Genome Sequence Analysis of the Oleaginous Yeast, Rhodotorula diobovata, and Comparison of the Carotenogenic and Oleaginous Pathway Genes and Gene Products with Other Oleaginous Yeasts
Abstract
1. Introduction
2. Materials and Methods
2.1. Microorganism and DNA Extraction
2.2. Genome Sequencing and Annotation
2.3. Comparative Genomics and Phylogenetic Analyses
3. Results and Discussion
3.1. General Features of the Rhodotorula Diobovata Genome
3.2. Analysis of Proteins Associated with De Novo Fatty Acid Biosynthesis
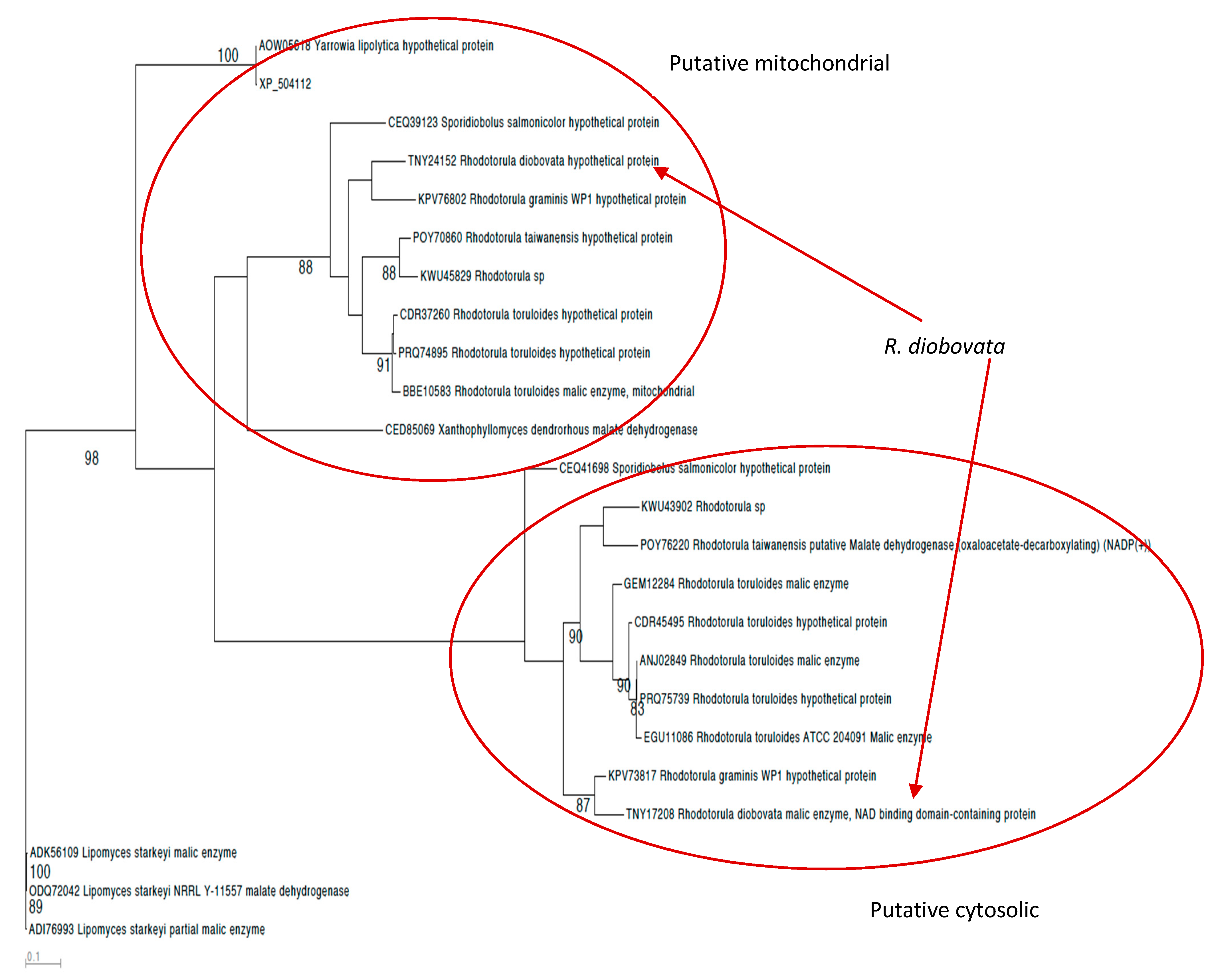
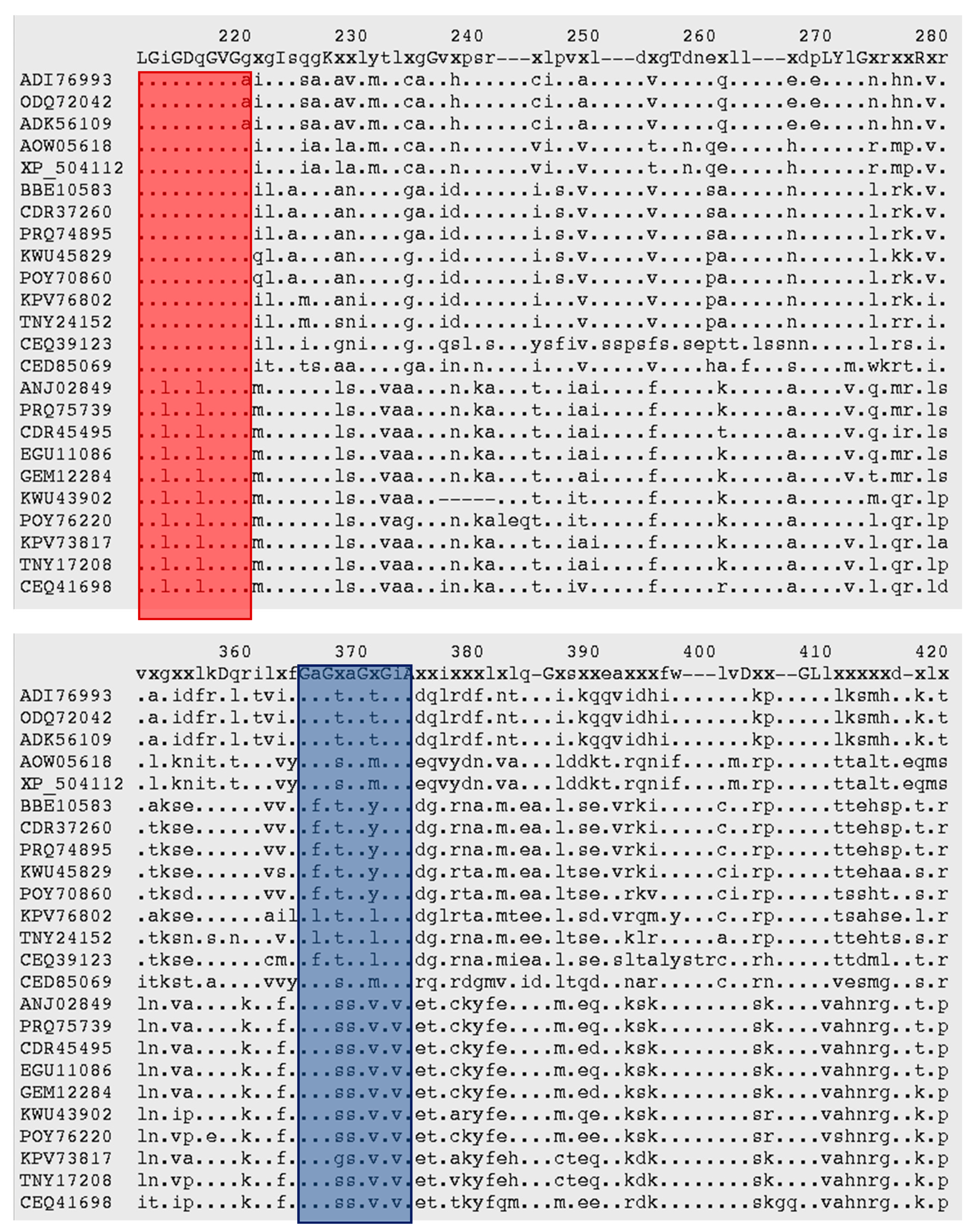
3.3. Malic Enzyme
3.4. ATP-Citrate Lyase
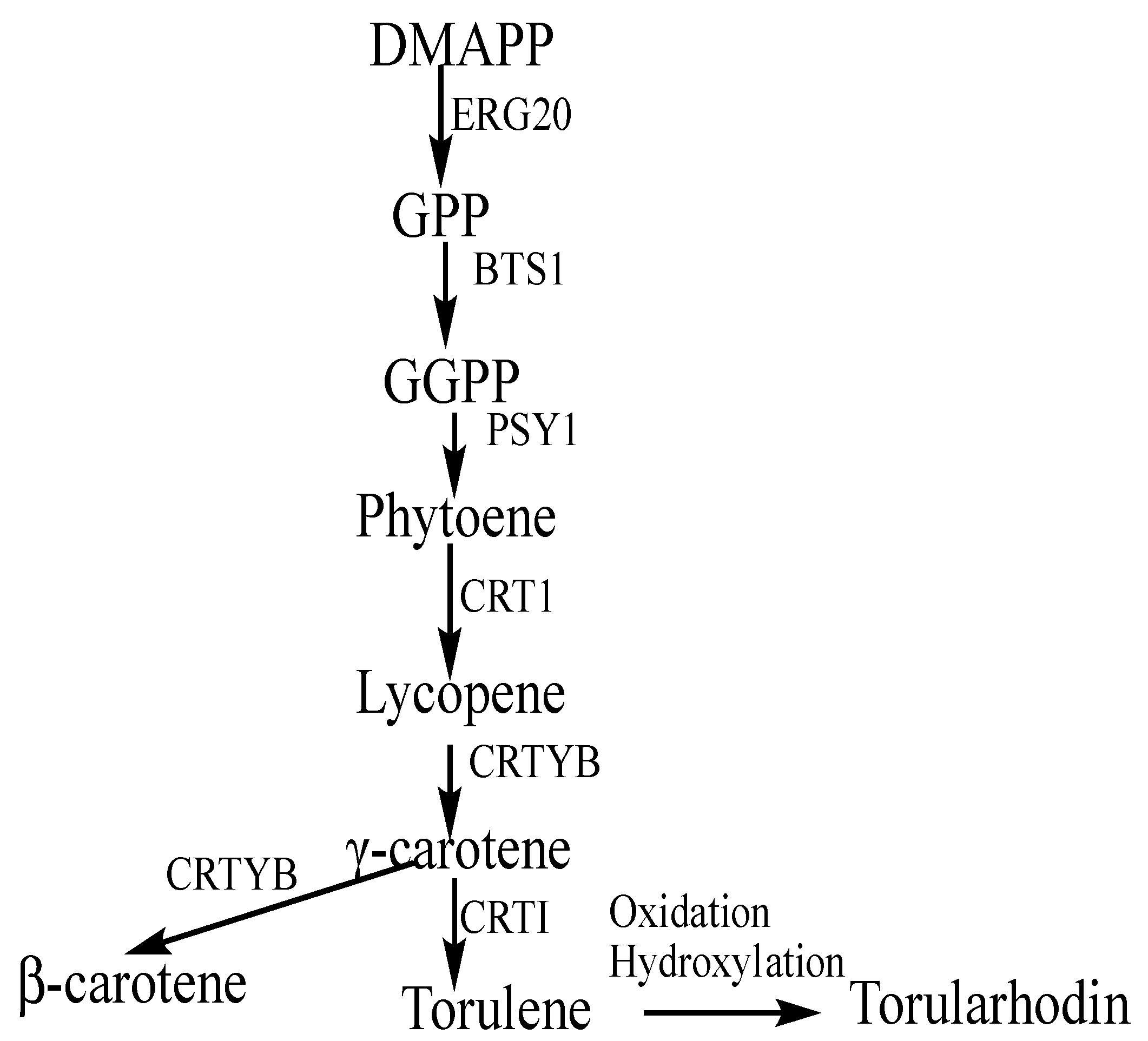
3.5. Analysis of Proteins Associated with Carotenoid Biosynthesis
3.6. Acetyl-CoA C-Acetyltransferase
3.7. Bifunctional Lycopene Cyclase/Phytoene Synthase
3.8. Phytoene Desaturase
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Kaneda, T.; Greenbaum, C.; Kline, K. Population Reference Bureau. 2020 World Population Data Sheet Shows Older Populations Growing, Total Fertility Rates Declining. 2020. Available online: https://www.prb.org/2020-world-population-data-sheet (accessed on 30 November 2020).
