, 127–128. [Google Scholar] [CrossRef]
- Fungi, D.B. Available online: https://fungidb.org/fungidb/app (accessed on 5 January 2021).
- Zabala, A.O.; Xu, W.; Chooi, Y.-H.; Tang, Y. Characterization of a Silent Azaphilone Gene Cluster from Aspergillus niger ATCC 1015 Reveals a Hydroxylation-Mediated Pyran-Ring Formation. Chem. Biol. 2012, 19, 1049–1059. [Google Scholar] [CrossRef] [PubMed]
- Keller, N.P. Fungal secondary metabolism: Regulation, function and drug discovery. Nat. Rev. Genet. 2019, 17, 167–180. [Google Scholar] [CrossRef] [PubMed]
- Lin, T.; Chen, B.; Chiang, Y.; Wang, C.C.C. Discovery and Elucidation of the Biosynthesis of Aspernidgulenes: Novel Polyenes from Aspergillus Nidulans by Using Serial Promoter Replacement. ChemBioChem 2019, 20, 329–334. [Google Scholar] [CrossRef]
- Palys, S.; Pham, T.T.M.; Tsang, A. Biosynthesis of alkylcitric acids in Aspergillus niger involves both co-localized and un-linked genes. Front. Microbiol. 2020, 11, 1378. [Google Scholar] [CrossRef]
- Tsai, H.-F.; Wheeler, M.H.; Chang, Y.C.; Kwon-Chung, K.J. A Developmentally Regulated Gene Cluster Involved in Conidial Pigment Biosynthesis in Aspergillus fumigatus. J. Bacteriol. 1999, 181, 6469–6477. [Google Scholar] [CrossRef]
- Kimura, N.; Tsuge, T. Gene cluster involved in melanin biosynthesis of the filamentous fungus Alternaria alternata. J. Bacteriol. 1993, 175, 4427–4435. [Google Scholar] [CrossRef]
- Jørgensen, T.R.; Park, J.; Arentshorst, M.; van Welzen, A.M.; Lamers, G.; Vankuyk, P.A.; Damveld, R.A.; Hondel, C.A.V.D.; Nielsen, K.F.; Frisvad, J.C.; et al. The molecular and genetic basis of conidial pigmentation in Aspergillus niger. Fungal Genet. Biol. 2011, 48, 544–553. [Google Scholar] [CrossRef]
- Vesth, T.C.; Nybo, J.L.; Theobald, S.; Frisvad, J.C.; Larsen, T.O.; Nielsen, K.F.; Hoof, J.B.; Brandl, J.; Salamov, A.; Riley, R.; et al. Investigation of inter- and intraspecies variation through genome sequencing of Aspergillus section Nigri. Nat. Genet. 2018, 50, 1688–1695. [Google Scholar] [CrossRef] [PubMed]
- Inglis, D.O.; Binkley, J.; Skrzypek, M.S.; Arnaud, M.B.; Cerqueira, G.C.; Shah, P.; Wymore, F.; Wortman, J.R.; Sherlock, G. Comprehensive annotation of secondary metabolite biosynthetic genes and gene clusters of Aspergillus nidulans, A. fumigatus, A. niger and A. oryzae. BMC Microbiol. 2013, 13, 91. [Google Scholar] [CrossRef] [PubMed]
- Almeida, H.; Palys, S.; Tsang, A.; Diallo, A.B. TOUCAN: A framework for fungal biosynthetic gene cluster discovery. NAR Genom. Bioinform. 2020, 2, lqaa098. [Google Scholar] [CrossRef] [PubMed]
- Kaper, J.B.; Nataro, J.P.; Mobley, H.L.T. Pathogenic Escherichia coli. Nat. Rev. Genet. 2004, 2, 123–140. [Google Scholar] [CrossRef]
- Tong, S.Y.C.; Davis, J.S.; Eichenberger, E.; Holland, T.L.; Jr, V.G.F. Staphylococcus aureus Infections: Epidemiology, Pathophysiology, Clinical Manifestations, and Management. Clin. Microbiol. Rev. 2015, 28, 603–661. [Google Scholar] [CrossRef] [PubMed]
- Su, Y.; Liu, C.; Fang, H.; Zhang, D. Bacillus subtilis: A universal cell factory for industry, agriculture, biomaterials and medicine. Microb. Cell Factories 2020, 19, 1–12. [Google Scholar] [CrossRef] [PubMed]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Evdokias, G.; Semper, C.; Mora-Ochomogo, M.; Di Falco, M.; Nguyen, T.T.M.; Savchenko, A.; Tsang, A.; Benoit-Gelber, I. Identification of a Novel Biosynthetic Gene Cluster in Aspergillus niger Using Comparative Genomics. J. Fungi 2021, 7, 374. https://doi.org/10.3390/jof7050374
Evdokias G, Semper C, Mora-Ochomogo M, Di Falco M, Nguyen TTM, Savchenko A, Tsang A, Benoit-Gelber I. Identification of a Novel Biosynthetic Gene Cluster in Aspergillus niger Using Comparative Genomics. Journal of Fungi. 2021; 7(5):374. https://doi.org/10.3390/jof7050374
Chicago/Turabian StyleEvdokias, Gregory, Cameron Semper, Montserrat Mora-Ochomogo, Marcos Di Falco, Thi Truc Minh Nguyen, Alexei Savchenko, Adrian Tsang, and Isabelle Benoit-Gelber. 2021. "Identification of a Novel Biosynthetic Gene Cluster in Aspergillus niger Using Comparative Genomics" Journal of Fungi 7, no. 5: 374. https://doi.org/10.3390/jof7050374
APA StyleEvdokias, G., Semper, C., Mora-Ochomogo, M., Di Falco, M., Nguyen, T. T. M., Savchenko, A., Tsang, A., & Benoit-Gelber, I. (2021). Identification of a Novel Biosynthetic Gene Cluster in Aspergillus niger Using Comparative Genomics. Journal of Fungi, 7(5), 374. https://doi.org/10.3390/jof7050374





