Enhanced Lignocellulolytic Enzyme Activities on Hardwood and Softwood during Interspecific Interactions of White- and Brown-Rot Fungi
Abstract
1. Introduction
2. Materials and Methods
2.1. Fungal Cultures
2.2. Enzyme Assays
2.3. Statistical Analysis and Fungal Carbohydrate-Active Enzymes
2.4. Substrate Composition Analysis
3. Results
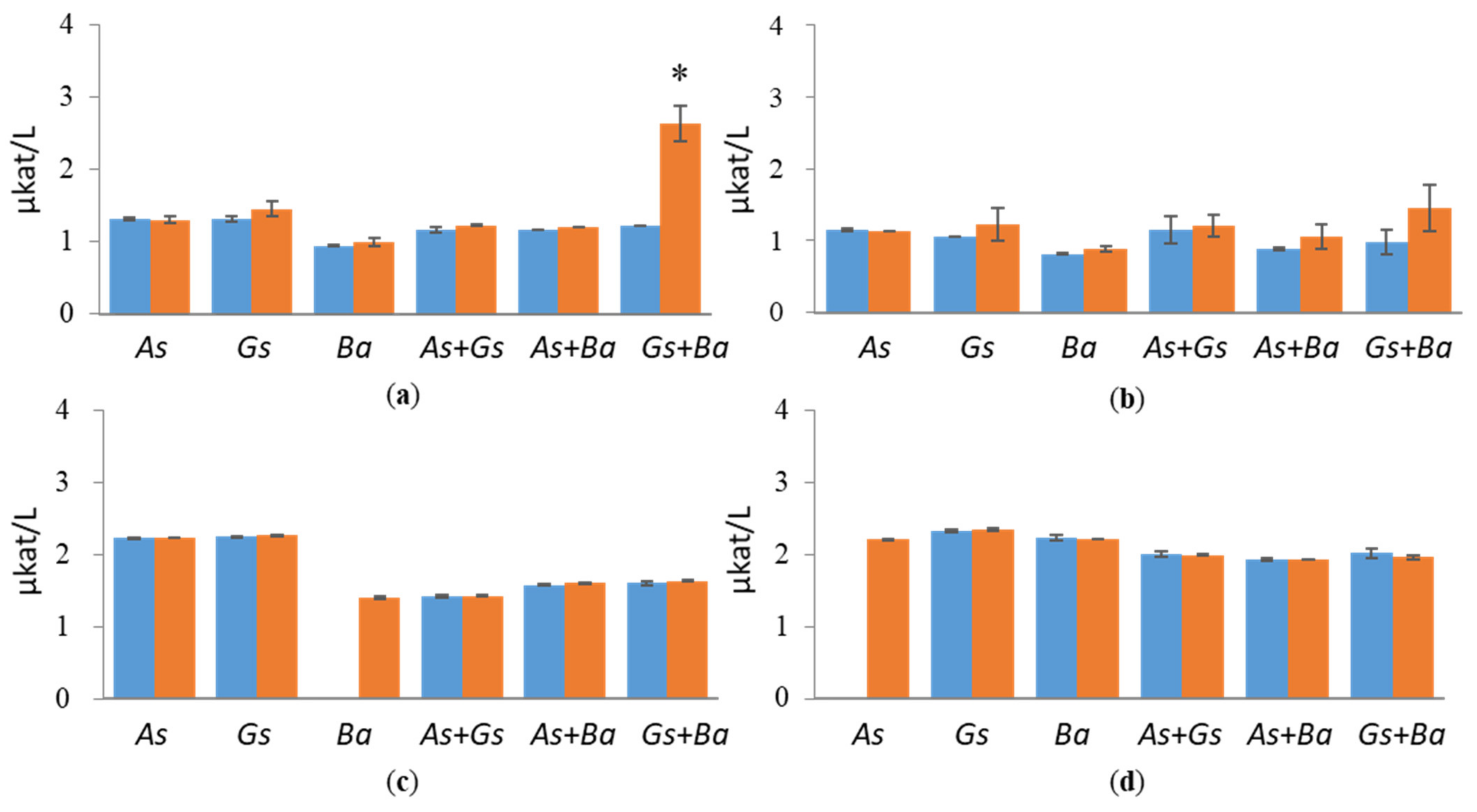
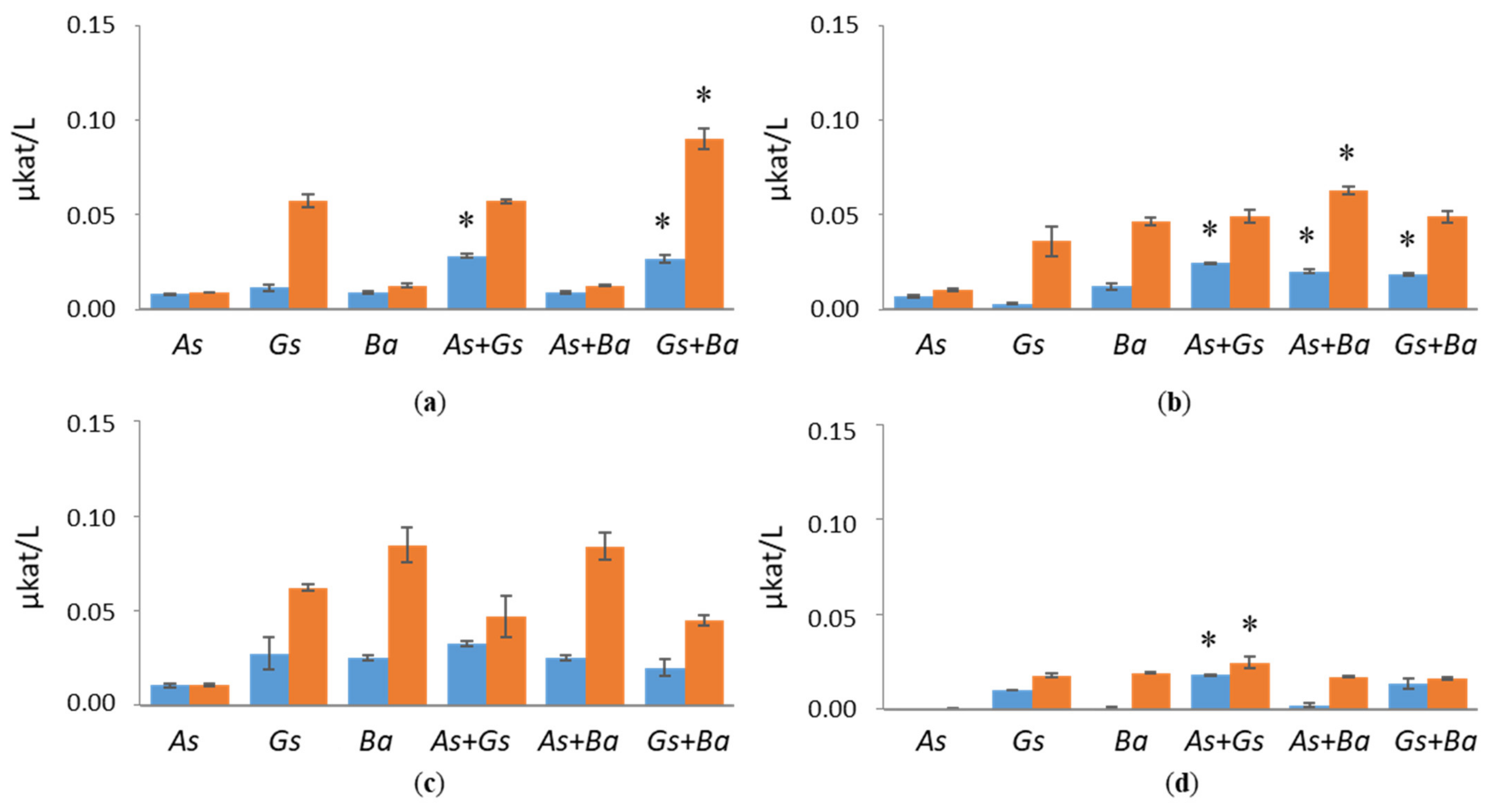
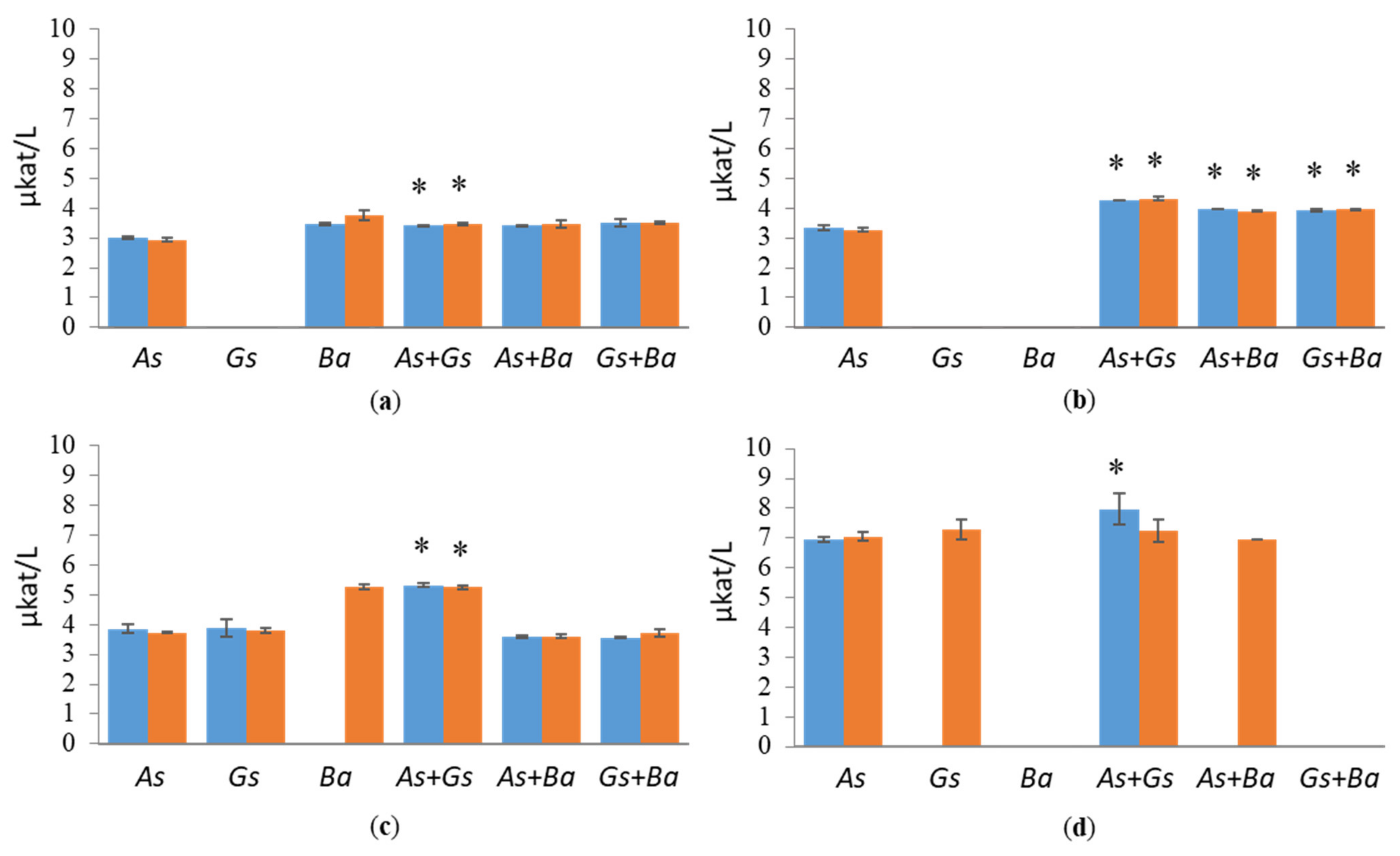
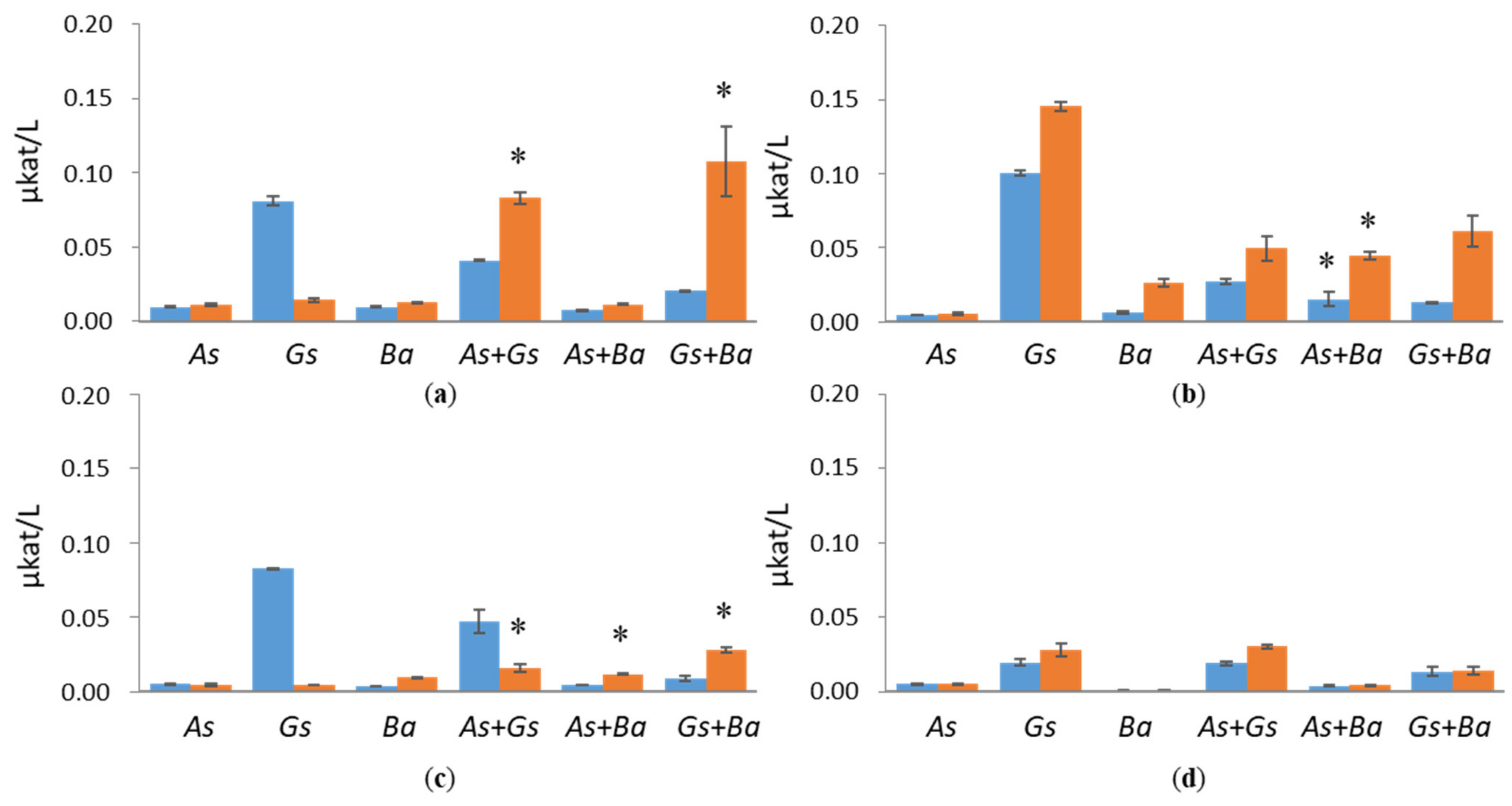
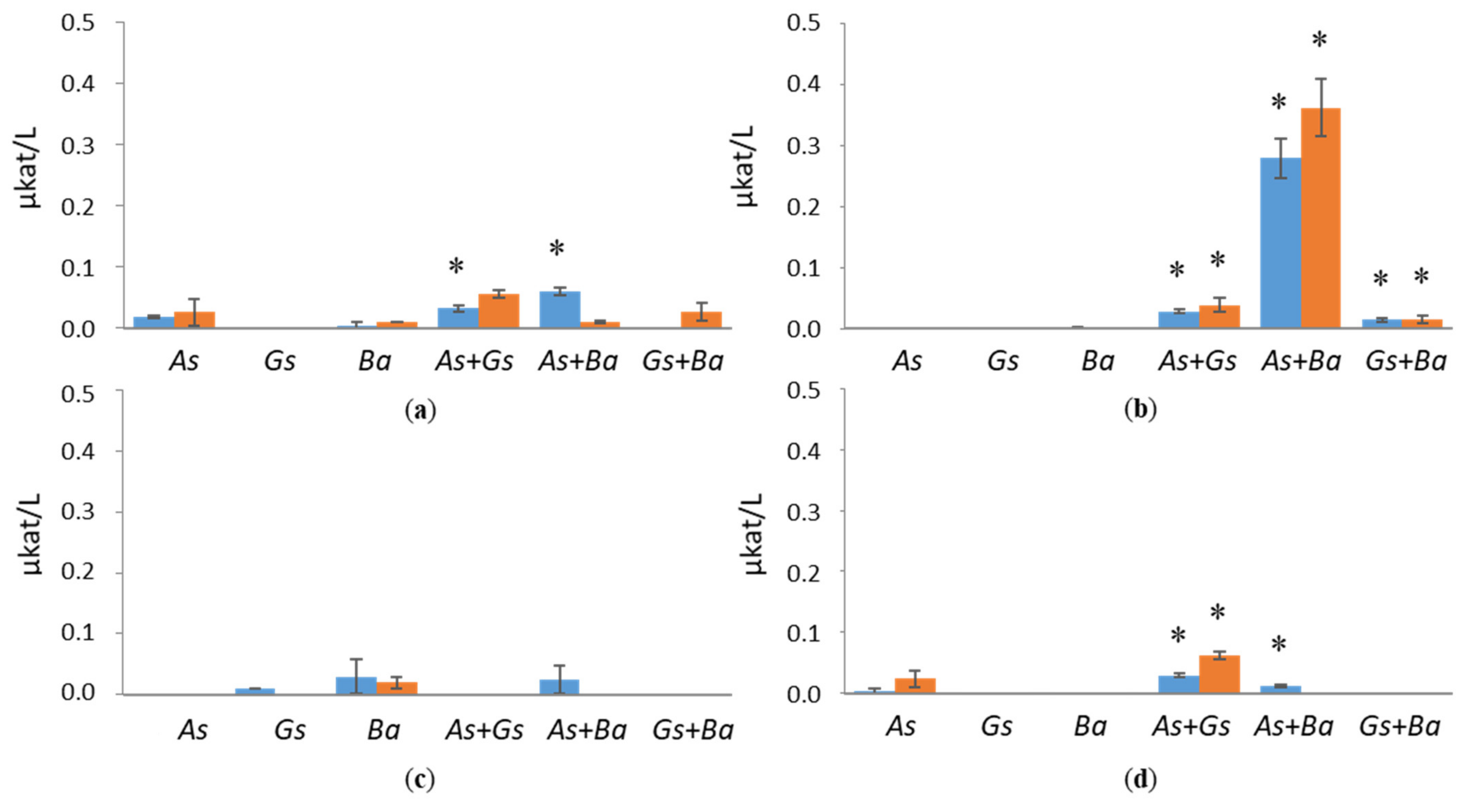
3.1. Hydrolytic Enzyme Activities
3.1.1. Cellulolytic Activities
3.1.2. Hemicellulolytic Activities
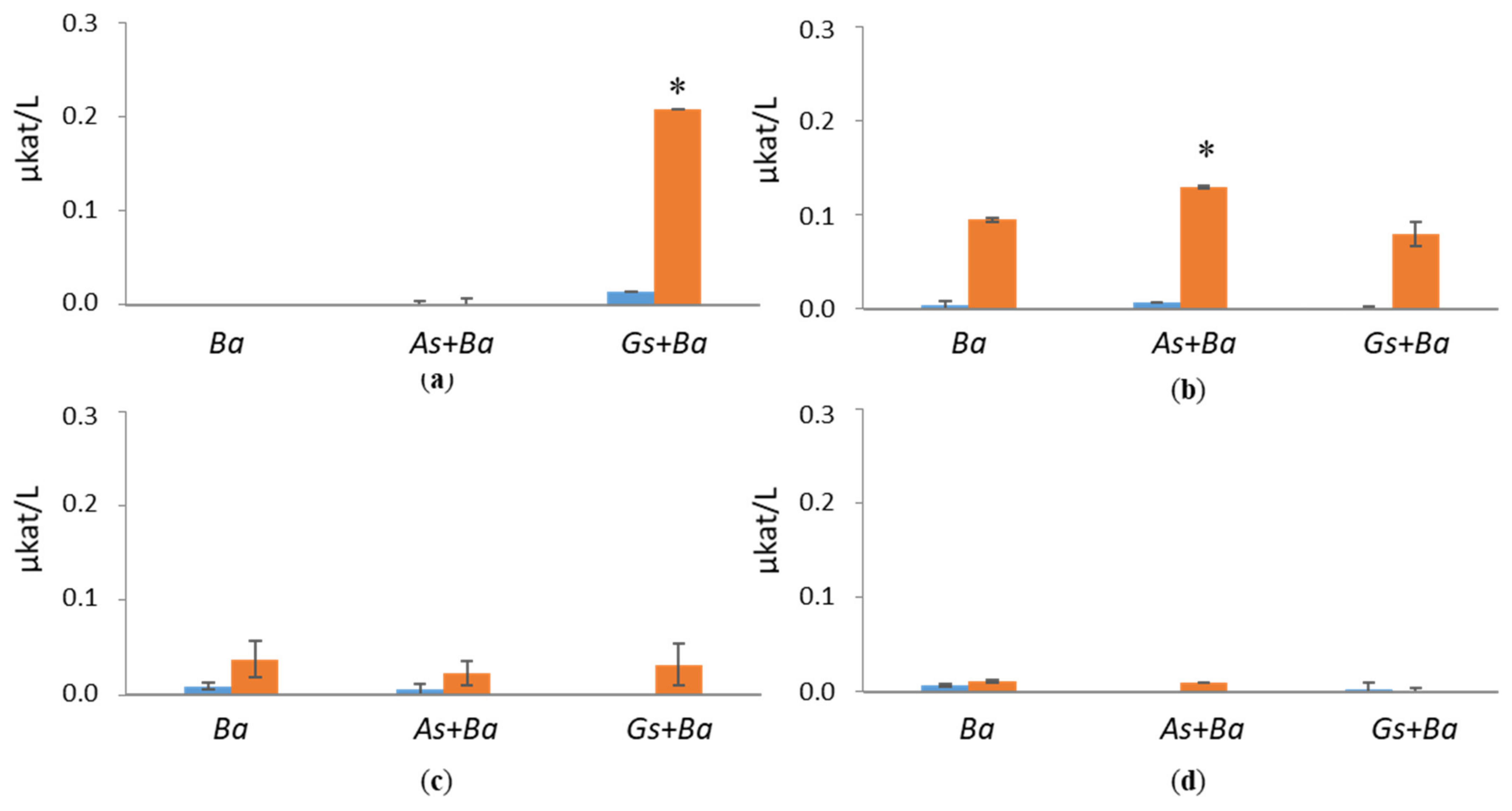
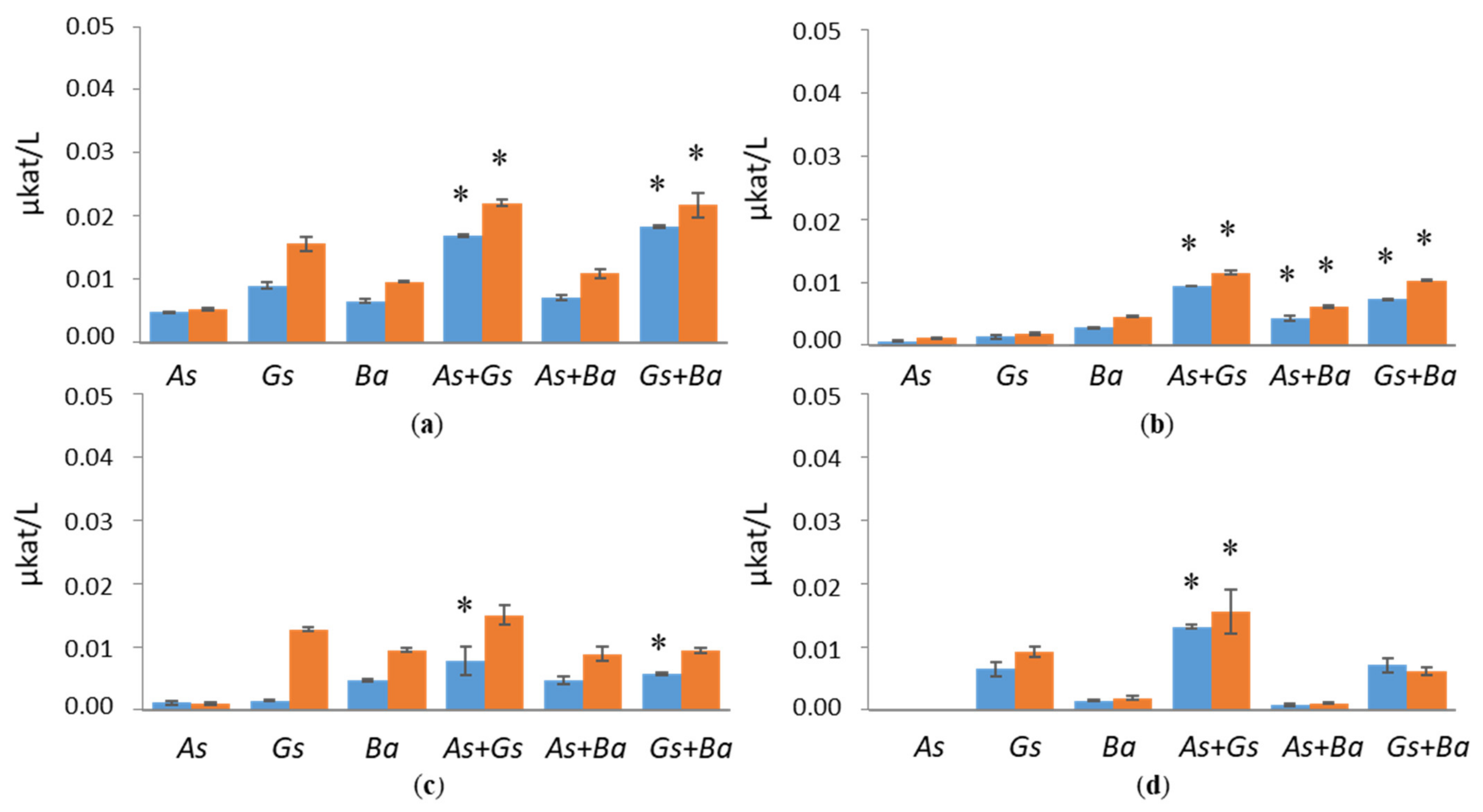
3.2. Activities of Lignin-Degrading Enzymes
3.3. Wood Composition after Fungal Treatment
4. Discussion
4.1. Co-Cultivation of White-Rot Fungus, B. adusta, and Brown-Rot Fungus, G. sepiarium
4.2. Co-Cultivation of White-Rot Fungus, B. adusta, and Brown-Rot Fungus, A. sinuosa
4.3. Co-Cultivation of Brown-Rot Fungi, G. sepiarium and A. sinuosa
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Hatakka, A.; Hammel, K.E. Fungal Biodegradation of Lignocelluloses. In Industrial Applications; Hofrichter, M., Ed.; Springer: Berlin/Heidelberg, Germany, 2011; pp. 319–340. ISBN 978-3-642-11458-8. [Google Scholar]
