Effect of Static Magnetic Field on Monascus ruber M7 Based on Transcriptome Analysis
Abstract
1. Introduction
2. Materials and Methods
2.1. Static Magnetic Field Device and Its Treatment on M. ruber M7
2.2. Strains and Culture Conditions
2.3. Morphological Analysis of M. ruber M7
2.4. Determination of MPs, CIT and ERG Produced by M. ruber M7
2.5. Transcriptome Analysis of M. ruber M7 Treated with 30 mT SMF
2.6. Statistical Analysis
3. Results
3.1. Effects of SMF on the Morphological Characteristics of M. ruber M7
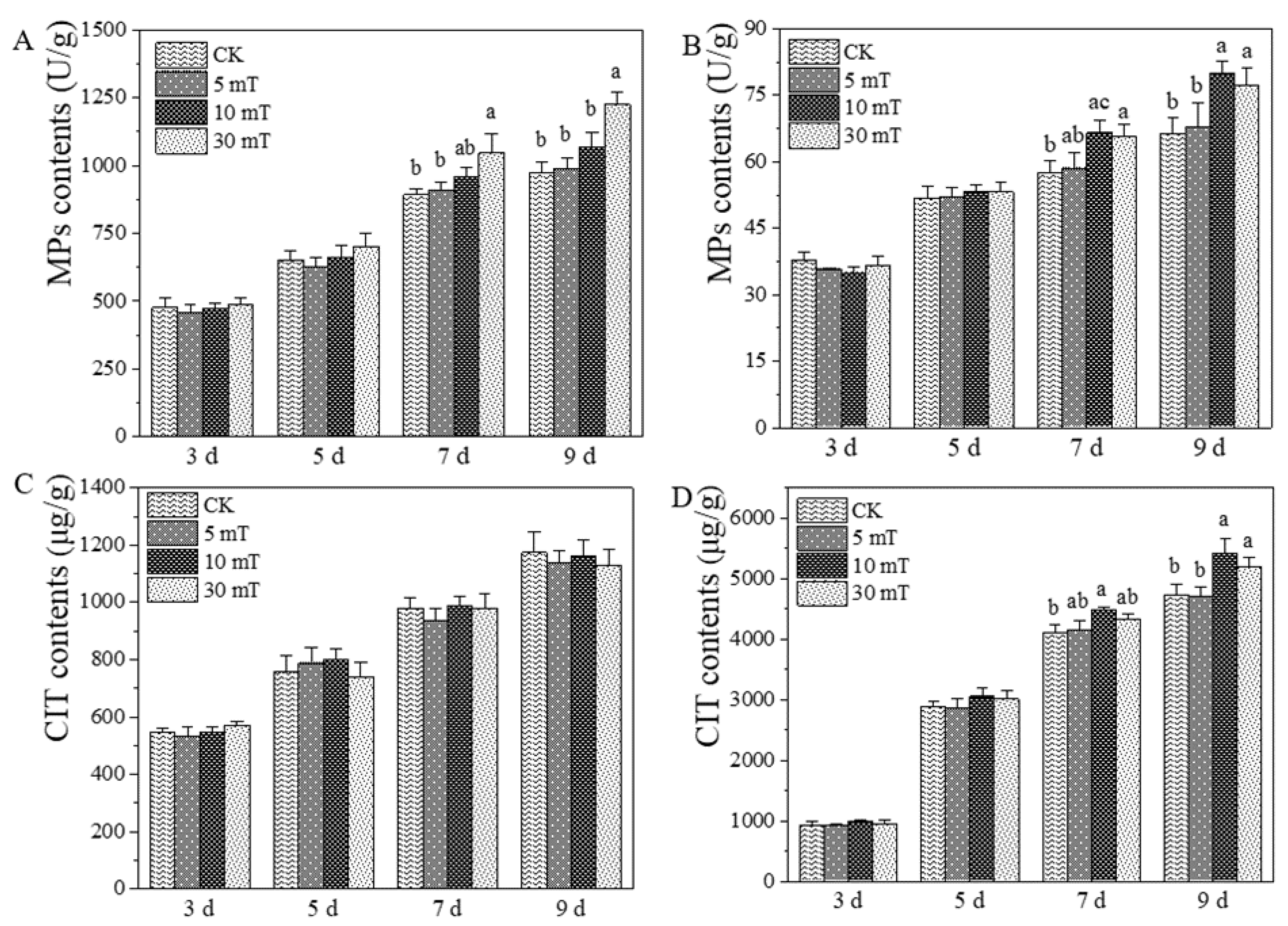
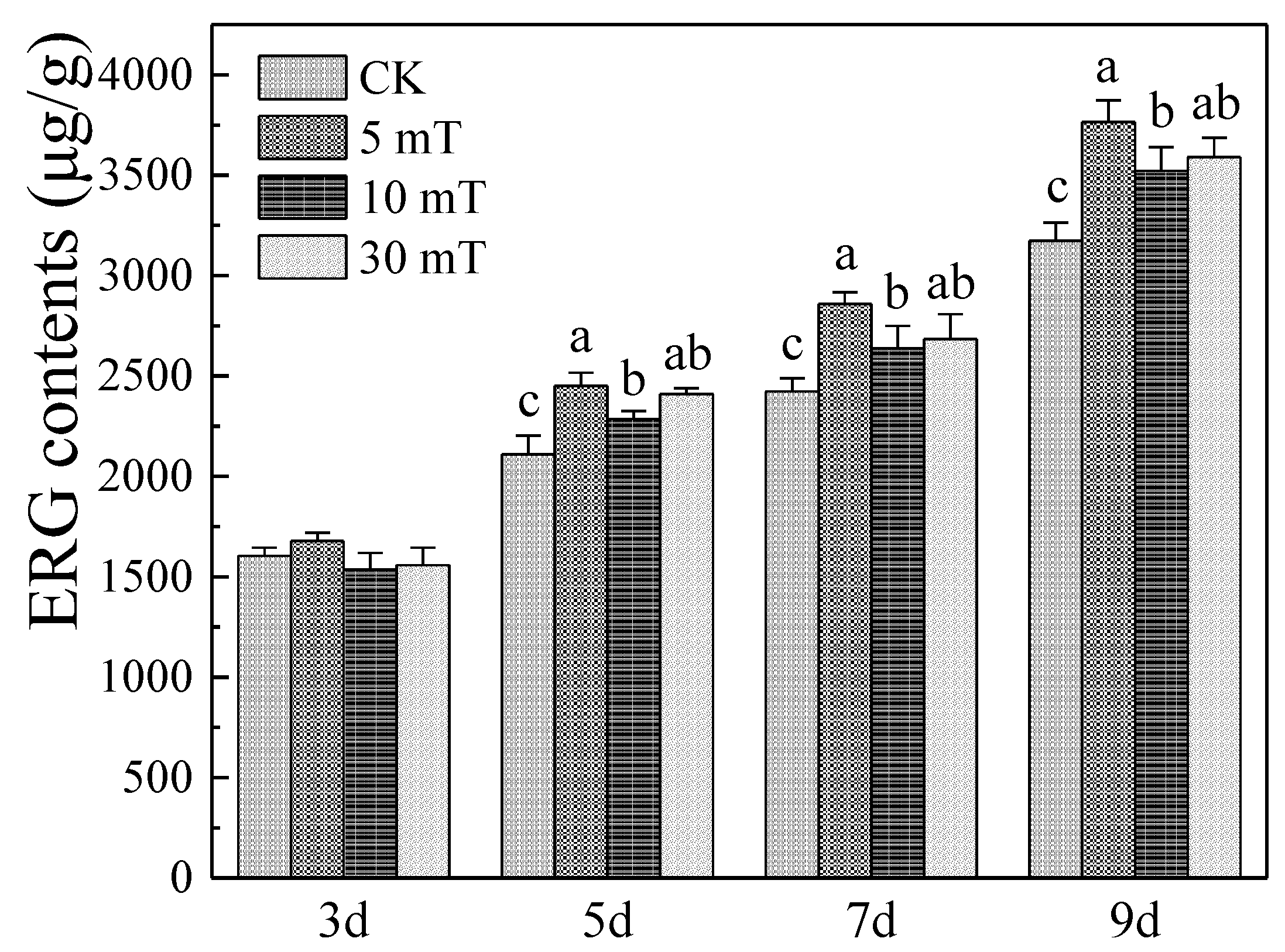
3.2. Effects of SMF on MPs, and CIT Produced by M. ruber M7
3.3. The Transcriptomic Analysis of M. ruber M7 under 30 mT SMF
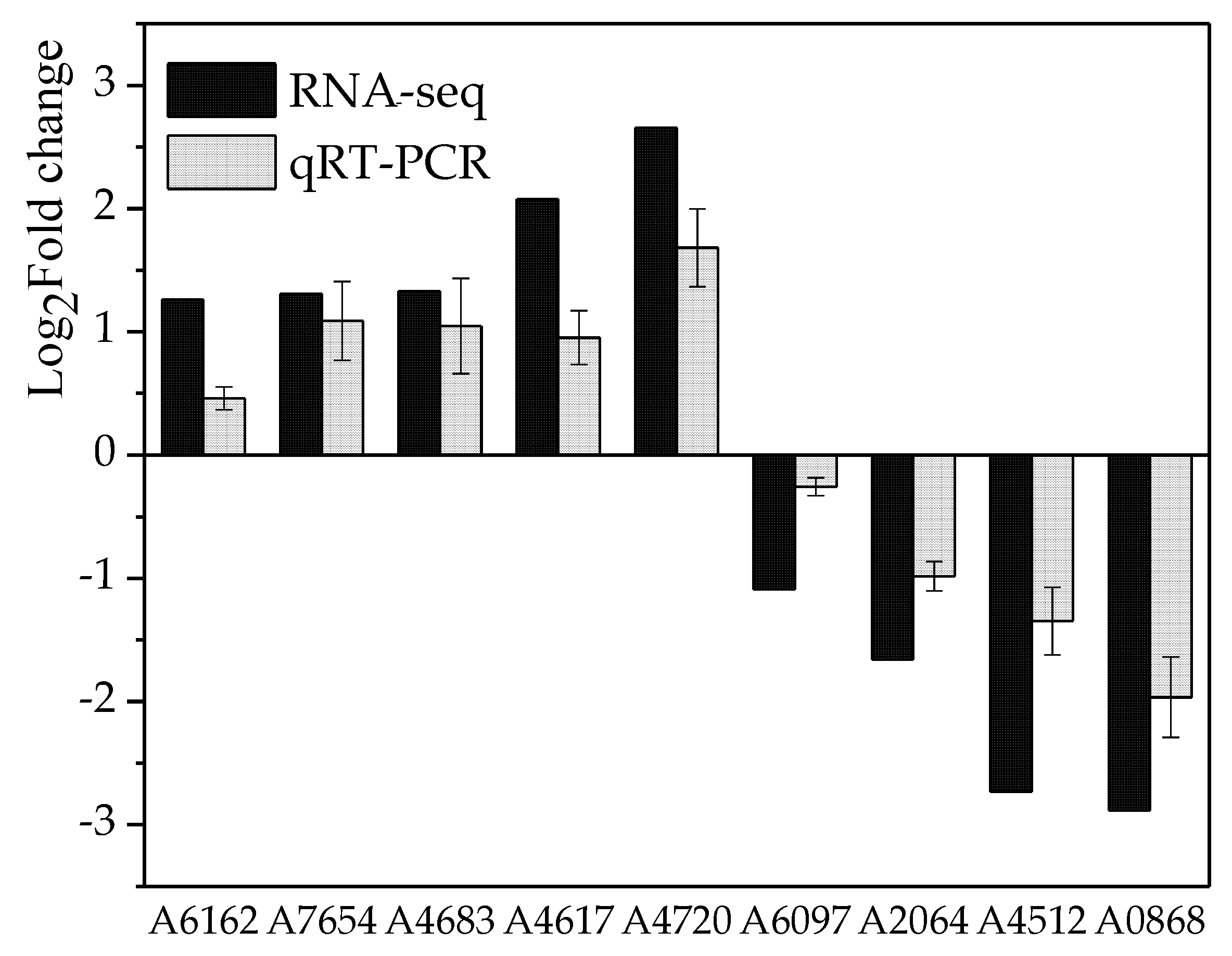
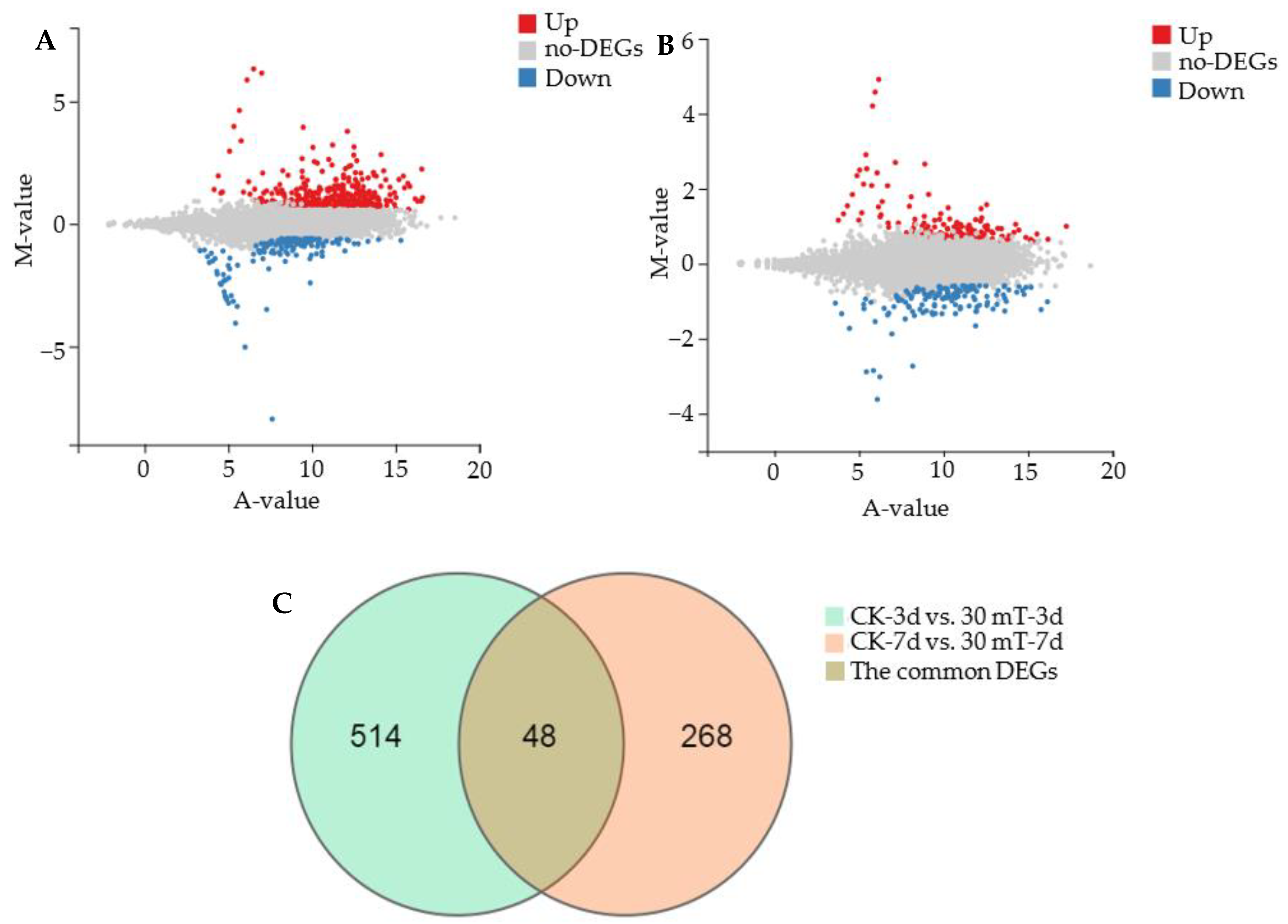
3.3.1. Analysis of Differentially Expressed Genes
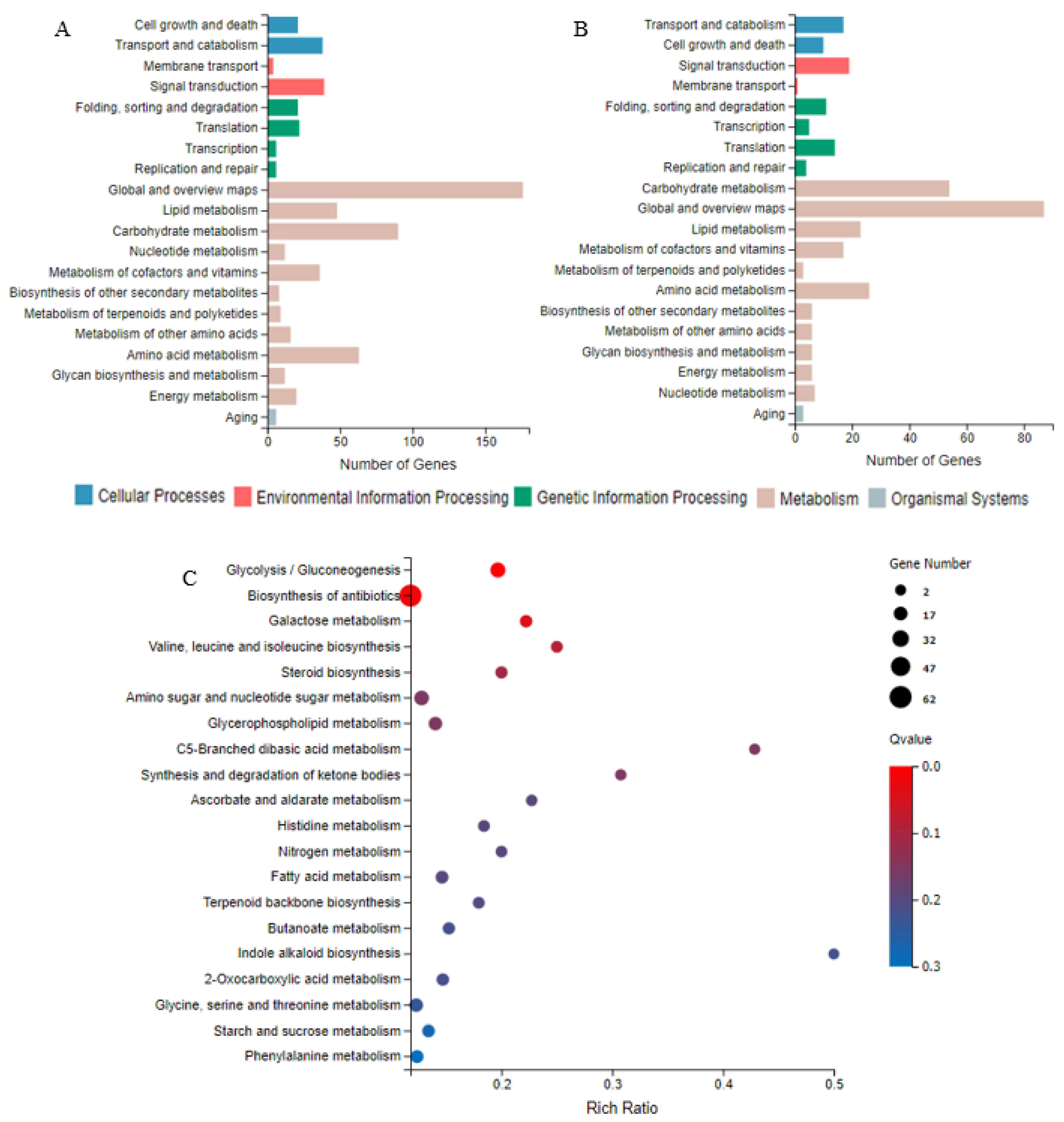
3.3.2. The KEGG Pathways and Their Enrichment Analyses for DEGs
3.3.3. Analysis of the DEGs Involved in the Primary Metabolism
3.3.4. Analysis of the DEGs Involved in the Secondary Metabolism
3.3.5. Analysis of the DEGs Involved in Signal Transduction Pathways
3.3.6. Effects of SMF on Transcriptional Factors
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Shi, Y.C.; Pan, T.M. Beneficial effects of Monascus purpureus NTU 568-fermented products: A review. Appl. Microbiol. Biotechnol. 2011, 90, 1207–1217. [Google Scholar] [CrossRef]
- Chen, W.P.; He, Y.; Zhou, Y.X.; Shao, Y.C.; Feng, Y.L.; Li, M.; Chen, F.S. Edible filamentous fungi from the species Monascus: Early traditional fermentations, modern molecular biology, and future genomics. Compr. Rev. Food Sci. Food Saf. 2015, 14, 555–567. [Google Scholar] [CrossRef]
- Wang, T.H.; Lin, T.F. Monascus Rice Products. Adv. Food Nutr. Res. 2007, 53, 123–159. [Google Scholar]
- Endo, A. Compactin (ML-236B) and related compounds as potential cholesterol-lowering agents that inhibit HMG-CoA reductase. J. Med. Chem. 1985, 28, 401–405. [Google Scholar] [CrossRef] [PubMed]
- Diana, M.; Quílez, J.; Rafecas, M. Gamma-aminobutyric acid as a bioactive compound in foods: A review. J. Funct. Food. 2014, 10, 407–420. [Google Scholar] [CrossRef]