- Donot, F.; Fontana, A.; Baccou, J.C.; Strub, C.; Schorr-Galindo, S. Single Cell Oils (SCOs) from Oleaginous Yeasts and Moulds: Production and Genetics. Biomass Bioenergy 2014, 68, 135–150. [Google Scholar] [CrossRef]
- Ochsenreither, K.; Glück, C.; Stressler, T.; Fischer, L.; Syldatk, C. Production Strategies and Applications of Microbial Single Cell Oils. Front. Microbiol. 2016, 7, 1539. [Google Scholar] [CrossRef]
- Yurkov, A.M.; Vustin, M.M.; Tyaglov, B.V.; Maksimova, I.A.; Sineokiy, S.P. Pigmented Basidiomycetous Yeasts Are a Promising Source of Carotenoids and Ubiquinone Q10. Microbiology 2008, 77, 1–6. [Google Scholar] [CrossRef]
- Park, Y.-K.; Nicaud, J.-M.; Ledesma-Amaro, R. The Engineering Potential of Rhodosporidium Toruloides as a Workhorse for Biotechnological Applications. Trends Biotechnol. 2018, 36, 304–317. [Google Scholar]
- Nasirian, N.; Mirzaie, M.; Cicek, N.; Levin, D.B. Lipid and Carotenoid Synthesis by Rhodosporidium Diobovatum, Grown on Glucose versus Glycerol, and Its Biodiesel Properties. Can. J. Microbiol. 2018, 64, 277–289. [Google Scholar] [CrossRef]
- Munch, G.; Sestric, R.; Sparling, R.; Levin, D.B.; Cicek, N. Lipid Production in the Under-Characterized Oleaginous Yeasts, Rhodosporidium babjevae and Rhodosporidium diobovatum, from Biodiesel-Derived Waste Glycerol. Bioresour. Technol. 2015, 185, 49–55. [Google Scholar] [CrossRef]
- Civiero, E.; Pintus, M.; Ruggeri, C.; Tamburini, E.; Sollai, F.; Sanjust, E.; Zucca, P. Physiological and Phylogenetic Characterization of Rhodotorula Diobovata DSBCA06, a Nitrophilous Yeast. Biology 2018, 7, 39. [Google Scholar] [CrossRef]
- Newell, S.Y.; Hunter, I.L. Rhodosporidium diobovatum sp. n., the Perfect Form of an Asporogenous Yeast (Rhodotorula Sp.)1. J. Bacteriol. 1970, 104, 503–508. [Google Scholar] [CrossRef]
- Fakankun, I.; Mirzaei, M.; Levin, D.B. Impact of Culture Conditions on Neutral Lipid Production by Oleaginous Yeast. Methods Mol. Biol. 2019, 1995, 311–325. [Google Scholar]
- Kelley, D.R.; Schatz, M.C.; Salzberg, S.L. Quake: Quality-Aware Detection and Correction of Sequencing Errors. Genome Biol. 2010, 11, R116. [Google Scholar] [CrossRef]
- Marinier, E.; Brown, D.G.; McConkey, B.J. Pollux: Platform Independent Error Correction of Single and Mixed Genomes. BMC Bioinform. 2015, 16, 10. [Google Scholar] [CrossRef]
- Nordberg, H.; Cantor, M.; Dusheyko, S.; Hua, S.; Poliakov, A.; Shabalov, I.; Smirnova, T.; Grigoriev, I.V.; Dubchak, I. The Genome Portal of the Department of Energy Joint Genome Institute: 2014 Updates. Nucleic Acids Res. 2014, 42, D26–D31. [Google Scholar] [CrossRef] [PubMed]
- Carver, T.; Harris, S.R.; Berriman, M.; Parkhill, J.; McQuillan, J.A. Artemis: An Integrated Platform for Visualization and Analysis of High-Throughput Sequence-Based Experimental Data. Bioinformatics 2012, 28, 464–469. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Zhang, L.; Chen, H.; Chen, Y.Q.; Ratledge, C.; Song, Y.; Chen, W. Regulatory Properties of Malic Enzyme in the Oleaginous Yeast, Yarrowia Lipolytica, and Its Non-Involvement in Lipid Accumulation. Biotechnol. Lett. 2013, 35, 2091–2098. [Google Scholar] [CrossRef] [PubMed]