- Lundell, T.K.; Mäkelä, M.R.; de Vries, R.P.; Hildén, K.S. Genomics, lifestyles and future prospects of wood-decay and litter-decomposing basidiomycota. Adv. Bot. Res. 2014, 70, 329–370. [Google Scholar] [CrossRef]
- Rytioja, J.; Hildén, K.; Yuzon, J.; Hatakka, A.; de Vries, R.P.; Mäkelä, M.R. Plant-polysaccharide-degrading enzymes from basidiomycetes. Microbiol. Mol. Biol. Rev. 2014, 78, 614–649. [Google Scholar] [CrossRef] [PubMed]
- Sjöström, E. Wood Chemistry: Fundamentals and Applications; Sjöström, E., Alén, R., Eds.; Academic Press: San Diego, CA, USA, 1993. [Google Scholar]
- Mäkelä, M.R.; Hildén, K.S.; de Vries, R.P. Degradation and modification of plant biomass by Fungi. In Fungal Genomics; Nowrousian, M., Ed.; Springer: Berlin/Heidelberg, Germany, 2014; pp. 175–208. ISBN 978-3-642-45218-5. [Google Scholar]
- Riley, R.; Salamov, A.A.; Brown, D.W.; Nagy, L.G.; Floudas, D.; Held, B.W.; Levasseur, A.; Lombard, V.; Morin, E.; Otillar, R.; et al. Extensive sampling of basidiomycete genomes demonstrates inadequacy of the white-rot/brown-rot paradigm for wood decay fungi. Proc. Natl. Acad. Sci. USA 2014, 111, 9923–9928. [Google Scholar] [CrossRef] [PubMed]
- Koenigs, J.W. Production of hydrogen peroxide by wood-rotting fungi in wood and its correlation with weight loss, depolymerization, and pH changes. Arch. Microbiol. 1974, 99, 129–145. [Google Scholar] [CrossRef]