- Cheng, M.J.; Wu, M.D.; Chen, I.S.; Chen, C.Y.; Lo, W.L.; Yuan, G.F. Secondary metabolites from the red mould rice of Monascus purpureus BCRC 38113. Nat. Prod. Res. 2010, 24, 1719–1725. [Google Scholar] [CrossRef] [PubMed]
- Chen, W.P.; Feng, Y.L.; Molnar, I.; Chen, F.S. Nature and nurture: Confluence of pathway determinism with metabolic and chemical serendipity diversifies Monascus azaphilone pigments. Nat. Prod. Rep. 2019, 36, 561–572. [Google Scholar] [CrossRef] [PubMed]
- Blanc, P.J.; Laussac, J.P.; Bars, J.L.; Bars, P.L.; Loret, M.O.; Pareilleux, A.; Prome, D.; Prome, J.C.; Santerre, A.L.; Goma, G. Characterization of monascidin A from Monascus as citrinin. Int. J. Food Microbiol. 1995, 27, 201–213. [Google Scholar] [CrossRef]
- Zhang, X.; Yarema, K.; Xu, A. Impact of static magnetic field (SMF) on microorganisms, plants and animals. In Biological Effects of Static Magnetic Fields; Springer: Singapore, 2017; pp. 133–172. [Google Scholar]
- Wan, G.J.; Jiang, S.L.; Zhao, Z.C.; Xu, J.J.; Tao, X.R.; Sword, G.A.; Gao, Y.B.; Pan, W.D.; Chen, F.J. Bio-effects of near-zero magnetic fields on the growth, development and reproduction of small brown planthopper, Laodelphax striatellus and brown plant hopper, Nilaparvata lugens. J. Insect Physiol. 2014, 68, 7–15. [Google Scholar] [CrossRef] [PubMed]
- Kataria, S.; Baghel, L.; Guruprasad, K.N. Pre-treatment of seeds with static magnetic field improves germination and early growth characteristics under salt stress in maize and soybean. Biocatal. Biotransform. 2017, 10, 83–90. [Google Scholar] [CrossRef]
- Chen, C.; Chen, L.; Wang, P.; Wu, L.F.; Song, T. Magnetically-induced elimination of Staphylococcus aureus by magnetotactic bacteria under a swing magnetic field. Nanomed. Nanotechnol. Biol. Med. 2017, 13, 363–370. [Google Scholar] [CrossRef]
- Gould, L.J. Magnetic field sensitivity in animals. Annu. Rev. Physiol. 1984, 46, 585–598. [Google Scholar] [CrossRef] [PubMed]
- Rakoczy, R.; Konopacki, M.; Fijakowski, K. The influence of a ferrofluid in the presence of an external rotating magnetic field on the growth rate and cell metabolic activity of a wine yeast strain. Biochem. Eng. J. 2016, 109, 43–50. [Google Scholar] [CrossRef]
