Recent Advances in Genome Editing Tools in Medical Mycology Research
Abstract
1. Introduction
2. Genome Editing Technologies
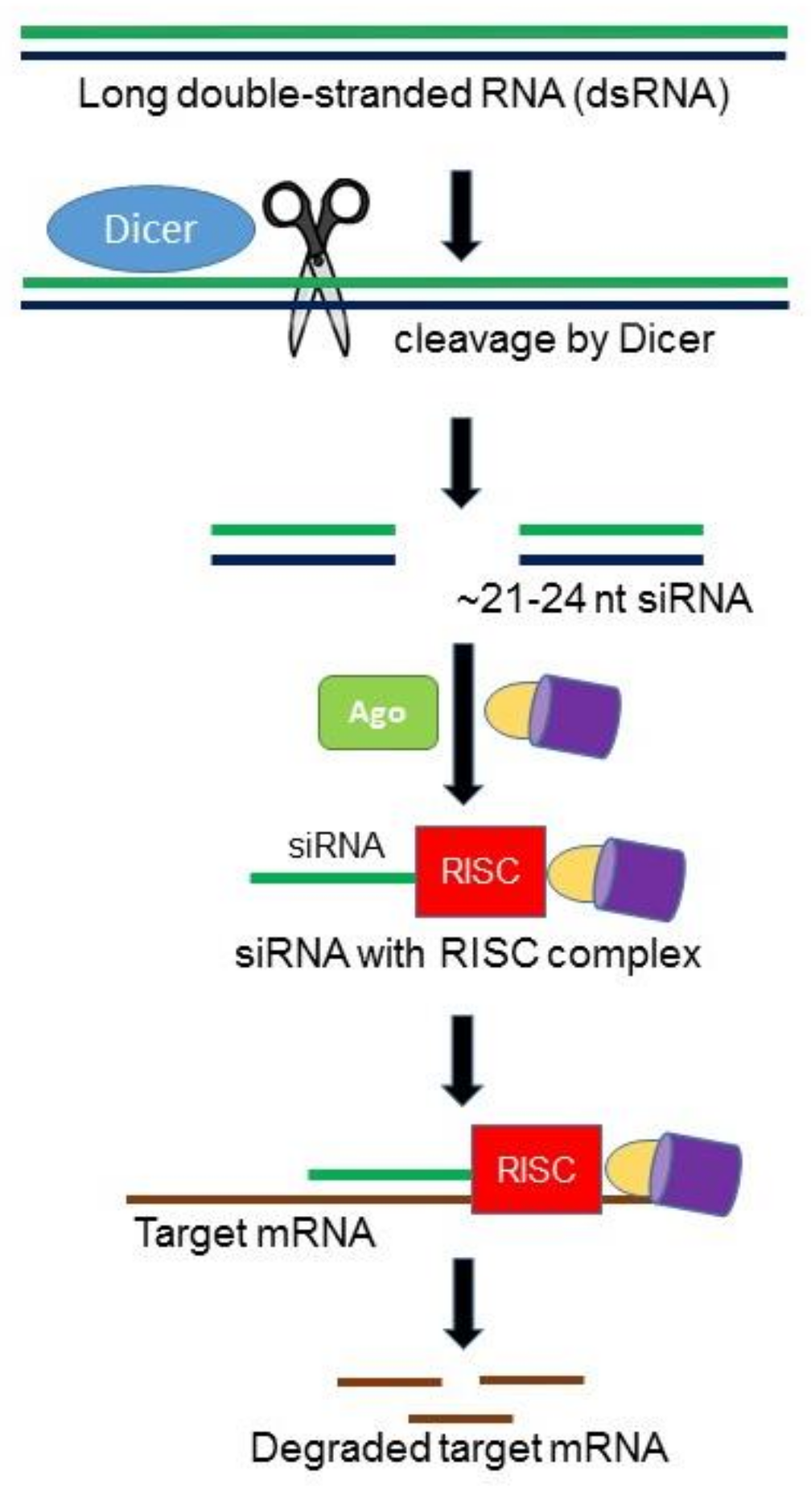
2.1. RNA Interference (RNAi)
2.2. Restriction Enzymes
2.3. Zinc-Finger Nucleases (ZFNs)
2.4. Transcription Activator-Like Effector Nucleases (TALENs)
2.5. Clustered Regularly Interspaced Short Palindromic Repeats (CRISPR)-CRISPR Associated Protein 9 (CRISPR-Cas9)
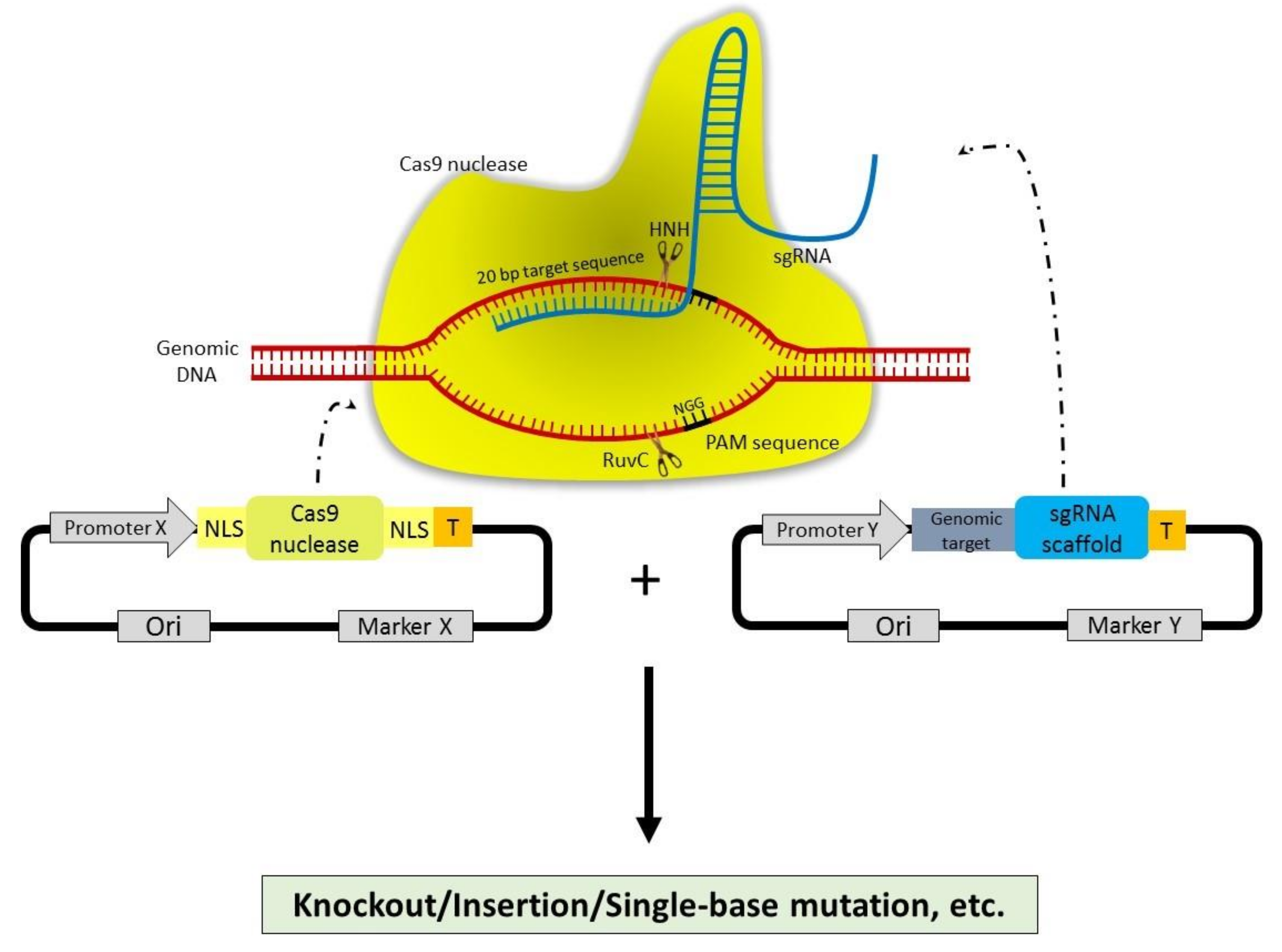
3. Mechanism of Action of CRISPR-Cas9
4. Applications of CRISPR-Cas9 Genome Editing Tool in Medically Important Fungi
4.1. Clinically Relevant Yeasts
4.2. Filamentous Fungi
5. Conclusions and Future Prospects
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| Ago | Argonaute |
| AIDS | acquired immunodeficiency syndrome |
| CRISPR | clustered regularly interspaced short palindromic repeats |
| CRISPR-Cas9 | clustered regularly interspaced short palindromic repeats associated protein 9 |
| Cas9-NLS | cas9 with a nuclear localization signal |
| crRNA | CRISPR RNA |
| dCas9 | dead version of Cas9 |
| DSB | double stranded break |
| dsRNA | double-stranded RNA |
| HR | homologous recombination |
| HDR | homology directed repair |
| IDT | integrated DNA technologies |
| MIC | minimum inhibitory concentration |
| mRNA | messenger RNA |
| NHEJ | non-homologous end joining |
| NGG | nucleotide- guanine-guanine |
| PAM | protospacer adjacent motif |
| PKS | polyketide synthase |
| REMI | restriction enzyme mediated integration |
| RISC | RNA- induced silencing complex |
| RVD | repeat variable di-residues |
| SAT1-FLP | SAT1 flipper |
| siRNA | small interfering RNA |
| sgRNA | single guide RNA |
| SNP | single nucleotide polymorphism |
| TALEN | transcription activator-like effector nucleases |
| tracrRNA | trans-activating crRNA |
| ZFN | zinc-finger nuclease |
References
- Lin, X.; Alspaugh, J.A.; Liu, H.; Harris, S. Fungal Morphogenesis. Cold Spring Harb. Perspect. Med. 2014, 5, a019679. [Google Scholar] [CrossRef]
- Schmiedel, Y.; Zimmerli, S. Common invasive fungal diseases: An overview of invasive candidiasis, aspergillosis, cryptococcosis and Pneumocystis pneumonia. Swiss Med. Wkly. 2016, 146, 14281. [Google Scholar] [CrossRef] [PubMed]
- Ramana, K.V.; Kandi, S.P.V.B.; Sharada, C.V.; Rao, R.; Mani, R.; Rao, S.D. Invasive Fungal Infections: A Comprehensive Review. Am. J. Infect. Dis. Microbiol. 2013, 1, 64–69. [Google Scholar] [CrossRef][Green Version]
- Wang, Q.; Zhong, C.; Xiao, H. Genetic Engineering of Filamentous Fungi for Efficient Protein Expression and Secretion. Front. Bioeng. Biotechnol. 2020, 8, 293. [Google Scholar] [CrossRef] [PubMed]
- Hittinger, C.T.; Alexander, W.G. Constructs and Methods for Genome Editing and Genetic Engineering of Fungi and Protists. U.S. Patent 9879270, 30 January 2018. [Google Scholar]
- Perez-Nadales, E.; Nogueira, M.F.A.; Baldin, C.; Castanheira, S.; El Ghalid, M.; Grund, E.; Lengeler, K.; Marchegiani, E.; Mehrotra, P.V.; Moretti, M.; et al. Fungal model systems and the elucidation of pathogenicity determinants. Fungal Genet. Biol. 2014, 70, 42–67. [Google Scholar] [CrossRef] [PubMed]
- Dudakova, A.; Spiess, B.; Tangwattanachuleeporn, M.; Sasse, C.; Buchheidt, D.; Weig, M.; Groß, U.; Bader, O. Molecular Tools for the Detection and Deduction of Azole Antifungal Drug Resistance Phenotypes in Aspergillus Species. Clin. Microbiol. Rev. 2017, 30, 1065–1091. [Google Scholar] [CrossRef] [PubMed]
- Gaj, T.; Sirk, S.J.; Shui, S.-L.; Liu, J. Genome-Editing Technologies: Principles and Applications. Cold Spring Harb. Perspect. Biol. 2016, 8, a023754. [Google Scholar] [CrossRef] [PubMed]
- Hilton, I.B.; Gersbach, C.A. Enabling functional genomics with genome engineering. Genome Res. 2015, 25, 1442–1455. [Google Scholar] [CrossRef] [PubMed]
- Chang, S.-S.; Zhang, Z.; Liu, Y. RNA Interference Pathways in Fungi: Mechanisms and Functions. Annu. Rev. Microbiol. 2012, 66, 305–323. [Google Scholar] [CrossRef]
- Lax, C.; Tahiri, G.; Patiño-Medina, J.A.; Cánovas-Márquez, J.T.; Pérez-Ruiz, J.A.; Osorio-Concepción, M.; Navarro, E.; Calo, S. The Evolutionary Significance of RNAi in the Fungal Kingdom. Int. J. Mol. Sci. 2020, 21, 9348. [Google Scholar] [CrossRef]
- Mousavi, B.; Hedayati, M.T.; Teimoori-Toolabi, L.; Guillot, J.; Alizadeh, A.; Badali, H. cyp51A gene silencing using RNA interference in azole-resistant Aspergillus fumigatus. Mycoses 2015, 58, 699–706. [Google Scholar] [CrossRef]