- Rättö, M.; Ritschkoff, A.C.; Viikari, L. The effect of oxidative pretreatment on cellulose degradation by Poria placenta and Trichoderma reesei cellulases. Appl. Microbiol. Biotechnol. 1997, 48, 53–57. [Google Scholar] [CrossRef]
- Renvall, P. Community structure and dynamics of wood-rotting basidiomycetes on decomposing conifer trunks in northern Finland. Karstenia 1995, 35, 1–51. [Google Scholar] [CrossRef]
- Lindblad, I. Wood-inhabiting fungi on fallen logs of Norway spruce: Relations to forest management and substrate quality. Nord. J. Bot. 1998, 18, 243–255. [Google Scholar] [CrossRef]
- Matheron, M.E.; Porchas, M.; Bigelow, D.M. Factors affecting the development of wood rot on lemon trees infected with Antrodia sinuosa, Coniophora eremophila, and a Nodulisporium sp. Plant Dis. 2006, 90, 554–558. [Google Scholar] [CrossRef] [PubMed]
- Jönsson, M.T.; Edman, M.; Jonsson, B.G. Colonization and extinction patterns of wood-decaying fungi in a boreal old-growth Picea abies forest. J. Ecol. 2008, 96, 1065–1075. [Google Scholar] [CrossRef]
- Olsson, J.; Jonsson, B.G. Restoration fire and wood-inhabiting fungi in a Swedish Pinus sylvestris forest. For. Ecol. Manag. 2010, 259, 1971–1980. [Google Scholar] [CrossRef]
- Haas, D.; Mayrhofer, H.; Habib, J.; Galler, H.; Reinthaler, F.F.; Fuxjäger, M.L.; Buzina, W. Distribution of building-associated wood-destroying fungi in the federal state of Styria, Austria. Eur. J. Wood Wood Prod. 2019, 77, 527–537. [Google Scholar] [CrossRef]