- Moreira Vieira, N.; Zandonade Ventorim, R.; de Moura Ferreira, M.A.; Barcelos de Souza, G.; Menezes de Almeida, E.L.; Pereira Vidigal, P.M.; Nunes Nesi, A.; Gomes Fietto, L.; Batista da Silveira, W. Insights into Oleaginous Phenotype of the Yeast Papiliotrema laurentii. Fungal Genet. Biol. 2020, 144, 103456. [Google Scholar] [CrossRef] [PubMed]
- Katoh, K.; Standley, D.M. MAFFT Multiple Sequence Alignment Software Version 7: Improvements in Performance and Usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef]
- Fristensky, B. BIRCH: A User-Oriented, Locally-Customizable, Bioinformatics System. BMC Bioinf. 2007, 8, 54. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Firrincieli, A.; Otillar, R.; Salamov, A.; Schmutz, J.; Khan, Z.; Redman, R.S.; Fleck, N.D.; Lindquist, E.; Grigoriev, I.V.; Doty, S.L. Genome Sequence of the Plant Growth Promoting Endophytic Yeast Rhodotorula Graminis WP1. Front. Microbiol. 2015, 6, 978. [Google Scholar] [CrossRef]
- Zhu, Z.; Zhang, S.; Liu, H.; Shen, H.; Lin, X.; Yang, F.; Zhou, Y.J.; Jin, G.; Ye, M.; Zou, H.; et al. A Multi-Omic Map of the Lipid-Producing Yeast Rhodosporidium Toruloides. Nat. Commun. 2012, 3, 1112. [Google Scholar] [CrossRef] [PubMed]
- Goordial, J.; Raymond-Bouchard, I.; Riley, R.; Ronholm, J.; Shapiro, N.; Woyke, T.; LaButti, K.M.; Tice, H.; Amirebrahimi, M.; Grigoriev, I.V.; et al. Improved High-Quality Draft Genome Sequence of the Eurypsychrophile rhodotorula sp. JG1b, Isolated from Permafrost in the Hyperarid Upper-Elevation McMurdo Dry Valleys, Antarctica. Genome Announc. 2016, 4. [Google Scholar] [CrossRef]
- Papanikolaou, S.; Aggelis, G. Lipids of Oleaginous Yeasts. Part II: Technology and Potential Applications. Eur. J. Lipid Sci. Technol. 2011, 113, 1052–1073. [Google Scholar] [CrossRef]
- Papanikolaou, S.; Aggelis, G. Lipids of Oleaginous Yeasts. Part I: Biochemistry of Single Cell Oil Production. Eur. J. Lipid Sci. Technol. 2011, 113, 1031–1051. [Google Scholar] [CrossRef]
- Ratledge, C. Microbial Lipids: Commercial Realities or Academic Curiosities. In Industrial Applications of Single Cell Oils; AOCS Publishing: Boulder Urbana, IL, USA, 1992. [Google Scholar]
- Akpinar-Bayizit, A. Fungal Lipids: The Biochemistry of Lipid Accumulation. Int. J. Chem. Eng. Appl. 2014, 5, 409–414. [Google Scholar] [CrossRef]
- Ratledge, C. The Role of Malic Enzyme as the Provider of NADPH in Oleaginous Microorganisms: A Reappraisal and Unsolved Problems. Biotechnol. Lett. 2014, 36, 1557–1568. [Google Scholar] [CrossRef]
- Morin, N.; Cescut, J.; Beopoulos, A.; Lelandais, G.; Le Berre, V.; Uribelarrea, J.-L.; Molina-Jouve, C.; Nicaud, J.-M. Transcriptomic Analyses during the Transition from Biomass Production to Lipid Accumulation in the Oleaginous Yeast Yarrowia lipolytica. PLoS ONE 2011, 6, e27966. [Google Scholar] [CrossRef] [PubMed]
- Shen, Q.; Chen, Y.; Jin, D.; Lin, H.; Wang, Q.; Zhao, Y.-H. Comparative Genome Analysis of the Oleaginous Yeast Trichosporon Fermentans Reveals Its Potential Applications in Lipid Accumulation. Microbiol. Res. 2016, 192, 203–210. [Google Scholar] [CrossRef]