- Martel, C.M.; Parker, J.E.; Bader, O.; Weig, M.; Gross, U.; Warrilow, A.G.S.; Kelly, D.E.; Kelly, S.L. A Clinical Isolate of Candida albicans with Mutations in ERG11 (Encoding Sterol 14α-Demethylase) and ERG5 (Encoding C22 Desaturase) is Cross Resistant to Azoles and Amphotericin B. Antimicrob. Agents Chemother. 2010, 54, 3578–3583. [Google Scholar] [CrossRef]
- Sanglard, D.; Ischer, F.; Parkinson, T.; Falconer, D.; Bille, J. Candida albicans Mutations in the Ergosterol Biosynthetic Pathway and Resistance to Several Antifungal Agents. Antimicrob. Agents Chemother. 2003, 47, 2404–2412. [Google Scholar] [CrossRef]
- Zhou, Y.; Liao, M.; Zhu, C.; Hu, Y.; Tong, T.; Peng, X.; Li, M.; Feng, M.; Cheng, L.; Ren, B.; et al. ERG3 and ERG11 genes are critical for the pathogenesis of Candida albicans during the oral mucosal infection. Int. J. Oral Sci. 2018, 10, 1–8. [Google Scholar] [CrossRef]
- Chang, Z.; Yadav, V.; Lee, S.C.; Heitman, J. Epigenetic mechanisms of drug resistance in fungi. Fungal Genet. Biol. 2019, 132, 103253. [Google Scholar] [CrossRef]
- Poças-Fonseca, M.J.; Cabral, C.G.; Manfrão-Netto, J.H.C. Epigenetic manipulation of filamentous fungi for biotechnological applications: A systematic review. Biotechnol. Lett. 2020, 42, 885–904. [Google Scholar] [CrossRef]
- Brandão, F.; Esher, S.K.; Ost, K.S.; Pianalto, K.; Nichols, C.B.; Fernandes, L.; Bocca, A.L.; Poças-Fonseca, M.J.; Alspaugh, J.A. HDAC genes play distinct and redundant roles in Cryptococcus neoformans virulence. Sci. Rep. 2018, 8, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Brandão, F.A.; Derengowski, L.S.; Albuquerque, P.; Nicola, A.M.; Silva-Pereira, I.; Poças-Fonseca, M.J. Histone deacetylases inhibitors effects on Cryptococcus neoformans major virulence phenotypes. Virulence 2015, 6, 618–630. [Google Scholar] [CrossRef]
- Elbashir, S.M.; Harborth, J.; Weber, K.; Tuschl, T. Analysis of gene function in somatic mammalian cells using small interfering RNAs. Methods 2002, 26, 199–213. [Google Scholar] [CrossRef]
- Martinez, J.; Patkaniowska, A.; Elbashir, S.M.; Harborth, J.; Hossbach, M.; Urlaub, H.; Meyer, J.; Weber, K.; VanDenburgh, K.; Manninga, H.; et al. Analysis of mammalian gene function using small interfering RNAs. Nucleic Acids Symp. Ser. 2003, 3, 333. [Google Scholar] [CrossRef] [PubMed]
- Alic, N.; Hoddinott, M.P.; Foley, A.; Slack, C.; Piper, M.D.W.; Partridge, L. Detrimental Effects of RNAi: A Cautionary Note on its use in Drosophila Ageing Studies. PLoS ONE 2012, 7, e45367. [Google Scholar] [CrossRef]
- Roberts, R.J.; Murray, K. Restriction Endonuclease. CRC Crit. Rev. Biochem. 1976, 4, 123–164. [Google Scholar] [CrossRef]
- Brown, D.H.; Slobodkin, I.V.; Kumamoto, C.A. Stable transformation and regulated expression of an inducible reporter construct in Candida albicans using restriction enzyme-mediated integration. Mol. Genet. Genom. 1996, 251, 75–80. [Google Scholar] [CrossRef]
- Brown, J.S.; Aufauvre-Brown, A.; Holden, D.W. Insertional mutagenesis of Aspergillus fumigatus. Mol. Genet. Genom. 1998, 259, 327–335. [Google Scholar] [CrossRef]
- Warrilow, A.G.S.; Parker, J.E.; Price, C.L.; Nes, W.D.; Kelly, S.L.; Kelly, D.E. In Vitro Biochemical Study of CYP51-Mediated Azole Resistance in Aspergillus fumigatus. Antimicrob. Agents Chemother. 2015, 59, 7771–7778. [Google Scholar] [CrossRef] [PubMed]
- Jiang, C.; Dong, D.; Yu, B.; Cai, G.; Wang, X.; Ji, Y.; Peng, Y. Mechanisms of azole resistance in 52 clinical isolates of Candida tropicalis in China. J. Antimicrob. Chemother. 2012, 68, 778–785. [Google Scholar] [CrossRef] [PubMed]
- Lartigue, C.; Vashee, S.; Algire, M.A.; Chuang, R.-Y.; Benders, G.A.; Ma, L.; Noskov, V.N.; Denisova, E.A.; Gibson, D.G.; Assad-Garcia, N.; et al. Creating Bacterial Strains from Genomes that Have Been Cloned and Engineered in Yeast. Science 2009, 325, 1693–1696. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.G.; Cha, J.; Chandrasegaran, S. Hybrid restriction enzymes: Zinc finger fusions to Fok I cleavage domain. Proc. Natl. Acad. Sci. USA 1996, 93, 1156. [Google Scholar] [CrossRef] [PubMed]
- Urnov, F.D.; Rebar, E.J.; Holmes, M.C.; Zhang, H.S.; Gregory, P.D. Genome editing with engineered zinc finger nucleases. Nat. Rev. Genet. 2010, 11, 636–646. [Google Scholar] [CrossRef]
- Carroll, D. Genome engineering with zinc-finger nucleases. Genetics 2011, 188, 773–782. [Google Scholar] [CrossRef]