- Sugano, J.; Linnakoski, R.; Huhtinen, S.; Pappinen, A.; Niemelä, P.; Asiegbu, F.O. Cellulolytic activity of brown-rot Antrodia sinuosa at the initial stage of cellulose degradation. Holzforschung 2019, 73, 673–680. [Google Scholar] [CrossRef]
- Garcia-Sandoval, R.; Wang, Z.; Binder, M.; Hibbett, D.S. Molecular phylogenetics of the Gloeophyllales and relative ages of clades of Agaricomycotina producing a brown rot. Mycologia 2011, 103, 510–524. [Google Scholar] [CrossRef] [PubMed]
- Floudas, D.; Binder, M.; Riley, R.; Barry, K.; Blanchette, R.A.; Henrissat, B.; Martínez, A.T.; Otillar, R.; Spatafora, J.W.; Yadav, J.S.; et al. The paleozoic origin of enzymatic lignin decomposition reconstructed from 31 fungal genomes. Science 2012, 336, 1715–1719. [Google Scholar] [CrossRef]
- Presley, G.N.; Zhang, J.; Purvine, S.O.; Schilling, J.S. Functional genomics, transcriptomics, and proteomics reveal distinct combat strategies between lineages of wood-degrading fungi with redundant wood decay mechanisms. Front. Microbiol. 2020, 11, 1646. [Google Scholar] [CrossRef] [PubMed]
- Owens, E.M.; Reddy, C.A.; Grethlein, H.E. Outcome of interspecific interactions among brown-rot and white-rot wood decay fungi. FEMS Microbiol. Ecol. 1994, 14, 19–24. [Google Scholar] [CrossRef]