- Börsch, D.; Westhoff, P. Primary Structure of NADP-Dependent Malic Enzyme in the Dicotyledonous C4 Plant Flaveria trinervia. FEBS Lett. 1990, 273, 111–115. [Google Scholar] [CrossRef]
- Zhang, Y.; Adams, I.P.; Ratledge, C. Malic Enzyme: The Controlling Activity for Lipid Production? Overexpression of Malic Enzyme in Mucor circinelloides Leads to a 2.5-Fold Increase in Lipid Accumulation. Microbiology 2007, 153, 2013–2025. [Google Scholar] [CrossRef]
- Ratledge, C. Regulation of Lipid Accumulation in Oleaginous Micro-Organisms. Biochem. Soc. Trans. 2002, 30, 1047–1050. [Google Scholar] [CrossRef] [PubMed]
- Sun, T.; Hayakawa, K.; Bateman, K.S.; Fraser, M.E. Identification of the Citrate-Binding Site of Human ATP-Citrate Lyase Using X-ray Crystallography. J. Biol. Chem. 2010, 285, 27418–27428. [Google Scholar] [CrossRef] [PubMed]
- Fabiszewska, A.; Misiukiewicz-Stępień, P.; Paplińska-Goryca, M.; Zieniuk, B.; Białecka-Florjańczyk, E. An Insight into Storage Lipid Synthesis by Yarrowia lipolytica Yeast Relating to Lipid and Sugar Substrates Metabolism. Biomolecules 2019, 9, 685. [Google Scholar] [CrossRef] [PubMed]
- Lu, S.; Wang, J.; Chitsaz, F.; Derbyshire, M.K.; Geer, R.C.; Gonzales, N.R.; Gwadz, M.; Hurwitz, D.I.; Marchler, G.H.; Song, J.S.; et al. CDD/SPARCLE: The Conserved Domain Database in 2020. Nucleic Acids Res. 2020, 48, D265–D268. [Google Scholar] [CrossRef] [PubMed]
- Goodwin, T.W. [29] Biosynthesis of Carotenoids: An Overview. In Methods in Enzymology; Elsevier: Amsterdam, The Netherlands, 1993; pp. 330–340. [Google Scholar]
- Li, L.; Liu, Z.; Jiang, H.; Mao, X. Biotechnological Production of Lycopene by Microorganisms. Appl. Microbiol. Biotechnol. 2020, 104, 10307–10324. [Google Scholar] [CrossRef] [PubMed]
- Albertyn, J.; Pohl, C.H.; Viljoen, B.C. Rhodotorula. In Encyclopedia of Food Microbiology; Elsevier: Amsterdam, The Netherlands, 2014; pp. 291–295. [Google Scholar]
- Buzzini, P.; Innocenti, M.; Turchetti, B.; Libkind, D.; van Broock, M.; Mulinacci, N. Carotenoid Profiles of Yeasts Belonging to the Genera Rhodotorula, Rhodosporidium, Sporobolomyces, and Sporidiobolus. Can. J. Microbiol. 2007, 53, 1024–1031. [Google Scholar] [CrossRef] [PubMed]
- Heath, R.J.; Rock, C.O. The Claisen Condensation in Biology. Nat. Prod. Rep. 2002, 19, 581–596. [Google Scholar] [CrossRef] [PubMed]
- Chevillard, G.; Clémencet, M.-C.; Latruffe, N.; Nicolas-Francès, V. Targeted Disruption of the Peroxisomal Thiolase B Gene in Mouse: A New Model to Study Disorders Related to Peroxisomal Lipid Metabolism. Biochimie 2004, 86, 849–856. [Google Scholar] [CrossRef]
- Guo, W.; Tang, H.; Zhang, L. Lycopene Cyclase and Phytoene Synthase Activities in the Marine Yeast Rhodosporidium Diobovatum Are Encoded by a Single Gene CrtYB: Lycopene Cyclase/Phytoene Synthase of Rhodosporidium diobovatum. J. Basic Microbiol. 2014, 54, 1053–1061. [Google Scholar] [CrossRef] [PubMed]