- Joung, J.K.; Sander, J.D. TALENs: A widely applicable technology for targeted genome editing. Nat. Rev. Mol. Cell Biol. 2012, 14, 49–55. [Google Scholar] [CrossRef] [PubMed]
- Gaj, T.; Gersbach, C.A.; Barbas, C.F. Zfn, Talen and Crispr/Cas-based methods for genome engineering. Trends Biotechnol. 2013, 31, 397–405. [Google Scholar] [CrossRef] [PubMed]
- Cong, L.; Ran, F.A.; Cox, D.; Lin, S.; Barretto, R.; Habib, N.; Hsu, P.D.; Wu, X.; Jiang, W.; Marraffini, L.A.; et al. Multiplex Genome Engineering using CRISPR/Cas Systems. Science 2013, 339, 819–823. [Google Scholar] [CrossRef]
- Mali, P.; Yang, L.; Esvelt, K.M.; Aach, J.; Guell, M.; DiCarlo, J.E.; Norville, J.E.; Church, G.M. RNA-Guided Human Genome Engineering via Cas9. Science 2013, 339, 823–826. [Google Scholar] [CrossRef]
- Dicarlo, J.E.; Norville, J.E.; Mali, P.; Rios, X.; Aach, J.; Church, G.M. Genome engineering in Saccharomyces cerevisiae using CRISPR-Cas systems. Nucleic Acids Res. 2013, 41, 4336–4343. [Google Scholar] [CrossRef]
- Kennedy, E.M.; Bassit, L.C.; Mueller, H.; Kornepati, A.V.; Bogerd, H.P.; Nie, T.; Chatterjee, P.; Javanbakht, H.; Schinazi, R.F.; Cullen, B.R. Suppression of hepatitis B virus DNA accumulation in chronically infected cells using a bacterial CRISPR/Cas RNA-guided DNA endonuclease. Virology 2015, 476, 196–205. [Google Scholar] [CrossRef]
- Ran, F.A.; Hsu, P.D.; Wright, J.; Agarwala, V.; Scott, D.A.; Zhang, F. Genome engineering using the CRISPR-Cas9 system. Nat. Protoc. 2013, 8, 2281–2308. [Google Scholar] [CrossRef]
- Li, Y.; Li, S.; Wang, J.; Liu, G. CRISPR/Cas Systems towards Next-Generation Biosensing. Trends Biotechnol. 2019, 37, 730–743. [Google Scholar] [CrossRef]
- Wang, J.; Lu, A.; Bei, J.; Zhao, G.; Wang, J. CRISPR/ddCas12a-based programmable and accurate gene regulation. Cell Discov. 2019, 5, 15. [Google Scholar] [CrossRef] [PubMed]
- Vercoe, R.B.; Chang, J.T.; Dy, R.L.; Taylor, C.; Gristwood, T.; Clulow, J.S.; Richter, C.; Przybilski, R.; Pitman, A.R.; Fineran, P.C. Cytotoxic Chromosomal Targeting by CRISPR/Cas Systems Can Reshape Bacterial Genomes and Expel or Remodel Pathogenicity Islands. PLoS Genet. 2013, 9, e1003454. [Google Scholar] [CrossRef] [PubMed]
- Farboud, B.; Jarvis, E.; Roth, T.L.; Shin, J.; Corn, J.E.; Marson, A.; Meyer, B.J.; Patel, N.H.; Hochstrasser, M.L. Enhanced Genome Editing with Cas9 Ribonucleoprotein in Diverse Cells and Organisms. J. Vis. Exp. 2018, 135, e57350. [Google Scholar] [CrossRef]
- Rahimi, H.; Salehiabar, M.; Barsbay, M.; Ghaffarlou, M.; Kavetskyy, T.; Sharafi, A.; Davaran, S.; Chauhan, S.C.; Danafar, H.; Kaboli, S.; et al. Crispr Systems for COVID-19 Diagnosis. ACS Sensors 2021. [Google Scholar] [CrossRef] [PubMed]
- Sasano, Y.; Nagasawa, K.; Kaboli, S.; Sugiyama, M.; Harashima, S. CRISPR-PCS: A powerful new approach to inducing multiple chromosome splitting in Saccharomyces cerevisiae. Sci. Rep. 2016, 6, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Kuivanen, J.; Wang, Y.-M.J.; Richard, P. Engineering Aspergillus niger for galactaric acid production: Elimination of galactaric acid catabolism by using RNA sequencing and CRISPR/Cas9. Microb. Cell Fact. 2016, 15, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Umeyama, T.; Hayashi, Y.; Shimosaka, H.; Inukai, T.; Yamagoe, S.; Takatsuka, S.; Hoshino, Y.; Nagi, M.; Nakamura, S.; Kamei, K.; et al. CRISPR/Cas9 Genome Editing to Demonstrate the Contribution of Cyp51A Gly138Ser to Azole Resistance in Aspergillus fumigatus. Antimicrob. Agents Chemother. 2018, 62. [Google Scholar] [CrossRef]
- Gu, Y.; Gao, J.; Cao, M.; Dong, C.; Lian, J.; Huang, L.; Cai, J.; Xu, Z. Construction of a series of episomal plasmids and their application in the development of an efficient CRISPR/Cas9 system in Pichia pastoris. World J. Microbiol. Biotechnol. 2019, 35, 79. [Google Scholar] [CrossRef] [PubMed]
- Al Abdallah, Q.; Ge, W.; Fortwendel, J.R. A Simple and Universal System for Gene Manipulation in Aspergillus fumigatus: In Vitro-Assembled Cas9-Guide RNA Ribonucleoproteins Coupled with Microhomology Repair Templates. Msphere 2017, 2, e00446. [Google Scholar] [CrossRef] [PubMed]
- Gong, Z.; Karlsson, A.J. Translocation of cell-penetrating peptides into Candida fungal pathogens. Protein Sci. 2017, 26, 1714–1725. [Google Scholar] [CrossRef]
- Farkhani, S.M.; Valizadeh, A.; Karami, H.; Mohammadi, S.; Sohrabi, N.; Badrzadeh, F. Cell penetrating peptides: Efficient vectors for delivery of nanoparticles, nanocarriers, therapeutic and diagnostic molecules. Peptides 2014, 57, 78–94. [Google Scholar] [CrossRef]