- Boddy, L. Interspecific combative interactions between wood-decaying basidiomycetes. FEMS Microbiol. Ecol. 2000, 31, 185–194. [Google Scholar] [CrossRef]
- Hiscox, J.; Savoury, M.; Müller, C.T.; Lindahl, B.D.; Rogers, H.J.; Boddy, L. Priority effects during fungal community establishment in beech wood. ISME J. 2015, 14, 24–32. [Google Scholar] [CrossRef]
- Kubartová, A.; Ottosson, E.; Dahlberg, A.; Stenlid, J. Patterns of fungal communities among and within decaying logs, revealed by 454 sequencing. Mol. Ecol. 2012, 21, 4514–4532. [Google Scholar] [CrossRef]
- Ovaskainen, O.; Hottola, J.; Shtonen, J. Modeling species co-occurrence by multivariate logistic regression generates new hypotheses on fungal interactions. Ecology 2010, 91, 2514–2521. [Google Scholar] [CrossRef]
- Ottosson, E.; Nordén, J.; Dahlberg, A.; Edman, M.; Jönsson, M.; Larsson, K.H.; Olsson, J.; Penttilä, R.; Stenlid, J.; Ovaskainen, O. Species associations during the succession of wood-inhabiting fungal communities. Fungal Ecol. 2014, 11, 17–28. [Google Scholar] [CrossRef]
- Baldrian, P. Increase of laccase activity during interspecific interactions of white-rot fungi. FEMS Microbiol. Ecol. 2004, 50, 245–253. [Google Scholar] [CrossRef] [PubMed]
- Toljander, Y.K.; Lindahl, B.D.; Holmer, L.; Högberg, N.O.S. Environmental fluctuations facilitate species co-existence and increase decomposition in communities of wood decay fungi. Oecologia 2006, 148, 625–631. [Google Scholar] [CrossRef] [PubMed]