- Fojt, L.; Klapetek, P.; Strasák, L.; Vetterl, V. 50 Hz magnetic field effect on the morphology of bacteria. Micron 2009, 40, 918–922. [Google Scholar] [CrossRef] [PubMed]
- Tang, H.; Wang, P.; Wang, H.; Fang, Z.; Yang, Q.; Ni, W.; Sun, X.; Liu, H.; Wang, L.; Zhao, G.; et al. Effect of static magnetic field on morphology and growth metabolism of flavobacterium sp. m1-14. Bioprocess Biosyst. Eng. 2019, 42, 1923–1933. [Google Scholar] [CrossRef] [PubMed]
- Kthiri, A.; Hidouri, S.; Wiem, T.; Jeridi, R.; Sheehan, D.; Landouls, A. Biochemical and biomolecular effects induced by a static magnetic field in Saccharomyces cerevisiae: Evidence for oxidative stress. PLoS ONE 2019, 14, e0209843. [Google Scholar] [CrossRef] [PubMed]
- Silva, P.G.P.D.; Júnior, D.P.; Sala, L.; Burkert, J.F.D.M.; Santos, L.O. Magnetic field as a trigger of carotenoid production by Phaffia rhodozyma. Process Biochem. 2020, 98, 131–138. [Google Scholar] [CrossRef]
- Drozd, R.; Szymańska, M.; Żywicka, A.; Kowalska, U.; Rakoczy, R.; Kordas, M.; Konopacki, M.; Junka, A.F.; Fijałkowski, K. Exposure to non-continuous rotating magnetic field induces metabolic strain-specific response of Komagataeibacter xylinus. Biochem. Eng. J. 2020, 166, 107855. [Google Scholar] [CrossRef]
- Wang, X.; Wang, Y.; Ning, S.; Shi, S.; Tan, L. Improving azo dye decolorization performance and halotolerance of Pichia occidentalis A2 by static magnetic field and possible mechanisms through comparative transcriptome analysis. Front. Microbiol. 2020, 11, 712. [Google Scholar] [CrossRef]
- Ahmad, A.M.; Yahya, A.G.I.; Jabir, A.W.S. Effect of magnetic field energy on growth of Aspergillus flavus and aflatoxins production. J. Al-Nahrain Univ. Sci. 2013, 16, 180–187. [Google Scholar] [CrossRef]
- Mateescu, C.; Burunea, N.; Stancu, N. Investigation of Aspergillus niger growth and activity in a static magnetic flux density field. Rom. Biotechnol. Lett. 2011, 16, 6364–6368. [Google Scholar]
- Gao, M.; Zhang, J.; Feng, H. Extremely low frequency magnetic field effects on metabolite of Aspergillus niger. Bioelectromagnetics 2011, 32, 73–78. [Google Scholar] [CrossRef]
- Zhang, J.L.; Zeng, D.J.; Xu, C.; Gao, M.X. Effect of low-frequency magnetic field on formation of pigments of Monascus purpureus. Eur. Food Res. Technol. 2015, 240, 577–582. [Google Scholar] [CrossRef]