- Idnurm, A.; Meyer, V. The CRISPR revolution in fungal biology and biotechnology, and beyond. Fungal Biol. Biotechnol. 2018, 5, 19. [Google Scholar] [CrossRef]
- Jinek, M.; Chylinski, K.; Fonfara, I.; Hauer, M.; Doudna, J.A.; Charpentier, E. A Programmable Dual-RNA-Guided DNA Endonuclease in Adaptive Bacterial Immunity. Science 2012, 337, 816–821. [Google Scholar] [CrossRef] [PubMed]
- Makarova, K.S.; Koonin, E.V. Annotation and Classification of CRISPR-Cas Systems. In Methods in Molecular Biology; Springer Science and Business Media: Berlin/Heidelberg, Germany, 2015; Volume 1311, pp. 47–75. [Google Scholar]
- Horvath, P.; Barrangou, R. CRISPR/Cas the Immune System of Bacteria and Archaea. Science 2010, 327, 167–170. [Google Scholar] [CrossRef] [PubMed]
- Jiang, F.; Doudna, J.A. CRISPR–Cas9 Structures and Mechanisms. Annu. Rev. Biophys. 2017, 46, 505–529. [Google Scholar] [CrossRef] [PubMed]
- Brandsma, I.; Van Gent, D.C. Pathway choice in DNA double strand break repair: Observations of a balancing act. Genome Integr. 2012, 3, 1–10. [Google Scholar] [CrossRef]
- Vyas, V.K.; Barrasa, M.I.; Fink, G.R. A Candida albicans CRISPR system permits genetic engineering of essential genes and gene families. Sci. Adv. 2015, 1, e1500248. [Google Scholar] [CrossRef]
- Nguyen, N.; Quail, M.M.F.; Hernday, A.D. An Efficient, Rapid and Recyclable System for CRISPR-Mediated Genome Editing in Candida albicans. Msphere 2017, 2, e00149. [Google Scholar] [CrossRef]
- Min, K.; Ichikawa, Y.; Woolford, C.A.; Mitchell, A.P. Candida albicans Gene Deletion with a Transient CRISPR-Cas9 System. Msphere 2016, 1, e00130. [Google Scholar] [CrossRef]
- Min, K.; Biermann, A.; Hogan, D.A.; Konopka, J.B. Genetic Analysis ofNDT80Family Transcription Factors in Candida albicans using new CRISPR-Cas9 Approaches. Msphere 2018, 3, e00545. [Google Scholar] [CrossRef]
- Enkler, L.; Richer, D.; Marchand, A.L.; Ferrandon, D.; Jossinet, F. Genome engineering in the yeast pathogen Candida glabrata using the CRISPR-Cas9 system. Sci. Rep. 2016, 6, 35766. [Google Scholar] [CrossRef]
- Cen, Y.; Timmermans, B.; Souffriau, B.; Thevelein, J.M.; Van Dijck, P. Comparison of genome engineering using the CRISPR-Cas9 system in C. glabrata wild-type and lig4 strains. Fungal Genet. Biol. 2017, 107, 44–50. [Google Scholar] [CrossRef]
- Shapiro, R.S.; Chavez, A.; Porter, C.B.M.; Hamblin, M.; Kaas, C.S.; Dicarlo, J.E.; Zeng, G.; Xu, X.; Revtovich, A.V.; Kirienko, N.V.; et al. A CRISPR-Cas9-based gene drive platform for genetic interaction analysis in Candida albicans. Nat. Microbiol. 2018, 3, 73–82. [Google Scholar] [CrossRef] [PubMed]
- Ng, H.; Dean, N. Dramatic Improvement of CRISPR/Cas9 Editing in Candida albicans by Increased Single Guide RNA Expression. Msphere 2017, 2. [Google Scholar] [CrossRef] [PubMed]
- Halder, V.; Porter, C.B.M.; Chavez, A.; Shapiro, R.S. Design, execution and analysis of CRISPR-Cas9-based deletions and genetic interaction networks in the fungal pathogen Candida albicans. Nat. Protoc. 2019, 14, 955–975. [Google Scholar] [CrossRef] [PubMed]
- Lombardi, L.; Turner, S.A.; Zhao, F.; Butler, G. Gene editing in clinical isolates of Candida parapsilosis using CRISPR/Cas9. Sci. Rep. 2017, 7, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Lombardi, L.; Oliveira-Pacheco, J.; Butler, G. Plasmid-Based CRISPR-Cas9 Gene Editing in Multiple Candida Species. Msphere 2019, 4, e00125. [Google Scholar] [CrossRef] [PubMed]
- Wensing, L.; Sharma, J.; Uthayakumar, D.; Proteau, Y.; Chavez, A.; Shapiro, R.S. A CRISPR Interference Platform for Efficient Genetic Repression in Candida albicans. Msphere 2019, 4, e00002. [Google Scholar] [CrossRef]
- Elhariri, M.; Hamza, D.; Elhelw, R.; Refai, M. Eucalyptus Tree: A Potential Source of Cryptococcus neoformansin Egyptian Environment. Int. J. Microbiol. 2016, 2016, 1–5. [Google Scholar] [CrossRef]
- Andreou, M.; Cogliati, M.; Kolonitsiou, F.; Stroumpos, C.; Stamouli, V.; Ravazoula, P.; Siagris, D.; Papadogeorgaki, H.; Christofidou, M.; Lekkou, A. Cryptococcus gattii infection in an immunocompetent host in Greece. Med Mycol. Case Rep. 2020, 27, 1–3. [Google Scholar] [CrossRef]
- Arras, S.D.M.; Chua, S.M.H.; Wizrah, M.S.I.; Faint, J.A.; Yap, A.S.; Fraser, J.A. Targeted Genome Editing via CRISPR in the Pathogen Cryptococcus neoformans. PLoS ONE 2016, 11, e0164322. [Google Scholar] [CrossRef]