- Van Der Wal, A.; Ottosson, E.; De Boer, W. Neglected role of fungal community composition in explaining variation in wood decay rates. Ecology 2015, 96, 124–133. [Google Scholar] [CrossRef]
- Noll, L.; Leonhardt, S.; Arnstadt, T.; Hoppe, B.; Poll, C.; Matzner, E.; Hofrichter, M.; Kellner, H. Fungal biomass and extracellular enzyme activities in coarse woody debris of 13 tree species in the early phase of decomposition. For. Ecol. Manag. 2016, 378, 181–192. [Google Scholar] [CrossRef]
- Mali, T.; Kuuskeri, J.; Shah, F.; Lundell, T.K. Interactions affect hyphal growth and enzyme profiles in combinations of coniferous wood-decaying fungi of Agaricomycetes. PLoS ONE 2017, 12, e0185171. [Google Scholar] [CrossRef]
- Mali, T.; Mäki, M.; Hellén, H.; Heinonsalo, J.; Bäck, J.; Lundell, T. Decomposition of spruce wood and release of volatile organic compounds depend on decay type, fungal interactions and enzyme production patterns. FEMS Microbiol. Ecol. 2019, 95, fiz135. [Google Scholar] [CrossRef]
- Sperandio, G.B.; Ferreira Filho, E.X. Fungal co-cultures in the lignocellulosic biorefinery context: A review. Int. Biodeterior. Biodegrad. 2019, 142, 109–123. [Google Scholar] [CrossRef]
- Sugano, J.; Awan, H.U.M.; Asiegbu, F.O. Effect of interspecific fungal interactions on wood decomposition. Manuscript in preparation.
