Recent Molecular Tools for the Genetic Manipulation of Highly Industrially Important Mucoromycota Fungi
Abstract
1. Introduction
- (1)
- the Glomeromycotina, which form arbuscular mycorrhizae,
- (2)
- the Mortierellomycotina, which are typically root endophytes, and,
- (3)
- the Mucoromycotina, which include the orders Mucorales, Umbelopsidales, and Endogonales [1].
2. Attributes of Model Organisms for Genetic Manipulation
- -
- Genetic model organisms are species that can be genetically modified, and large-scale genetic crosses are possible with them;
- -
- Genomic model organisms have a particular genomic size or composition that can be used, such as pufferfish;
- -
- Experimental model organisms may not be amenable to genetic modification; however, other advantages are specific to the experiment and the traits being sought.
2.1. Mucor circinelloides
2.2. Rhizopus arrhizus
2.3. Mortierella alpina
2.4. Cunninghamella spp.
3. Genetic Tools and Approaches for Genetic Modification in Mucorales
3.1. Restriction Enzymes
3.2. Genetic Markers
3.3. Random Mutagenesis
3.4. Gene Knockout
3.5. Gene Silencing Using RNA Interference


3.6. CRISPR-Cas9 Based Genome Modification
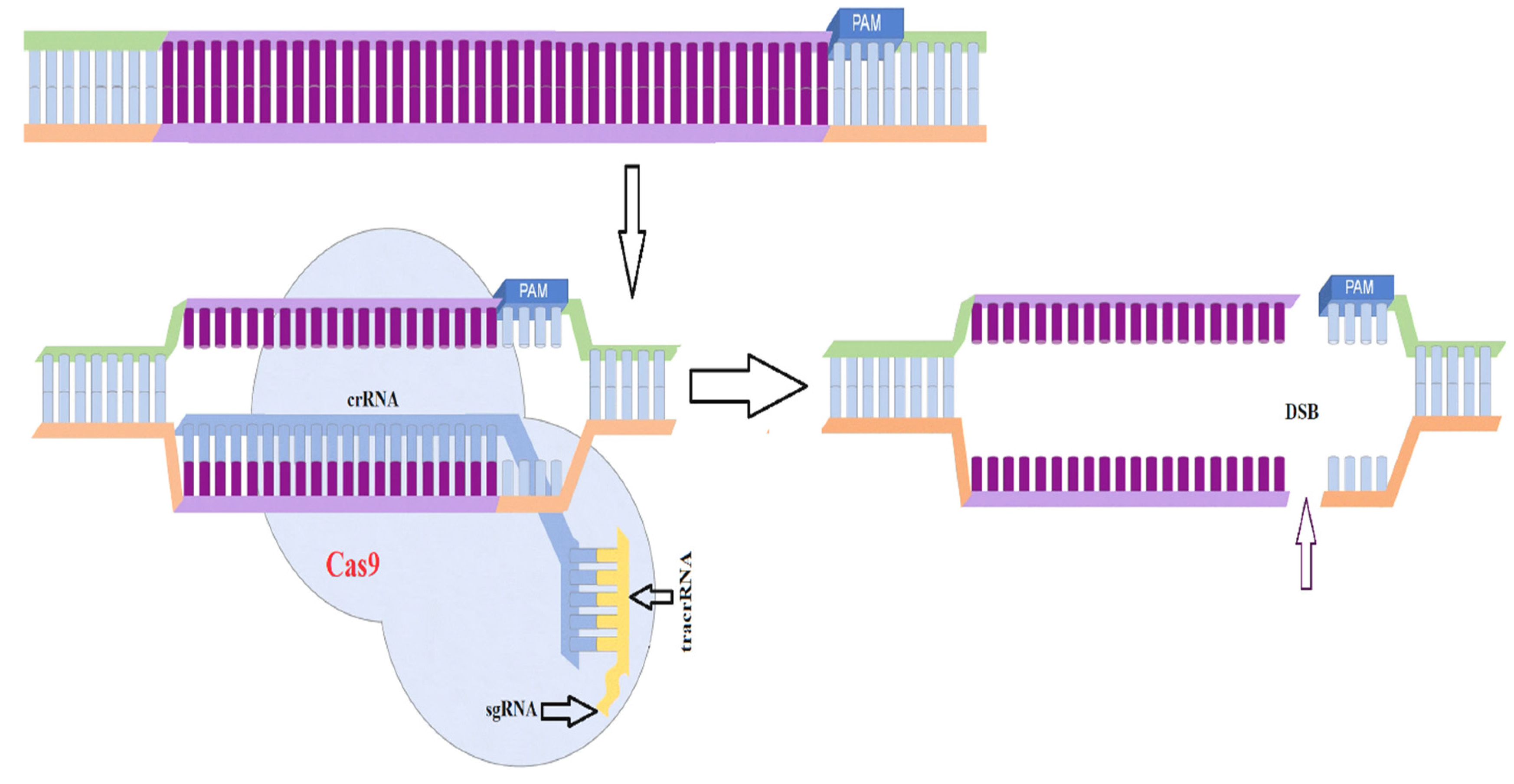
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Conflicts of Interest
References
- Walther, G.; Wagner, L.; Kurzai, O. Updates on the taxonomy of mucorales with an emphasis on clinically important taxa. J. Fungi 2019, 5, 106. [Google Scholar] [CrossRef] [PubMed]
- Benny, G.L.; Humber, R.A.; Morton, J.B. Zygomycota: Zygomycetes. In Systematics and Evolution; Springer: Berlin/Heidelberg, Germany, 2001; pp. 113–146. [Google Scholar]
- Hibbett, D.S.; Binder, M.; Bischoff, J.F.; Blackwell, M.; Cannon, P.F.; Eriksson, O.E.; Huhndorf, S.; James, T.; Kirk, P.M.; Lücking, R.; et al. A higher-level phylogenetic classification of the Fungi. Mycol. Res. 2007, 111, 509–547. [Google Scholar] [CrossRef] [PubMed]
- Spatafora, J.W.; Chang, Y.; Benny, G.L.; Lazarus, K.; Smith, M.E.; Berbee, M.L.; Bonito, G.; Corradi, N.; Grigoriev, I.; Gryganskyi, A.; et al. A phylum-level phylogenetic classification of zygomycete fungi based on genome-scale data. Mycologia 2016, 108, 1028–1046. [Google Scholar] [CrossRef] [PubMed]
- Morin-Sardin, S.; Nodet, P.; Coton, E.; Jany, J.L. Mucor: A Janus-faced fungal genus with human health impact and industrial applications. Fungal Biol. Rev. 2017, 31, 12–32. [Google Scholar] [CrossRef]
- Lübbehüsen, T.L.; Nielsen, J.; McIntyre, M. Characterization of the Mucor circinelloides life cycle by on-line image analysis. J. Appl. Microbiol. 2003, 95, 1152–1160. [Google Scholar] [CrossRef]
- Lee, S.C.; Li, A.; Calo, S.; Heitman, J. Calcineurin Plays Key Roles in the Dimorphic Transition and Virulence of the Human Pathogenic Zygomycete Mucor circinelloides. PLoS Pathog. 2013, 9, e1003625. [Google Scholar] [CrossRef]