- Wang, Y.; Wei, D.; Zhu, X.; Pan, J.; Zhang, P.; Huo, L.; Zhu, X. A ‘suicide’ CRISPR-Cas9 system to promote gene deletion and restoration by electroporation in Cryptococcus neoformans. Sci. Rep. 2016, 6, 31145. [Google Scholar] [CrossRef]
- Fan, Y.; Lin, X. Multiple Applications of a Transient CRISPR-Cas9 Coupled with Electroporation (TRACE) System in the Cryptococcus neoformans Species Complex. Genetics 2018, 208, 1357–1372. [Google Scholar] [CrossRef] [PubMed]
- Wang, P. Two Distinct Approaches for CRISPR-Cas9-Mediated Gene Editing in Cryptococcus neoformans and Related Species. Msphere 2018, 3, e00208. [Google Scholar] [CrossRef]
- Pagano, L.; Girmenia, C.; Mele, L.; Ricci, P.E.; Tosti, M.; Nosari, A.; Buelli, M.; Picardi, M.; Allione, B.; Corvatta, L.; et al. Infections caused by filamentous fungi in patients with hematologic malignancies. A report of 391 cases by GIMEMA Infection Program. Haematology 2001, 86, 862–870. [Google Scholar]
- Pihet, M.; Carrere, J.; Cimon, B.; Chabasse, D.; Delhaes, L.; Symoens, F.; Bouchara, J.-P. Occurrence and relevance of filamentous fungi in respiratory secretions of patients with cystic fibrosis—A review. Med. Mycol. 2009, 47, 387–397. [Google Scholar] [CrossRef]
- Slavin, M.; Van Hal, S.; Sorrell, T.; Lee, A.; Marriott, D.; Daveson, K.; Kennedy, K.; Hajkowicz, K.; Halliday, C.; Athan, E.; et al. Invasive infections due to filamentous fungi other than Aspergillus: Epidemiology and determinants of mortality. Clin. Microbiol. Infect. 2015, 21. [Google Scholar] [CrossRef] [PubMed]
- Egbuta, M.A.; Mwanza, M.; Babalola, O.O. Health Risks Associated with Exposure to Filamentous Fungi. Int. J. Environ. Res. Public Health 2017, 14, 719. [Google Scholar] [CrossRef] [PubMed]
- Lamoth, F.; Calandra, T. Let’s add invasive aspergillosis to the list of influenza complications. Lancet Respir. Med. 2018, 6, 733–735. [Google Scholar] [CrossRef]
- Lim, S.-Y.; Son, Y.-E.; Lee, D.-H.; Eom, T.-J.; Kim, M.-J.; Park, H.-S. Function of crzA in Fungal Development and Aflatoxin Production in Aspergillus flavus. Toxins 2019, 11, 567. [Google Scholar] [CrossRef]
- Langfelder, K.; Jahn, B.; Gehringer, H.; Schmidt, A.; Wanner, G.; Brakhage, A.A. Identification of a polyketide synthase gene (pksP) of Aspergillus fumigatus involved in conidial pigment biosynthesis and virulence. Med. Microbiol. Immunol. 1998, 187, 79–89. [Google Scholar] [CrossRef]
- Yang, G.; Rose, M.S.; Turgeon, B.G.; Yoder, O.C. A polyketide synthase is required for fungal virulence and production of the polyketide T-toxin. Plant Cell 1996, 8, 2139–2150. [Google Scholar] [CrossRef]
- Fuller, K.K.; Chen, S.; Loros, J.J.; Dunlap, J.C. Development of the CRISPR/Cas9 System for Targeted Gene Disruption in Aspergillus fumigatus. Eukaryot. Cell 2015, 14, 1073–1080. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Meng, X.; Wei, X.; Lu, L. Highly efficient CRISPR mutagenesis by microhomology-mediated end joining in Aspergillus fumigatus. Fungal Genet. Biol. 2016, 86, 47–57. [Google Scholar] [CrossRef] [PubMed]
- Nagy, G.; Szebenyi, C.; Csernetics, A.; Vaz, A.G.; Toth, E.J.; Vagvolgyi, C.; Papp, T. Development of a plasmid free CRISPR-Cas9 system for the genetic modification of Mucor circinelloides. Sci. Rep. 2017, 7, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Bruni, G.O.; Zhong, K.; Lee, S.C.; Wang, P. Crispr-Cas9 induces point mutation in the mucormycosis fungus Rhizopus delemar. Fungal Genet. Biol. 2019, 124, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Agrawal, N.; Dasaradhi, P.V.; Mohmmed, A.; Malhotra, P.; Bhatnagar, R.K.; Mukherjee, S.K. RNA interference: Biology, mechanism, and applications. Microbiol. Mol. Biol. Rev. 2003, 67, 657–685. [Google Scholar] [CrossRef]
- Boettcher, M.; McManus, M.T. Choosing the Right Tool for the Job: RNAi, TALEN or CRISPR. Mol. Cell 2015, 58, 575–585. [Google Scholar] [CrossRef]
- Zheng, X.; Zheng, P.; Zhang, K.; Cairns, T.C.; Meyer, V.; Sun, J.; Ma, Y. 5S rRNA Promoter for Guide RNA Expression Enabled Highly Efficient CRISPR/Cas9 Genome Editing in Aspergillus niger. ACS Synth. Biol. 2019, 8, 1568–1574. [Google Scholar] [CrossRef]
- Weninger, A.; Fischer, J.E.; Raschmanová, H.; Kniely, C.; Vogl, T.; Glieder, A. Expanding the CRISPR/Cas9 toolkit for Pichia pastoris with efficient donor integration and alternative resistance markers. J. Cell. Biochem. 2018, 119, 3183–3198. [Google Scholar] [CrossRef]