- Liao, Q.; Liu, Y.; Zhang, J.; Li, L.; Gao, M. A low-frequency magnetic field regulates Monascus pigments synthesis via reactive oxygen species in M. purpureus. Process Biochem. 2019, 86, 16–24. [Google Scholar] [CrossRef]
- Zhang, J.L.; Liu, Y.B.; Li, L.; Gao, M.X. iTRAQ-Based quantitative proteomic analysis reveals changes in metabolite biosynthesis in Monascus purpureus in response to a low-frequency magnetic field. Toxins 2018, 10, 440. [Google Scholar] [CrossRef]
- Wan, Y.L.; Zhang, J.L.; Han, H.X.; Li, L.; Liu, Y.B.; Gao, M.X. Citrinin-producing capacity of Monascus purpureus in response to low-frequency magnetic fields. Process Biochem. 2017, 53, 25–29. [Google Scholar] [CrossRef]
- He, Y.; Cox, R.J. The molecular steps of citrinin biosynthesis in fungi. Chem. Sci. 2016, 7, 2119–2127. [Google Scholar] [CrossRef] [PubMed]
- Chen, F.S.; Hu, X.Q. Study on red fermented rice with high concentration of monacolin K and low concentration of citrinin. Int. J. Food Microbiol. 2005, 103, 331–337. [Google Scholar] [CrossRef]
- Zhou, H.Y.; Yang, S.Y.; Chen, F.S. The magnetic receptor of Monascus ruber M7: Gene clone and its heterologous expression in Escherichia coli. Front. Microbiol. 2020, 11, 1112. [Google Scholar] [CrossRef]
- Li, L.; Chen, F. Effects of mrpigG on Development and Secondary Metabolism of Monascus ruber M7. J. Fungi 2020, 6, 156. [Google Scholar] [CrossRef]
- Wang, L.; Dai, Y.; Chen, W.; Shao, Y.; Chen, F. Effects of light intensity and color on the biomass, extracellular red pigment and citrinin production of Monascus ruber. J. Agric. Food Chem. 2016, 64, 9506–9514. [Google Scholar] [CrossRef]
- Liu, Q.P.; Cai, L.; Shao, Y.C.; Zhou, Y.X.; Li, M.; Wang, X.H.; Chen, F.S. Inactivation of the global regulator LaeA in Monascus ruber results in a species-dependent response in sporulation and secondary metabolism. Fungal Biol. 2016, 120, 297–305. [Google Scholar] [CrossRef]
- Chang, C.Y.; Lue, M.Y.; Pan, T.M. Determination of adenosine, cordycepin and ergosterol contents in cultivated Antrodia camphorata by HPLC method. J. Food Drug Anal. 2005, 13, 338–342. [Google Scholar]