- Lee, S.C.; Li, A.; Calo, S.; Inoue, M.; Tonthat, N.K.; Bain, J.M.; Louw, J.; Shinohara, M.L.; Erwig, L.P.; Schumacher, M.A.; et al. Calcineurin orchestrates dimorphic transitions, antifungal drug responses and host-pathogen interactions of the pathogenic mucoralean fungus Mucor circinelloides. Mol. Microbiol. 2015, 97, 844–865. [Google Scholar] [CrossRef]
- Iwen, P.C.; Thapa, I.; Bastola, D. Review of methods for the identification of zygomycetes with an emphasis on advances in molecular diagnostics. Lab. Med. 2011, 42, 260–266. [Google Scholar] [CrossRef][Green Version]
- Thakur, A.; Pahwa, R.; Singh, S.; Gupta, R. Production, purification, and characterization of polygalacturonase from mucor circinelloides ITCC 6025. Enzyme Res. 2010, 2010, 1–7. [Google Scholar] [CrossRef]
- Magnuson, J.K.; Lasure, L.L. Organic Acid Production by Filamentous Fungi. In Advances in Fungal Biotechnology for Industry, Agriculture, and Medicine; Springer: Boston, MA, USA, 2004. [Google Scholar]
- Zhang, K.; Yang, S.T.; Chalmers, J.J.; Wood, D. Fumaric Acid Fermentation by Rhizopus oryzae with Integrated Separation Technologies; The Ohio State University: Columbus, OH, USA, 2012. [Google Scholar]
- Papp, T.; Nyilasi, I.; Csernetics, Á.; Nagy, G.; Takó, M.; Vágvölgyi, C. Improvement of Industrially Relevant Biological Activities in Mucoromycotina Fungi. In Gene Expression Systems in Fungi: Advancements and Applications. Fungal Biology; Schmoll, M., Dattenböck, C., Eds.; Springer: Cham, Switzerland, 2016; pp. 97–118. [Google Scholar]
- Naz, T.; Nosheen, S.; Li, S.; Nazir, Y.; Mustafa, K.; Liu, Q.; Garre, V.; Song, Y. Comparative analysis of β-carotene production by mucor circinelloides strains CBS 277.49 and WJ11 under light and dark conditions. Metabolites 2020, 10, 38. [Google Scholar] [CrossRef]
- Dinesh Babu, P.; Bhakyaraj, R.; Vidhyalakshmi, R. A Low Cost Nutritious Food “Tempeh”—A Review. World J. Dairy Food Sci. 2009, 4, 22–27. [Google Scholar]
- Riley, T.T.; Muzny, C.A.; Swiatlo, E.; Legendre, D.P. Breaking the Mold: A Review of Mucormycosis and Current Pharmacological Treatment Options. Ann. Pharmacother. 2016, 50, 747–757. [Google Scholar] [CrossRef]
- Corrochano, L.M.; Garre, V. Photobiology in the Zygomycota: Multiple photoreceptor genes for complex responses to light. Fungal Genet. Biol. 2010, 47, 893–899. [Google Scholar] [CrossRef]
- Ruiz-Vázquez, R.M.; Nicolás, F.E.; Torres-Martínez, S.; Garre, V. Distinct RNAi Pathways in the Regulation of Physiology and Development in the Fungus Mucor circinelloides. Adv. Genet. 2015, 91, 55–102. [Google Scholar] [CrossRef]
- Wang, L.; Lin, X. Morphogenesis in Fungal Pathogenicity: Shape, Size, and Surface. PLoS Pathog. 2012, 8, e1003027. [Google Scholar] [CrossRef]
- Wöstemeyer, J.; Schimek, C.; Wetzel, J.; Burmester, A.; Voigt, J.; Schulz, E.; Ellenberger, S.; Siegmund, L. 10 Pheromone Action in the Fungal Groups Chytridiomycetes and Zygomycetes and in the Oophytes. In Growth, Differentiation and Sexuality; Springer: Singapore, 2016. [Google Scholar]