- Nødvig, C.S.; Nielsen, J.B.; Kogle, M.E.; Mortensen, U.H. A CRISPR-Cas9 System for Genetic Engineering of Filamentous Fungi. PLoS ONE 2015, 10, e0133085. [Google Scholar] [CrossRef] [PubMed]
- Katayama, T.; Tanaka, Y.; Okabe, T.; Nakamura, H.; Fujii, W.; Kitamoto, K.; Maruyama, J.-I. Development of a genome editing technique using the CRISPR/Cas9 system in the industrial filamentous fungus Aspergillus oryzae. Biotechnol. Lett. 2016, 38, 637–642. [Google Scholar] [CrossRef]
- Weyda, I.; Yang, L.; Vang, J.; Ahring, B.K.; Lübeck, M.; Lübeck, P.S. A comparison of Agrobacterium-mediated transformation and protoplast-mediated transformation with CRISPR-Cas9 and bipartite gene targeting substrates, as effective gene targeting tools for Aspergillus carbonarius. J. Microbiol. Methods 2017, 135, 26–34. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Cobine, P.A.; Coleman, J.J. Efficient genome editing in Fusarium oxysporum based on CRISPR/Cas9 ribonucleoprotein complexes. Fungal Genet. Biol. 2018, 117, 21–29. [Google Scholar] [CrossRef]
- Wenderoth, M.; Pinecker, C.; Voß, B.; Fischer, R. Establishment of CRISPR/Cas9 in Alternaria alternata. Fungal Genet. Biol. 2017, 101, 55–60. [Google Scholar] [CrossRef] [PubMed]
- Schuster, M.; Schweizer, G.; Reissmann, S.; Kahmann, R. Genome editing in Ustilago maydis using the CRISPR-Cas system. Fungal Genet. Biol. 2016, 89, 3–9. [Google Scholar] [CrossRef] [PubMed]
- Zalatan, J.G.; Lee, M.E.; Almeida, R.; Gilbert, L.A.; Whitehead, E.H.; La Russa, M.; Tsai, J.C.; Weissman, J.S.; Dueber, J.E.; Qi, L.S.; et al. Engineering Complex Synthetic Transcriptional Programs with CRISPR RNA Scaffolds. Cell 2015, 160, 339–350. [Google Scholar] [CrossRef] [PubMed]
- Shi, S.; Liang, Y.; Zhang, M.M.; Ang, E.L.; Zhao, H. A highly efficient single-step, markerless strategy for multi-copy chromosomal integration of large biochemical pathways in Saccharomyces cerevisiae. Metab. Eng. 2016, 33, 19–27. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.-L.; Peng, Y.-Z.; Liu, D.; Liu, H.; Cao, Y.-X.; Li, B.-Z.; Li, C.; Yuan, Y.-J. Gene repression via multiplex gRNA strategy in Y. lipolytica. Microb. Cell Fact. 2018, 17, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Liu, R.; Chen, L.; Jiang, Y.; Zhou, Z.; Zou, G. Efficient genome editing in filamentous fungus Trichoderma reesei using the CRISPR/Cas9 system. Cell Discov. 2015, 1, 15007. [Google Scholar] [CrossRef] [PubMed]
- Katayama, T.; Nakamura, H.; Zhang, Y.; Pascal, A.; Fujii, W.; Maruyama, J.-I. Forced Recycling of an AMA1-Based Genome-Editing Plasmid Allows for Efficient Multiple Gene Deletion/Integration in the Industrial Filamentous Fungus Aspergillus oryzae. Appl. Environ. Microbiol. 2018, 85, e01896. [Google Scholar] [CrossRef]
- Ronda, C.; Maury, J.; Jakočiunas, T.; Jacobsen, S.A.B.; Germann, S.M.; Harrison, S.J.A.; Borodina, I.; Keasling, J.D.; Jensen, M.K.; Nielsen, A.T. CrEdit: CRISPR mediated multi-loci gene integration in Saccharomyces cerevisiae. Microb. Cell Fact. 2015, 14, 1–11. [Google Scholar] [CrossRef]
- Jakočiūnas, T.; Jensen, M.K.; Keasling, J.D. CRISPR/Cas9 advances engineering of microbial cell factories. Metab. Eng. 2016, 34, 44–59. [Google Scholar] [CrossRef]
- Adiego-Pérez, B.; Randazzo, P.; Daran, J.M.; Verwaal, R.A.; Roubos, J.; Daran-Lapujade, P.; Van Der Oost, J. Multiplex genome editing of microorganisms using CRISPR-Cas. FEMS Microbiol. Lett. 2019, 366, 86. [Google Scholar] [CrossRef] [PubMed]
- Ozaki, A.; Konishi, R.; Otomo, C.; Kishida, M.; Takayama, S.; Matsumoto, T.; Tanaka, T.; Kondo, A. Metabolic engineering of Schizosaccharomyces pombe via CRISPR-Cas9 genome editing for lactic acid production from glucose and cellobiose. Metab. Eng. Commun. 2017, 5, 60–67. [Google Scholar] [CrossRef]
- Lian, J.; Bao, Z.; Hu, S.; Zhao, H. Engineered CRISPR/Cas9 system for multiplex genome engineering of polyploid industrial yeast strains. Biotechnol. Bioeng. 2018, 115, 1630–1635. [Google Scholar] [CrossRef] [PubMed]
- Nielsen, J.C. Development of fungal cell factories for the production of secondary metabolites: Linking genomics and metabolism. Synth. Syst. Biotechnol. 2017, 2, 5–12. [Google Scholar] [CrossRef]
- Calderone, R.; Sun, N.; Gay-Andrieu, F.; Groutas, W.C.; Weerawarna, P.M.; Prasad, S.; Alex, D.; Li, D. Antifungal drug discovery: The process and outcomes. Futur. Microbiol. 2014, 9, 791–805. [Google Scholar] [CrossRef]
- Hara, S.; Jin, F.J.; Takahashi, T.; Koyama, Y. A further study on chromosome minimization by protoplast fusion in Aspergillus oryzae. Mol. Genet. Genom. 2011, 287, 177–187. [Google Scholar] [CrossRef]