- Kim, D.; Langmead, B.; Salzberg, S.L. HISAT: A fast spliced aligner with low memory requirements. Nat. Methods 2015, 12, 357–360. [Google Scholar] [CrossRef]
- Langmead, B.; Salzberg, S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 2012, 9, 357. [Google Scholar] [CrossRef] [PubMed]
- Li, B.; Dewey, C.N. RSEM: Accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinform. 2011, 12, 323. [Google Scholar] [CrossRef] [PubMed]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [PubMed]
- Storey, J.D.; Tibshirani, R. Statistical signicance for genome wide studies. Proc. Natl. Acad. Sci. USA 2003, 100, 9440–9445. [Google Scholar] [CrossRef] [PubMed]
- Bolstad, B.M. Low-Level Analysis of High-Density Oligonucleotide Array Data: Background, Normalization and Summarization. Ph.D. Thesis, University of California, Berkeley, CA, USA, 2004. [Google Scholar]
- Kozubowski, L.; Lee, S.C.; Heitman, J. Signalling pathways in the pathogenesis of Cryptococcus. Cell Microbiol. 2009, 11, 370–380. [Google Scholar] [CrossRef]
- Hamel, L.P.; Nicole, M.C.; Duplessis, S.; Ellis, B.E. Mitogen-activated protein kinase signaling in plant-interacting fungi: Distinct messages from conserved messengers. Plant Cell 2012, 24, 1327–1351. [Google Scholar] [CrossRef]
- Zhou, Q.; Liu, Z.L.; Ning, K.; Wang, A.H.; Zeng, X.W.; Xu, J. Genomic and transcriptome analyses reveal that MAPK- and phosphatidylinositol-signaling pathways mediate tolerance to 5-hydroxymethyl-2-furaldehyde for industrial yeast Saccharomyces cerevisiae. Sci. Rep. 2014, 4, 6556. [Google Scholar] [CrossRef] [PubMed]
- Lin, C.H.; Yang, S.L.; Wang, N.Y.; Chung, K.R. The FUS3 MAPK signaling pathway of the citrus pathogen Alternaria alternata functions independently or cooperatively with the fungal redox-responsive AP1 regulator for diverse developmental, physiological and pathogenic processes. Fungal Genet. Biol. 2010, 47, 381–391. [Google Scholar] [CrossRef]
- Grabon, A.; Bankaitis, V.A.; Mcdermott, M.I. The interface between phosphatidylinositol transfer protein function and phosphoinositide signaling in higher eukaryotes. J. Lipid Res. 2019, 60, 242–268. [Google Scholar] [CrossRef]