- Hatakka, A.I.; Uusi-Rauva, A.K. Degradation of 14C-labelled poplar wood lignin by selected white-rot fungi. Eur. J. Appl. Microbiol. Biotechnol. 1983, 17, 235–242. [Google Scholar] [CrossRef]
- Wood, T.M.; Bhat, K.M. Methods for measuring cellulase activities. In Methods in Enzymology; Academic Press: Cambridge, MA, USA, 1988; Volume 160, pp. 87–112. ISBN 0076-6879. [Google Scholar]
- Bailey, M.J.; Nevalainen, K.M.H. Induction, isolation and testing of stable Trichoderma reesei mutants with improved production of solubilizing cellulase. Enzyme Microb. Technol. 1981, 3, 153–157. [Google Scholar] [CrossRef]
- Ko, K.C.; Han, Y.; Choi, J.H.; Kim, G.J.; Lee, S.G.; Song, J.J. A novel bifunctional endo-/exo-type cellulase from an anaerobic ruminal bacterium. Appl. Microbiol. Biotechnol. 2011, 89, 1453–1462. [Google Scholar] [CrossRef]
- Benoit, I.; Culleton, H.; Zhou, M.; DiFalco, M.; Aguilar-Osorio, G.; Battaglia, E.; Bouzid, O.; Brouwer, C.P.J.M.; El-Bushari, H.B.O.; Coutinho, P.M.; et al. Closely related fungi employ diverse enzymatic strategies to degrade plant biomass. Biotechnol. Biofuels 2015, 8, 107. [Google Scholar] [CrossRef]
- Presley, G.N.; Panisko, E.; Purvine, S.O.; Schilling, J.S. Coupling secretomics with enzyme activities to compare the temporal processes of wood metabolism among white and brown rot fungi. Appl. Environ. Microbiol. 2018, 84, e00159-18. [Google Scholar] [CrossRef]
- Slomczynski, D.; Nakas, J.P.; Tanenbaum, S.W. Production and characterization of laccase from Botrytis cinerea 61–34. Appl. Environ. Microbiol. 1995, 61, 907–912. [Google Scholar] [CrossRef] [PubMed]
- Wariishi, H.; Valli, K.; Gold, M.H. Manganese(II) oxidation by manganese peroxidase from the basidiomycete Phanerochaete chrysosporium: Kinetic mechanism and role of chelators. J. Biol. Chem. 1992, 267, 23688–23695. [Google Scholar] [CrossRef]
- R Foundation for Statistical Computing. A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2018; ISBN 3900051070. [Google Scholar]
- Laine, C.; Tamminen, T.; Vikkula, A.; Vuorinen, T. Methylation analysis as a tool for structural analysis of wood polysaccharides. Holzforschung 2002, 56, 607–614. [Google Scholar] [CrossRef]
- Sundberg, A.; Sundberg, K.; Lillandt, C.; Holmbom, B. Determination of hemicelluloses and pectins in wood and pulp fibres by acid methanolysis and gas chromatography. Nord. Pulp Pap. Res. J. 1996, 11, 216–219. [Google Scholar] [CrossRef]
- Kirk, T.K.; Farrell, R.L. Enzymatic “combustion”: The microbial degradation of lignin. Annu. Rev. Microbiol. 1987, 41, 465–505. [Google Scholar] [CrossRef]
- Stokland, J.N.; Siitonen, J.; Jonsson, B.G. Biodiversity in Dead Wood; Cambridge University Press: Cambridge, UK, 2012; pp. 18–20. ISBN 9781139025843. [Google Scholar]
- Dorado, J.; Van Beek, T.A.; Claassen, F.W.; Sierra-Alvarez, R. Degradation of lipophilic wood extractive constituents in Pinus sylvestris by the white-rot fungi Bjerkandera sp. and Trametes versicolor. Wood Sci. Technol. 2001, 35, 117–125. [Google Scholar] [CrossRef]
- Lindhe, A.; Åsenblad, N.; Toresson, H.G. Cut logs and high stumps of spruce, birch, aspen and oak—Nine years of saproxylic fungi succession. Biol. Conserv. 2004, 119, 443–454. [Google Scholar] [CrossRef]
- Hakala, T.K.; Maijala, P.; Konn, J.; Hatakka, A. Evaluation of novel wood-rotting polypores and corticioid fungi for the decay and biopulping of Norway spruce (Picea abies) wood. Enzyme Microb. Technol. 2004, 34, 255–263. [Google Scholar] [CrossRef]
- Quiroz-Castañeda, R.E.; Pérez-Mejía, N.; Martínez-Anaya, C.; Acosta-Urdapilleta, L.; Folch-Mallol, J. Evaluation of different lignocellulosic substrates for the production of cellulases and xylanases by the basidiomycete fungi Bjerkandera adusta and Pycnoporus sanguineus. Biodegradation 2011, 22, 565–572. [Google Scholar] [CrossRef]
- Moody, S.C.; Dudley, E.; Hiscox, J.; Boddy, L.; Eastwood, D.C. Interdependence of primary metabolism and xenobiotic mitigation characterizes the proteome of Bjerkandera adusta during wood decomposition. Appl. Environ. Microbiol. 2018, 84, e01401-17. [Google Scholar] [CrossRef] [PubMed]
- Villavicencio, E.V.; Mali, T.; Mattila, H.K.; Lundell, T. Enzyme activity profiles produced on wood and straw by four fungi of different decay strategies. Microorganisms 2020, 8, 73. [Google Scholar] [CrossRef]
- Kuuskeri, J.; Mäkelä, M.R.; Isotalo, J.; Oksanen, I.; Lundell, T. Lignocellulose-converting enzyme activity profiles correlate with molecular systematics and phylogeny grouping in the incoherent genus Phlebia (Polyporales, Basidiomycota) Ecological and evolutionary microbiology. BMC Microbiol. 2015, 15, 217. [Google Scholar] [CrossRef] [PubMed]
- Casado López, S.; Theelen, B.; Manserra, S.; Issak, T.Y.; Rytioja, J.; Mäkelä, M.R.; de Vries, R.P. Functional diversity in Dichomitus squalens monokaryons. IMA Fungus 2017, 8, 17–25. [Google Scholar] [CrossRef] [PubMed]