- Roemer, T.; Boone, C.W. Systems-level antimicrobial drug and drug synergy discovery. Nat. Chem. Biol. 2013, 9, 222–231. [Google Scholar] [CrossRef] [PubMed]
- Qi, L.S.; Larson, M.H.; Gilbert, L.A.; Doudna, J.A.; Weissman, J.S.; Arkin, A.P.; Lim, W.A. Repurposing CRISPR as an RNA-Guided Platform for Sequence-Specific Control of Gene Expression. Cell 2013, 152, 1173–1183. [Google Scholar] [CrossRef]
- Kang, J.G.; Park, J.S.; Ko, J.-H.; Kim, Y.-S. Regulation of gene expression by altered promoter methylation using a CRISPR/Cas9-mediated epigenetic editing system. Sci. Rep. 2019, 9, 1–12. [Google Scholar] [CrossRef]
- Kozel, T.R.; Wickes, B. Fungal Diagnostics. Cold Spring Harb. Perspect. Med. 2014, 4, a019299. [Google Scholar] [CrossRef] [PubMed]
- Wickes, B.L.; Wiederhold, N.P. Molecular diagnostics in medical mycology. Nat. Commun. 2018, 9, 5135. [Google Scholar] [CrossRef] [PubMed]
- Matsoukas, I.G. Prime Editing: Genome Editing for Rare Genetic Diseases without Double-Strand Breaks or Donor DNA. Front. Genet. 2020, 11, 528. [Google Scholar] [CrossRef] [PubMed]
- Anzalone, A.V.; Randolph, P.B.; Davis, J.R.; Sousa, A.A.; Koblan, L.W.; Levy, J.M.; Chen, P.J.; Wilson, C.; Newby, G.A.; Raguram, A.; et al. Search-and-replace genome editing without double-strand breaks or donor DNA. Nat. Cell Biol. 2019, 576, 149–157. [Google Scholar] [CrossRef] [PubMed]
- Abdullah; Jiang, Z.; Hong, X.; Zhang, S.; Yao, R.; Xiao, Y. CRISPR base editing and prime editing: DSB and template-free editing systems for bacteria and plants. Synth. Syst. Biotechnol. 2020, 5, 277–292. [Google Scholar] [CrossRef]
| Organism | CAS9 Expression Module | GRNA Expression Module | Target Gene (S) | Purpose of Application | Editing Rate and Result | References |
|---|---|---|---|---|---|---|
| C. albicans | Candida/Saccharomyces codon–optimized version of Cas9 (CaCas9)/the ENO1 promoter | The RNA polymerase III (Pol III) promoter SNR52 | ADE2, CDR1/CDR2 | To generate homozygous mutations in one transformation by Duet and Solo system | Duet system showed 20–40% mutagenesis efficiency, and Solo system enabled 60–80% targeting | Vyas et al. (2015) [57] |
| C. albicans | Transient CRISPR-Cas9 system by using a SAT1-FLP system | SNR52P/TENO1 | NDT80, REP1, and RON1 | To better understand role of target genes (single or in combination) in virulence | Single, double, and triple deletion strains were successfully constructed | Min et al. (2018) [60] |
| C. albicans | US-pENO1 ˃ Cas9-NAT | NAT-pSNR52-gRNA-DS | ADE2, URA3, WOR1,WOR, and CZF1 | To develop a marker less system without need for molecular cloning step | 80% single gene deletion, 20% double genes deletion and ˃50% integration efficiency | Nguyen et al. (2017) [58] |
| C. albicans | CIp-ARG4-PTEF CaCAS9 | PADH1-tRNA-driven gRNA expression | RFP | To optimize gRNA intracellular expression | Increase the gene editing efficiency by 10-fold | Ng et al. (2017) [64] |
| C. albicans | CaCas9 into the C. albicans genome at the NEUT5L locus | 5′ homology arm–SNR52 promoter–gRNA1–gRNA2-3′ homology arm | antifungal efflux and biofilm adhesion factors | To develop a gene drive array system for the generation of combinatorial deletion mutants | Two larges pairwise gene deletion mutants were successfully generated | Shapiro et al. (2018) [63] |
| C. albicans | the ENO1 promoter/Cas9 (CaCas9)/TCYC1 | SNR52P/TENO1 | ADE2 | To describe a transient CRISPR-Cas9 system for efficient gene deletion | Homozygous deletions by introduction of CaCas9 transiently | Min et al. (2016) [59] |
| C. parapsilosis | TEF1p-CAS9-TEF1t | pCpSNR52-sgRNA-SUP4t and cpGAPDHp-HH-sgRNA-HDV-GAPDHt | ADE2, CPAR2_101060 and URA3 | To apply gene manipulation in single transformation step which can be used for editing of any number of target genes | The system yielded up to 100% efficiency across a panel of 20 clinical isolates | Lombardi et al. (2017) [66] |
| C. glabrata | pTEF1-Cas9-tCYC1/pCYC1-Cas9-tCYC1 | pSNR52-sgRNA-tTY2/pRNAH1-sgRNA-tTY2 | ADE2, VPK1 and YPS11 | To establish a loss-of-function mutation through the NHEJ repair pathway | High | Enkler et al. (2016) [61] |
| C. glabrata | pTEF-Cas9-KanMX | pSNR52-sgRNA-CYC1t | ADE2, MET15 and SOK2 | To compare genome modifications in C. glabrata wild type and lig4 strains | Targeting efficiency in the lig4Δ mutant was higher than in the wild type strain | Cen et al. (2017) [62] |