- Zolov, S.N.; Bridges, D.; Zhang, Y.L.; Lee, W.W.; Riehle, E.; Verma, R.; Lenk, G.M.; Converso-Baran, K.; Weide, T.; Albin, R.L.; et al. In vivo, Pikfyve generates PI(3,5)P2, which serves as both a signaling lipid and the major precursor for PI5P. Proc. Natl Acad. Sci. USA 2012, 109, 17472–17477. [Google Scholar] [CrossRef]
- Kuo, H.F.; Hsu, Y.Y.; Lin, W.C.; Chen, K.Y.; Munnik, T.; Brearley, C.A.; Chiou, T.J. Arabidopsis inositol phosphate kinases, IPK1 and ITPK1, constitute a metabolic pathway in maintaining phosphate homeostasis. Plant J. 2018, 95, 613–630. [Google Scholar] [CrossRef]
- Shelest, E. Transcription factors in fungi: TFome dynamics, three major families, and dual-specificity TFs. Front. Genet. 2017, 8, 53. [Google Scholar] [CrossRef] [PubMed]
- Shelest, E. Transcription factors in fungi. FEMS Microbiol. Lett. 2008, 286, 145–151. [Google Scholar] [CrossRef]
- Masood, S.; Saleem, I.; Smith, D.; Chu, W.K. Growth Pattern of Magnetic Field-Treated Bacteria. Curr. Microbiol. 2020, 77, 194–203. [Google Scholar] [CrossRef] [PubMed]
- Nyakane, N.E.; Markus, E.D.; Sedibe, E.D.M. The effects of magnetic fields on plants growth: A comprehensive review. Int. J. Food Eng. 2019, 5, 79–87. [Google Scholar] [CrossRef]
- Mouritsen, H. Long-distance navigation and magnetoreception in migratory animals. Nature 2018, 558, 50–59. [Google Scholar] [CrossRef]
- Shang, W.; Chen, G.; Li, Y.; Zhuo, Y.; Wang, Y.; Fang, Z.; Yu, Y.; Ren, H. Static magnetic field accelerates diabetic wound healing by facilitating resolution of inflammation. J. Diabetes Res. 2019, 2019, 5641271. [Google Scholar] [CrossRef]
- Geng, S.; Fu, W.; Chen, W.; Zheng, S.; Gao, Q.; Wang, J.; Ge, X. Effects of an external magnetic field on microbial functional genes and metabolism of activated sludge based on metagenomic sequencing. Sci. Rep. 2020, 10, 8818. [Google Scholar] [CrossRef] [PubMed]
- Tan, L.; Shao, Y.; Mu, G.; Ning, S.; Shi, S. Enhanced azo dye biodegradation performance and halotolerance of Candida tropicalis SYF-1 by static magnetic field (SMF). Bioresour. Technol. 2020, 295, 122283. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.Y.; Zeng, X.B.; Guo, S.Y.; Li, Z.T. Effects of magnetic field on the antioxidant defense system of recirculation-cultured Chlorella vulgaris. Bioelectromagnetics 2008, 29, 39–46. [Google Scholar] [CrossRef]