- Ibrahim, A.S.; Gebremariam, T.; Lin, L.; Luo, G.; Husseiny, M.I.; Skory, C.D.; Fu, Y.; French, S.W.; Edwards, J.E.; Spellberg, B. The high affinity iron permease is a key virulence factor required for Rhizopus oryzae pathogenesis. Mol. Microbiol. 2010, 77, 587–604. [Google Scholar] [CrossRef]
- Gebremariam, T.; Liu, M.; Luo, G.; Bruno, V.; Phan, Q.T.; Waring, A.J.; Edwards, J.E.; Filler, S.G.; Yeaman, M.R.; Ibrahim, A.S. CotH3 mediates fungal invasion of host cells during mucormycosis. J. Clin. Investig. 2014, 124, 237–250. [Google Scholar] [CrossRef]
- Zhang, Y.; Navarro, E.; Cánovas-Márquez, J.T.; Almagro, L.; Chen, H.; Chen, Y.Q.; Zhang, H.; Torres-Martínez, S.; Chen, W.; Garre, V. A new regulatory mechanism controlling carotenogenesis in the fungus Mucor circinelloides as a target to generate β-carotene over-producing strains by genetic engineering. Microb. Cell Fact. 2016, 15, 1–14. [Google Scholar] [CrossRef]
- Kosa, G.; Zimmermann, B.; Kohler, A.; Ekeberg, D.; Afseth, N.K.; Mounier, J.; Shapaval, V. High-throughput screening of Mucoromycota fungi for production of low- and high-value lipids. Biotechnol. Biofuels 2018, 11, 66. [Google Scholar] [CrossRef]
- Orlowski, M. Mucor dimorphism. Microbiol. Rev. 1991, 55, 234–258. [Google Scholar] [CrossRef]
- Wolff, A.M.; Appel, K.F.; Petersen, J.B.; Poulsen, U.; Arnau, J. Identification and analysis of genes involved in the control of dimorphism in Mucor circinelloides (syn. racemosus). FEMS Yeast Res. 2002, 2, 203–213. [Google Scholar] [CrossRef]
- Patiño-Medina, J.A.; Maldonado-Herrera, G.; Pérez-Arques, C.; Alejandre-Castañeda, V.; Reyes-Mares, N.Y.; Valle-Maldonado, M.I.; Campos-García, J.; Ortiz-Alvarado, R.; Jácome-Galarza, I.E.; Ramírez-Díaz, M.I.; et al. Control of morphology and virulence by ADP-ribosylation factors (Arf) in Mucor circinelloides. Curr. Genet. 2018, 64, 853–869. [Google Scholar] [CrossRef]
- Garcia, A.; Vellanki, S.; Lee, S.C. Genetic Tools for Investigating Mucorales Fungal Pathogenesis. Curr. Clin. Microbiol. Rep. 2018, 5, 173–180. [Google Scholar] [CrossRef]
- Ankeny, R.A.; Leonelli, S. What’s so special about model organisms? Stud. Hist. Philos. Sci. Part A 2011, 42, 313–323. [Google Scholar] [CrossRef]
- Wilson, R.K. How the worm was won: The C. elegans genome sequencing project. Trends Genet. 1999, 15, 51–58. [Google Scholar] [CrossRef]
- Clendinen, C.S.; Stupp, G.S.; Wang, B.; Garrett, T.J.; Edison, A.S. Current Metabolomics BENTHAM SCIENCE Send Orders for Reprints to reprints@benthamscience.ae 13 C Metabolomics: NMR and IROA for Unknown Identification. Curr. Metab. 2016, 4, 116–120. [Google Scholar]
- Uppuluri, P.; Alqarihi, A.; Ibrahim, A.S. Mucormycoses. In Encyclopedia of Mycology; Zaragoza, Ó., Casadevall, A.B.T.-E., Eds.; Elsevier: Oxford, UK, 2021; pp. 600–612. ISBN 978-0-323-85180-0. [Google Scholar]