- Presley, G.N.; Schilling, J.S. Distinct growth and secretome strategies for two taxonomically divergent brown rot fungi. Appl. Environ. Microbiol. 2017, 83, e02987-16. [Google Scholar] [CrossRef] [PubMed]
- Arantes, V.; Goodell, B. Current understanding of brown-rot fungal biodegradation mechanisms: A review. ACS Symp. Ser. 2014, 1158, 3–21. [Google Scholar] [CrossRef]
- Aguiar, A.; Gavioli, D.; Ferraz, A. Extracellular activities and wood component losses during Pinus taeda biodegradation by the brown-rot fungus Gloeophyllum trabeum. Int. Biodeterior. Biodegrad. 2013, 82, 187–191. [Google Scholar] [CrossRef]
- Qi-He, C.; Krügener, S.; Hirth, T.; Rupp, S.; Zibek, S. Co-cultured production of lignin-modifying enzymes with white-rot fungi. Appl. Biochem. Biotechnol. 2011, 165, 700–718. [Google Scholar] [CrossRef]
- Tomšovský, M.; Popelářová, P.; Baldrian, P. Production and regulation of lignocellulose-degrading enzymes of Poria-like wood-inhabiting basidiomycetes. Folia Microbiol. 2009, 54, 74–80. [Google Scholar] [CrossRef] [PubMed]
- Krah, F.S.; Bässler, C.; Heibl, C.; Soghigian, J.; Schaefer, H.; Hibbett, D.S. Evolutionary dynamics of host specialization in wood-decay fungi. BMC Evol. Biol. 2018, 18, 119. [Google Scholar] [CrossRef] [PubMed]
- Hiscox, J.; Boddy, L. Armed and dangerous—Chemical warfare in wood decay communities. Fungal Biol. Rev. 2017, 31, 169–184. [Google Scholar] [CrossRef]
- Hiscox, J.; Baldrian, P.; Rogers, H.J.; Boddy, L. Changes in oxidative enzyme activity during interspecific mycelial interactions involving the white-rot fungus Trametes versicolor. Fungal Genet. Biol. 2010, 47, 562–571. [Google Scholar] [CrossRef]
- Huisman, J.; Weissing, F.J. Fundamental unpredictability in multispecies competition. Am. Nat. 2001, 157, 488–494. [Google Scholar] [CrossRef] [PubMed]



| Man | Glu | Xyl | Gal | GalA | Ara | Rha | GlcA | ||
|---|---|---|---|---|---|---|---|---|---|
| Pine wood | 10.76 | 9.98 | 5.72 | 2.31 | 1.37 | 1.36 | 0.44 | n.d | |
| Pine cultured with | A. sinuosa | 9.13 | 7.38 | 7.51 | 2.53 | 1.22 | 1.62 | 0.48 | n.d |
| G. sepiarium | 8.47 * | 7.30 * | 6.57 * | 2.15 | 1.13 * | 1.45 | 0.45 | n.d | |
| B. adusta | 6.22 * | 6.07 * | 5.77 | 2.22 | 1.11 * | 1.22 | 0.45 | n.d | |
| As + Gs | 8.03 * | 6.75 * | 6.20 | 2.37 | 1.07 * | 1.40 | 0.46 | n.d | |
| As + Ba | 6.53 * | 6.41 * | 5.99 | 3.19 * | 1.19 * | 1.23 | 0.46 | n.d | |
| Gs + Ba | 6.73 * | 5.68 * | 6.07 | 2.75 * | 1.12 * | 1.34 | 0.46 | n.d | |
| Spruce wood | 10.30 | 3.84 | 4.42 | 2.13 | 1.46 | 0.79 | 0.42 | n.d | |
| Spruce cultured with | A. sinuosa | 10.30 | 3.14 | 5.84 * | 1.70 | 1.72 | 0.96 | 0.66 * | n.d |
| G. sepiarium | 10.61 | 3.34 | 5.94 | 2.03 | 1.64 | 0.99 | 0.65 * | n.d | |
| B. adusta | 9.45 | 2.95 | 5.36 | 1.93 | 1.57 | 0.89 | 0.67 * | n.d | |
| As + Gs | 9.59 | 3.05 | 5.13 | 1.89 | 1.55 | 0.85 | 0.63 * | n.d | |
| As + Ba | 9.69 | 3.03 | 5.61 * | 1.78 | 1.58 | 0.99 | 0.63 * | n.d | |
| Gs + Ba | 9.42 | 3.06 | 5.49 | 1.95 | 1.62 | 0.94 | 0.64 * | n.d | |
| Birch wood | 0.98 | 7.56 | 21.44 | 1.17 | 2.16 | 0.53 | 0.68 | 0.35 | |
| Birch cultured with | A. sinuosa | 1.23 * | 5.28 * | 24.15 * | 1.74 | 2.11 | 0.69 * | 0.79 * | n.d |
| G. sepiarium | 1.17 * | 5.16 * | 22.18 | 1.46 * | 1.98 | 0.63 * | 0.74 * | n.d | |
| B. adusta | 1.74 * | 10.47 * | 13.19 * | 2.33 * | 1.85 | n.d | 0.60 | n.d | |
| As + Gs | 1.11 | 4.73 * | 18.34 | 1.93 * | 1.81 | 0.60 | 0.66 | n.d | |
| As + Ba | 1.85 * | 10.18 | 11.69 * | 3.86 * | 1.67 * | 0.58 | 0.56 * | n.d | |
| Gs + Ba | 1.34 | 4.83 | 18.04 | 2.91 | 2.08 | 0.63 | 0.70 | n.d | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sugano, J.; Maina, N.; Wallenius, J.; Hildén, K. Enhanced Lignocellulolytic Enzyme Activities on Hardwood and Softwood during Interspecific Interactions of White- and Brown-Rot Fungi. J. Fungi 2021, 7, 265. https://doi.org/10.3390/jof7040265
Sugano J, Maina N, Wallenius J, Hildén K. Enhanced Lignocellulolytic Enzyme Activities on Hardwood and Softwood during Interspecific Interactions of White- and Brown-Rot Fungi. Journal of Fungi. 2021; 7(4):265. https://doi.org/10.3390/jof7040265
Chicago/Turabian StyleSugano, Junko, Ndegwa Maina, Janne Wallenius, and Kristiina Hildén. 2021. "Enhanced Lignocellulolytic Enzyme Activities on Hardwood and Softwood during Interspecific Interactions of White- and Brown-Rot Fungi" Journal of Fungi 7, no. 4: 265. https://doi.org/10.3390/jof7040265
APA StyleSugano, J., Maina, N., Wallenius, J., & Hildén, K. (2021). Enhanced Lignocellulolytic Enzyme Activities on Hardwood and Softwood during Interspecific Interactions of White- and Brown-Rot Fungi. Journal of Fungi, 7(4), 265. https://doi.org/10.3390/jof7040265