- Rai, S. Causes and mechanism(s) of ner bioeffects. Electromagn. Biol. Med. 1997, 16, 59–67. [Google Scholar] [CrossRef]
- Veiga, M.C.; Fontoura, M.M.; Oliveira, M.G.D.; Costa, J.A.V.; Santos, L.O. Magnetic fields: Biomass potential of Spirulina sp. for food supplement. Bioprocess. Biosyst. Eng. 2020, 43, 1231–1240. [Google Scholar] [CrossRef] [PubMed]
- Beretta, G.; Mastorgio, A.F.; Pedrali, L.; Saponaro, S.; Sezenna, E. The effects of electric, magnetic and electromagnetic fields on microorganisms in the perspective of bioremediation. Rev. Environ. Sci. Bio-Technol. 2019, 18, 29–75. [Google Scholar] [CrossRef]
- Hong, J.L.; Wu, L.; Lu, J.Q.; Zhou, W.B.; Cao, Y.J.; Lv, W.L.; Liu, B.; Rao, P.F.; Ni, L.; Lv, X.C. Comparative transcriptomic analysis reveals the regulatory effects of inorganic nitrogen on the biosynthesis of Monascus pigments and citrinin. RSC Adv. 2020, 10, 5268–5282. [Google Scholar] [CrossRef]
- Chen, D.; Chen, M.H.; Wu, S.F.; Li, Z.J.; Yang, H.; Wang, C.L. The molecular mechanisms of Monascus purpureus M9 responses to blue light based on the transcriptome analysis. Sci. Rep. 2017, 7, 5537. [Google Scholar] [CrossRef]
- Chen, W.; Chen, R.; Liu, Q.; He, Y.; He, K.; Ding, X.; Kang, L.; Guo, X.; Xie, N.; Zhou, Y.; et al. Orange, red, yellow: Biosynthesis of azaphilone pigments in Monascus fungi. Chem. Sci. 2017, 8, 4917–4925. [Google Scholar] [CrossRef]
- Hynes, M.J.; Murray, S.L.; Duncan, A.; Khew, G.S.; Davis, M.A. Regulatory Genes Controlling Fatty Acid Catabolism and Peroxisomal Functions in the Filamentous Fungus Aspergillus nidulans. Eukaryot. Cell 2006, 5, 794–805. [Google Scholar] [CrossRef]
- Romana, R.; Gogala, N.; Jerman, I. Sinusoidal magnetic fields: Effects on the growth and ergosterol content in mycorrhizal fungi. Electromagn. Biol. Med. 2009, 16, 129–142. [Google Scholar]
- Zahumensky, J.; Malinsky, J. Role of MCC/Eisosome in fungal lipid homeostasis. Biomolecules 2019, 9, 305. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Dai, M.; Zhang, Y.; Lu, L. The sterol C-14 reductase Erg24 is responsible for ergosterol biosynthesis and ion homeostasis in Aspergillus fumigatus. Appl. Microbiol. Biotechnol. 2021, 105, 1253–1268. [Google Scholar] [CrossRef] [PubMed]