- Corrochano, L.M.; Kuo, A.; Marcet-Houben, M.; Polaino, S.; Salamov, A.; Villalobos-Escobedo, J.M.; Grimwood, J.; Álvarez, M.I.; Avalos, J.; Bauer, D.; et al. Expansion of Signal Transduction Pathways in Fungi by Extensive Genome Duplication. Curr. Biol. 2016, 26, 1577–1584. [Google Scholar] [CrossRef]
- McIntyre, M.; Breum, J.; Arnau, J.; Nielsen, J. Growth physiology and dimorphism of Mucor circinelloides (syn. racemosus) during submerged batch cultivation. Appl. Microbiol. Biotechnol. 2002, 58, 495–502. [Google Scholar] [CrossRef]
- Papp, T.; Velayos, A.; Bartók, T.; Eslava, A.P.; Vágvölgyi, C.; Iturriaga, E.A. Heterologous expression of astaxanthin biosynthesis genes in Mucor circinelloides. Appl. Microbiol. Biotechnol. 2006, 69, 526–531. [Google Scholar] [CrossRef]
- van Heeswijck, R.; Roncero, M.I.G. High frequency transformation of Mucor with recombinant plasmid DNA. Carlsberg Res. Commun. 1984, 49, 691–702. [Google Scholar] [CrossRef]
- Garre, V.; Barredo, J.L.; Iturriaga, E.A. Transformation of Mucor circinelloides f. lusitanicus Protoplasts. Fungal Biol. 2015, 49–59. [Google Scholar]
- Nyilasi, I.; Ács, K.; Papp, T.; Nagy, E.; Vágvölgyi, C. Agrobacterium tumefaciens-mediated transformation of Mucor circinelloides. Folia Microbiol. 2005, 50, 415–420. [Google Scholar] [CrossRef] [PubMed]
- González-Hernández, G.A.; Herrera-Estrella, L.; Rocha-Ramírez, V.; Roncero, M.I.G.; Gutierrez-Corona, J.F. Biolistic transformation of Mucor circinelloides. Mycol. Res. 1997, 101, 953–956. [Google Scholar] [CrossRef]
- Papp, T.; Csernetics, Á.; Nyilasi, I.; Ábrók, M.; Vágvólgyi, C. Genetic transformation of zygomycetes fungi. In Progress in Mycology; Springer: Singapore, 2010; ISBN 9789048137138. [Google Scholar]
- Papp, T.; Csernetics, Á.; Nagy, G.; Bencsik, O.; Iturriaga, E.A.; Eslava, A.P.; Vágvölgyi, C. Canthaxanthin production with modified Mucor circinelloides strains. Appl. Microbiol. Biotechnol. 2013, 97, 4937–4950. [Google Scholar] [CrossRef]
- Nicolás, F.E.; Vila, A.; Moxon, S.; Cascales, M.D.; Torres-Martínez, S.; Ruiz-Vázquez, R.M.; Garre, V. The RNAi machinery controls distinct responses to environmental signals in the basal fungus Mucor circinelloides. BMC Genom. 2015, 16, 237. [Google Scholar] [CrossRef]
- Li, C.H.; Cervantes, M.; Springer, D.J.; Boekhout, T.; Ruiz-Vazquez, R.M.; Torres-Martinez, S.R.; Heitman, J.; Lee, S.C. Sporangiospore size dimorphism is linked to virulence of mucor circinelloides. PLoS Pathog. 2011, 7, e1002086. [Google Scholar] [CrossRef]
- Nosheen, S.; Yang, J.; Naz, T.; Nazir, Y.; Ahmad, M.I.; Fazili, A.B.A.; Li, S.; Mustafa, K.; Song, Y. Annotation of AMP-activated protein kinase genes and its comparative transcriptional analysis between high and low lipid producing strains of Mucor circinelloides. Biotechnol. Lett. 2020, 43, 1–10. [Google Scholar] [CrossRef]