| C. albicans | Codon-optimized version of Cas9(CaCas9)-SV40NLS | SNR52 RNA polymerase III promoter | CDR1 and CDR2 | To present a modified gene-drive-based assay for gene manipulation | − | Halder et al. (2019) [65] |
| C. albicans | ACT1p-dCAS9-ACT1t | SNR52p-gRNA tail | ADE2 | To demonstrate a functional CRISPRi system for transcriptional repression | 20-fold repression of target gene achieved | Wensing et al. (2019) [68] |
| C. parapsilosis, C. orthopsilosis, C. metapsilosis and C. tropicalis | MgTEF1p-CAS9-MgTRP1t | pAgTEF1-sgRNA-HDV-ScCYC1t | ADE2 and CPAR2_101060 | To construct an autonomously replicating plasmid for markerless ediing in Candida spp. | Single gene distribution efficiency observed in C. parapsilosis (approximately 80%), C. meta psilosis (100%), C. tropicalis (88–100%) | Lombardi et al. (2019) [67] |
| Cryptococcus neoformance | TEF1p-Cas9-SV40NLS-TEF1t | pACT1-HH-gRNA-HDV-TRPt | ADE2 | To demonstrate the first proof of principle study | 70% | Arras et al. (2016) [71] |
| C. neoformans | ACT1P-SV40NLS-Cas9-NLS-bGHpAt | pCnU6-GN19-gRNA-6Ts | ADE2 and Tsp2-1 | To develop a system for gene alterations by subsequent complementation and off-target effects reduction | Frequency of gene deletion was over 80%, indel efficiency and HR rates were 40–90% and 20–90%, respectively | Wang et al. (2016) [72] |
| C. neoformans | GPD1p–Cas9-GPD1 t | pCnU6-sgRNA-6-Tt | ADE2 | To generate a TRACE system as an cost-effective and efficient strategy for genetic modifications | Up to 90% gene disruption rate | Fan et al. (2018) [73] |
| C. neoformans | pTEF-Cas9-FLAG-NLS | ptRNA-sgRNA-NLS | GIB2 | To deliver a preassembled RNP via electroporation to accelerate of gene editing | Approach is sufficient to induce gene modification | Wang P. (2018) [74] |
| Organism | CAS9 Expression Module | GRNA Expression Module | Target Gene (S) | Purpose of Application | Editing Rate and Result | References |
|---|---|---|---|---|---|---|
| A. fumigatus | p-hph-Ptef1-cas9 | p426-SNR52p-gRNA.CAN1.Y-SUP4t | PKSP | To test CRISPR-CAS9 method in this organism | High gene targeting efficiency reached 25–53% | Fuller et al. (2015) [83] |
| A. fumigatus | Gpdap-3xFLAG-NLS-Cas9-NLS-TRPCt | U6-3-gRNA | pksP and cnaA genes | To establish the system for mutagenesis using MMEJ process | Approximately 95–100% rate of mutagenesis obtained | Zhang et al. (2016) [84] |
| A. fumigatus | Alt-R-CRISPR-Cas9 components from integrated DNA technologies (IDT) | cr5 = pksP and cr3 = pksP | PKSP | An in vitro assembly of RNP demonstrated to eliminate the strain construction step | Gene deletion efficiency was close to 100% | Al-Abdallah et al. (2017) [48] |
| A. fumigatus | Cas9-NLS | T7-sgRNA | CYP51A | To investigate the mechanisms of azole resistance via cyp51A alteration | Site-directed mutagenesis successfully performed using CRISPR-CAS9 system | Umeyama et al. (2018) [46] |
| Mucor circinelloides | Alt-R CRISPR-Cas9 tracrRNA | Alt-R CRISPR crRNA | CARB and HMGR2 | To obtain mitotically stable mutants, a plasmid free CRISPR-Cas9 approach demonstrated | Targeting efficiency of NHEJ and HR reach to 100% | Nagy et al. (2017) [85] |
| Rhizopus delemar | pmCas9: tRNA-gRNA | pmCas9: tRNA-gRNA | PYRF | For investigating molecular pathogenesis mechanisms, point mutation introduced | Efficiency of 36% to 59% | Bruni et al. (2019) [86] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nargesi, S.; Kaboli, S.; Thekkiniath, J.; Heidari, S.; Keramati, F.; Seyedmousavi, S.; Hedayati, M.T. Recent Advances in Genome Editing Tools in Medical Mycology Research. J. Fungi 2021, 7, 257. https://doi.org/10.3390/jof7040257
Nargesi S, Kaboli S, Thekkiniath J, Heidari S, Keramati F, Seyedmousavi S, Hedayati MT. Recent Advances in Genome Editing Tools in Medical Mycology Research. Journal of Fungi. 2021; 7(4):257. https://doi.org/10.3390/jof7040257
Chicago/Turabian StyleNargesi, Sanaz, Saeed Kaboli, Jose Thekkiniath, Somayeh Heidari, Fatemeh Keramati, Seyedmojtaba Seyedmousavi, and Mohammad Taghi Hedayati. 2021. "Recent Advances in Genome Editing Tools in Medical Mycology Research" Journal of Fungi 7, no. 4: 257. https://doi.org/10.3390/jof7040257
APA StyleNargesi, S., Kaboli, S., Thekkiniath, J., Heidari, S., Keramati, F., Seyedmousavi, S., & Hedayati, M. T. (2021). Recent Advances in Genome Editing Tools in Medical Mycology Research. Journal of Fungi, 7(4), 257. https://doi.org/10.3390/jof7040257