- Mascaraque, V.; Hernáez, M.L.; Jiménez-Sánchez, M.; Hansen, R.; Gil, C.; Martín, H.; Cid, V.J.; Molina, M. Phosphoproteomic analysis of protein kinase C signaling in Saccharomyces cerevisiae reveals Slt2 mitogen-activated protein kinase (MAPK)-dependent phosphorylation of eisosome core components. Mol. Cell. Proteom. 2013, 12, 557–574. [Google Scholar] [CrossRef]

| Samples | Total Raw Reads (M) | Total Clean Reads (M) | Clean Reads Ratio (%) | Q20 (%) | Q30 (%) | GC (%) | Genome Mapping Ratio (%) | Gene Mapping Ratio (%) |
|---|---|---|---|---|---|---|---|---|
| CK-3d-1 | 47.33 | 44.25 | 93.50 | 97.20 | 89.22 | 52.50 | 95.96 | 78.94 |
| CK-3d-2 | 45.57 | 42.58 | 93.43 | 97.20 | 89.22 | 52.10 | 95.94 | 78.49 |
| 30 mT-3d-1 | 45.57 | 42.26 | 92.72 | 96.36 | 87.67 | 53.20 | 94.83 | 78.13 |
| 30 mT-3d-2 | 45.57 | 42.34 | 92.91 | 96.30 | 87.49 | 53.70 | 95.10 | 78.77 |
| CK-7d-1 | 45.57 | 42.92 | 94.17 | 97.08 | 88.95 | 53.30 | 95.91 | 76.99 |
| CK-7d-2 | 47.33 | 44.10 | 93.19 | 97.19 | 89.20 | 53.00 | 95.54 | 76.88 |
| 30 mT-7d-1 | 47.33 | 44.21 | 93.40 | 97.05 | 88.74 | 53.20 | 95.56 | 77.18 |
| 30 mT-7d-2 | 47.33 | 44.47 | 93.97 | 97.29 | 89.55 | 52.70 | 96.09 | 78.01 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yang, S.; Zhou, H.; Dai, W.; Xiong, J.; Chen, F. Effect of Static Magnetic Field on Monascus ruber M7 Based on Transcriptome Analysis. J. Fungi 2021, 7, 256. https://doi.org/10.3390/jof7040256
Yang S, Zhou H, Dai W, Xiong J, Chen F. Effect of Static Magnetic Field on Monascus ruber M7 Based on Transcriptome Analysis. Journal of Fungi. 2021; 7(4):256. https://doi.org/10.3390/jof7040256
Chicago/Turabian StyleYang, Shuyan, Hongyi Zhou, Weihua Dai, Juan Xiong, and Fusheng Chen. 2021. "Effect of Static Magnetic Field on Monascus ruber M7 Based on Transcriptome Analysis" Journal of Fungi 7, no. 4: 256. https://doi.org/10.3390/jof7040256
APA StyleYang, S., Zhou, H., Dai, W., Xiong, J., & Chen, F. (2021). Effect of Static Magnetic Field on Monascus ruber M7 Based on Transcriptome Analysis. Journal of Fungi, 7(4), 256. https://doi.org/10.3390/jof7040256