- Hameed, A.; Hussain, S.A.; Ijaz, M.U.; Ullah, S.; Muhammad, Z.; Suleria, H.A.R.; Song, Y. Antioxidant activity of polyphenolic extracts of filamentous fungus Mucor circinelloides (WJ11): Extraction, characterization and storage stability of food emulsions. Food Biosci. 2020, 34, 100525. [Google Scholar] [CrossRef]
- Khan, M.A.K.; Yang, J.; Hussain, S.A.; Zhang, H.; Liang, L.; Garre, V.; Song, Y. Construction of DGLA producing cell factory by genetic modification of Mucor circinelloides. Microb. Cell Fact. 2019, 18, 64. [Google Scholar] [CrossRef]
- Csernetics, Á.; Tóth, E.; Farkas, A.; Nagy, G.; Bencsik, O.; Manikandan, P.; Vágvölgyi, C.; Papp, T. Expression of a bacterial ß-carotene hydroxylase in canthaxanthin producing mutant Mucor circinelloides strains. Acta Biol. Szeged. 2014, 58, 139–146. [Google Scholar]
- Csernetics, Á.; Tóth, E.; Farkas, A.; Nagy, G.; Bencsik, O.; Vágvölgyi, C.; Papp, T. Expression of Xanthophyllomyces dendrorhous cytochrome-P450 hydroxylase and reductase in Mucor circinelloides. World J. Microbiol. Biotechnol. 2015, 31, 321–336. [Google Scholar] [CrossRef]
- Meussen, B.J.; De Graaff, L.H.; Sanders, J.P.M.; Weusthuis, R.A. Metabolic engineering of Rhizopus oryzae for the production of platform chemicals. Appl. Microbiol. Biotechnol. 2012, 94, 875–886. [Google Scholar] [CrossRef]
- Roden, M.M.; Zaoutis, T.E.; Buchanan, W.L.; Knudsen, T.A.; Sarkisova, T.A.; Schaufele, R.L.; Sein, M.; Sein, T.; Chiou, C.C.; Chu, J.H.; et al. Epidemiology and outcome of zygomycosis: A review of 929 reported cases. Clin. Infect. Dis. 2005, 41, 634–656. [Google Scholar] [CrossRef]
- Oda, Y.; Yajima, Y.; Kinoshita, M.; Ohnishi, M. Differences of Rhizopus oryzae strains in organic acid synthesis and fatty acid composition. Food Microbiol. 2003, 20, 371–375. [Google Scholar] [CrossRef]
- Michielse, C.B.; Salim, K.; Ragas, P.; Ram, A.F.J.; Kudla, B.; Jarry, B.; Punt, P.J.; Van Den Hondel, C.A.M.J.J. Development of a system for integrative and stable transformation of the zygomycete Rhizopus oryzae by Agrobacterium-mediated DNA transfer. Mol. Genet. Genom. 2004, 271, 499–510. [Google Scholar] [CrossRef]
- Klein, T.M.; Sanford, J.C.; Wolf, E.D.; Wu, R. High-velocity microprojectiles for delivering nucleic acids into living cells. Nature 1987, 327, 70–73. [Google Scholar] [CrossRef]
- Tang, X.; Chang, L.; Gu, S.; Zhang, H.; Chen, Y.Q.; Chen, H.; Zhao, J.; Chen, W. Role of beta-isopropylmalate dehydrogenase in lipid biosynthesis of the oleaginous fungus Mortierella alpine. Fungal Genet. Biol. 2021, 152, 103572. [Google Scholar] [CrossRef]
- Totani, N.; Oba, K. The filamentous fungus mortierella alpina, high in arachidonic acid. Lipids 1987, 22, 1060–1062. [Google Scholar] [CrossRef]
- Ge, C.; Chen, H.; Mei, T.; Tang, X.; Chang, L.; Gu, Z.; Zhang, H.; Chen, W.; Chen, Y.Q. Application of a ω-3 desaturase with an arachidonic acid preference to eicosapentaenoic acid production in Mortierella alpina. Front. Bioeng. Biotechnol. 2018, 5, 89. [Google Scholar] [CrossRef]