- Pham, K.D.; Shida, Y.; Miyata, A.; Takamizawa, T.; Suzuki, Y.; Ara, S.; Yamazaki, H.; Masaki, K.; Mori, K.; Aburatani, S.; et al. Effect of Light on Carotenoid and Lipid Production in the Oleaginous Yeast Rhodosporidium Toruloides. Biosci. Biotechnol. Biochem. 2020, 84, 1501–1512. [Google Scholar] [CrossRef]
- Pan, X.; Wang, B.; Duan, R.; Jia, J.; Li, J.; Xiong, W.; Ling, X.; Chen, C.; Huang, X.; Zhang, G.; et al. Enhancing Astaxanthin Accumulation in Xanthophyllomyces Dendrorhous by a Phytohormone: Metabolomic and Gene Expression Profiles. Microb. Biotechnol. 2020, 13, 1446–1460. [Google Scholar] [CrossRef]
- Dhar, H.; Chaudhary, A.; Rana, V. Designer Microbes for Nutraceutical Application. In Advances in Agri-Food Biotechnology; Springer: Singapore, 2020; pp. 239–285. [Google Scholar]
- Chamovitz, D.; Sandmann, G.; Hirschberg, J. Molecular and Biochemical Characterization of Herbicide-Resistant Mutants of Cyanobacteria Reveals That Phytoene Desaturation Is a Rate-Limiting Step in Carotenoid Biosynthesis. J. Biol. Chem. 1993, 268, 17348–17353. [Google Scholar] [CrossRef]
- Guo, W.; Liu, Y.; Yan, X.; Liu, M.; Tang, H.; Liu, Z.; Zhang, L. Cloning and Characterization of a Phytoene Dehydrogenase Gene from Marine Yeast Rhodosporidium Diobovatum. Antonie Leeuwenhoek 2015, 107, 1017–1027. [Google Scholar] [CrossRef] [PubMed]
- Nekrutenko, A.; Lariviere, D.; Rasche, H. Making Sense of a Newly Assembled Genome (Galaxy Training Materials). 2020. Available online: https://training.galaxyproject.org/trainingmaterial/topics/assembly/tutorials/ecoli_comparison/tutorial.html (accessed on 3 December 2020).




| Genome assembly size (Mbp) | 21.14 |
| Genome coverage | 44× |
| Number of contigs | 614 |
| Number of scaffolds | 361 |
| Number of scaffolds ≥2Kbp | 336 |
| Scaffold N50 | 55 |
| Scaffold L50 (Mbp) | 0.12 |
| Number of gaps | 253 |
| Percentage of scaffold length in gaps | 0.10% |
| Three largest scaffolds (Mbp) | 0.56, 0.49, 0.41 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fakankun, I.; Fristensky, B.; Levin, D.B. Genome Sequence Analysis of the Oleaginous Yeast, Rhodotorula diobovata, and Comparison of the Carotenogenic and Oleaginous Pathway Genes and Gene Products with Other Oleaginous Yeasts. J. Fungi 2021, 7, 320. https://doi.org/10.3390/jof7040320
Fakankun I, Fristensky B, Levin DB. Genome Sequence Analysis of the Oleaginous Yeast, Rhodotorula diobovata, and Comparison of the Carotenogenic and Oleaginous Pathway Genes and Gene Products with Other Oleaginous Yeasts. Journal of Fungi. 2021; 7(4):320. https://doi.org/10.3390/jof7040320
Chicago/Turabian StyleFakankun, Irene, Brian Fristensky, and David B. Levin. 2021. "Genome Sequence Analysis of the Oleaginous Yeast, Rhodotorula diobovata, and Comparison of the Carotenogenic and Oleaginous Pathway Genes and Gene Products with Other Oleaginous Yeasts" Journal of Fungi 7, no. 4: 320. https://doi.org/10.3390/jof7040320
APA StyleFakankun, I., Fristensky, B., & Levin, D. B. (2021). Genome Sequence Analysis of the Oleaginous Yeast, Rhodotorula diobovata, and Comparison of the Carotenogenic and Oleaginous Pathway Genes and Gene Products with Other Oleaginous Yeasts. Journal of Fungi, 7(4), 320. https://doi.org/10.3390/jof7040320