- Ando, A.; Sumida, Y.; Negoro, H.; Suroto, D.A.; Ogawa, J.; Sakuradani, E.; Shimizu, S. Establishment of Agrobacterium tumefaciens-mediated transformation of an oleaginous fungus, Mortierella alpina 1S-4, and its application for eicosapentaenoic acid producer breeding. Appl. Environ. Microbiol. 2009, 75, 5529–5535. [Google Scholar] [CrossRef]
- Hao, G.; Chen, H.; Wang, L.; Gu, Z.; Song, Y.; Zhang, H.; Chen, W.; Chen, Y.Q. Role of malic enzyme during fatty acid synthesis in the oleaginous fungus Mortierella alpina. Appl. Environ. Microbiol. 2014, 80, 2672–2678. [Google Scholar] [CrossRef]
- Lu, H.; Chen, H.; Tang, X.; Yang, Q.; Zhang, H.; Chen, Y.Q.; Chen, W. Time-resolved multi-omics analysis reveals the role of nutrient stress-induced resource reallocation for TAG accumulation in oleaginous fungus Mortierella alpina. Biotechnol. Biofuels 2020, 13, 1–17. [Google Scholar] [CrossRef]
- Hao, G.; Chen, H.; Gu, Z.; Zhang, H.; Chen, W.; Chen, Y.Q. Metabolic engineering of Mortierella alpina for enhanced arachidonic acid production through the NADPH-supplying strategy. Appl. Environ. Microbiol. 2016, 82, 3280–3288. [Google Scholar] [CrossRef]
- Chen, H.; Hao, G.; Wang, L.; Wang, H.; Gu, Z.; Liu, L.; Zhang, H.; Chen, W.; Chen, Y.Q. Identification of a critical determinant that enables efficient fatty acid synthesis in oleaginous fungi. Sci. Rep. 2015, 5, 11247. [Google Scholar] [CrossRef]
- Vongsangnak, W.; Ruenwai, R.; Tang, X.; Hu, X.; Zhang, H.; Shen, B.; Song, Y.; Laoteng, K. Genome-scale analysis of the metabolic networks of oleaginous Zygomycete fungi. Gene 2013, 521, 180–190. [Google Scholar] [CrossRef]
- Kawashima, H.; Akimoto, K.; Higashiyama, K.; Fujikawa, S.; Shimizu, S. Industrial production of dihomo-γ-linolenic acid by a Δ5 desaturase-defective mutant of Mortierella alpina 1S-4 fungus. J. Am. Oil Chem. Soc. 2000, 77, 1135. [Google Scholar] [CrossRef]
- Li, X.; Liu, R.; Li, J.; Chang, M.; Liu, Y.; Jin, Q.; Wang, X. Enhanced arachidonic acid production from Mortierella alpina combining atmospheric and room temperature plasma (ARTP) and diethyl sulfate treatments. Bioresour. Technol. 2015, 177, 134–140. [Google Scholar] [CrossRef]
- Mackenzie, D.A.; Wongwathanarat, P.; Carter, A.T.; Archer, D.B. Isolation and use of a homologous histone H4 promoter and a ribosomal DNA region in a transformation vector for the oil-producing fungus Mortierella alpina. Appl. Environ. Microbiol. 2000, 66, 4655–4661. [Google Scholar] [CrossRef]
- Takeno, S.; Sakuradani, E.; Murata, S.; Inohara-Ochiai, M.; Kawashima, H.; Ashikari, T.; Shimizu, S. Establishment of an overall transformation system for an oil-producing filamentous fungus, Mortierella alpina 1S-4. Appl. Microbiol. Biotechnol. 2004, 65, 419–425. [Google Scholar] [CrossRef]
- Zhang, H.; Cui, Q.; Song, X. Research advances on arachidonic acid production by fermentation and genetic modification of Mortierella alpina. World J. Microbiol. Biotechnol. 2021, 37, 1–9. [Google Scholar] [CrossRef]
- Sakuradani, E. Advances in the production of various polyunsaturated fatty acids through oleaginous fungus Mortierella alpina breeding. Biosci. Biotechnol. Biochem. 2010, 74, 908–917. [Google Scholar] [CrossRef] [PubMed]
- Takeno, S.; Sakuradani, E.; Tomi, A.; Inohara-Ochiai, M.; Kawashima, H.; Shimizu, S. Transformation of oil-producing fungus, Mortierella alpina 1S-4, using Zeocin, and application to arachidonic acid production. J. Biosci. Bioeng. 2005, 100, 617–622. [Google Scholar] [CrossRef] [PubMed]
- Hao, G.; Chen, H.; Gu, Z.; Zhang, H.; Chen, W.; Chen, Y.Q. Metabolic engineering of Mortierella alpina for arachidonic acid production with glycerol as carbon source. Microb. Cell Fact. 2015, 14, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Feng, Y.; Cui, Q.; Song, X. Expression of Vitreoscilla hemoglobin enhances production of arachidonic acid and lipids in Mortierella alpina. BMC Biotechnol. 2017, 17, 68. [Google Scholar] [CrossRef]
- de Hoog, G.S.; Guarro, J.; Gené, J.; Figueras, M.J. Atlas of Clinical Fungi; Amer Society for Microbiology: Baarn, The Netherlands, 2000; ISBN 9070351439. [Google Scholar]
- Larone, D.H. Medically Important Fungi-A Guide to Identification, 3rd ed.; ASM Press: Washington, DC, USA, 1995. [Google Scholar]
- Sutton, D.A.; Fothergill, A.W.; Rinaldi, M.G. Guide to Clinically Significant Fungi; Williams & Wilkins: Baltimore, MD, USA, 1998; ISBN 0683182749. [Google Scholar]
- Asha, S.; Vidyavathi, M. Cunninghamella-A microbial model for drug metabolism studies—A review. Biotechnol. Adv. 2009, 27, 16–29. [Google Scholar] [CrossRef]
- Zhang, D.; Yang, Y.; Leakey, J.E.A.; Cerniglia, C.E. Phase I and phase II enzymes produced by Cunninghamella elegans for the metabolism of xenobiotics. FEMS Microbiol. Lett. 1996, 138, 221–226. [Google Scholar] [CrossRef][Green Version]
- Parshikov, I.A.; Muraleedharan, K.M.; Avery, M.A.; Williamson, J.S. Transformation of artemisinin by Cunninghamella elegans. Appl. Microbiol. Biotechnol. 2004, 64, 782–786. [Google Scholar] [CrossRef]
- Skinner, C.E.; Emmons, C.W.; Tsuchiya, H.M. Molds, Yeasts, and Actinomycetes. Soil Sci. 1947, 63, 418. [Google Scholar] [CrossRef]
- El-Morsy, E.S.M. Cunninghamella echinulata a new biosorbent of metal ions from polluted water in Egypt. Mycologia 2004, 96, 1183–1189. [Google Scholar] [CrossRef]
- Gema, H.; Kavadia, A.; Dimou, D.; Tsagou, V.; Komaitis, M.; Aggelis, G. Production of γ-linolenic acid by Cunninghamella echinulata cultivated on glucose and orange peel. Appl. Microbiol. Biotechnol. 2002, 58, 303–307. [Google Scholar] [CrossRef]
- Fakas, S.; Papanikolaou, S.; Galiotou-Panayotou, M.; Komaitis, M.; Aggelis, G. Organic nitrogen of tomato waste hydrolysate enhances glucose uptake and lipid accumulation in Cunninghamella echinulata. J. Appl. Microbiol. 2008, 105, 1062–1070. [Google Scholar] [CrossRef]
- Papanikolaou, S.; Galiotou-Panayotou, M.; Fakas, S.; Komaitis, M.; Aggelis, G. Lipid production by oleaginous Mucorales cultivated on renewable carbon sources. Eur. J. Lipid Sci. Technol. 2007, 109, 1060–1070. [Google Scholar] [CrossRef]
- Chen, H.C.; Liu, T.M. Inoculum effects on the production of γ-linolenic acid by the shake culture of Cunninghamella echinulata CCRC 31840. Enzyme Microb. Technol. 1997, 21, 137–142. [Google Scholar] [CrossRef]
- Freitag, D.G.; Foster, R.T.; Coutts, R.T.; Pickard, M.A.; Pasutto, F.M. Stereoselective metabolism of rac-mexiletine by the fungus Cunninghamella echinulata yields the major human metabolites hydroxymethylmexiletine and p-hydroxymexiletine. Drug Metab. Dispos. 1997, 25, 685–692. [Google Scholar]
- Wan, X.; Zhang, Y.; Wang, P.; Huang, F.; Chen, H.; Jiang, M. Production of gamma-linolenic acid in Pichia pastoris by expression of a delta-6 desaturase gene from Cunninghamella echinulata. J. Microbiol. Biotechnol. 2009, 1098–1102. [Google Scholar] [CrossRef]
- LeBlanc, R.E.; Meriden, Z.; Sutton, D.A.; Thompson, E.H.; Neofytos, D.; Zhang, S.X. Cunninghamella echinulata causing fatally invasive fungal sinusitis. Diagn. Microbiol. Infect. Dis. 2013, 76, 506–509. [Google Scholar] [CrossRef]
- Manosroi, J.; Chisti, Y.; Manosroi, A. Biotransformation of cortexolone to hydrocortisone by molds using a rapid color-development assay. Appl. Biochem. Microbiol. 2006, 42, 479–483. [Google Scholar] [CrossRef]
- Seigle-Murandi, F.M.; Krivobok, S.M.A.; Steiman, R.L.; Benoit-Guyod, J.L.A.; Thiault, G.A. Biphenyl Oxide Hydroxylation by Cunninghamella echinulata. J. Agric. Food Chem. 1991, 39, 428–430. [Google Scholar] [CrossRef]
- Pekala, E.; Kubowicz, P.; Lazewska, D. Cunninghamella as a microbiological model for metabolism of histamine H3 receptor antagonist 1-[3-(4-tert-butylphenoxy)propyl]piperidine. Appl. Biochem. Biotechnol. 2012, 168, 1584–1593. [Google Scholar] [CrossRef]
- Zhang, Y.; Luan, X.; Zhang, H.; Garre, V.; Song, Y.; Ratledge, C. Improved γ-linolenic acid production in Mucor circinelloides by homologous overexpressing of delta-12 and delta-6 desaturases. Microb. Cell Fact. 2017, 16, 113. [Google Scholar] [CrossRef]
- Yang, J.; Li, S.; Kabir Khan, M.A.; Garre, V.; Vongsangnak, W.; Song, Y. Increased lipid accumulation in Mucor circinelloides by overexpression of mitochondrial citrate transporter genes. Ind. Eng. Chem. Res. 2019, 58, 2125–2134. [Google Scholar] [CrossRef]
- Nosheen, S.; Naz, T.; Yang, J.; Hussain, S.A.; Fazili, A.B.A.; Nazir, Y.; Li, S.; Mohamed, H.; Yang, W.; Mustafa, K. Role of Snf-β in lipid accumulation in the high lipid-producing fungus Mucor circinelloides WJ11. Microb. Cell Fact. 2021, 20, 1–11. [Google Scholar] [CrossRef]
- Csernetics, Á.; Nagy, G.; Iturriaga, E.A.; Szekeres, A.; Eslava, A.P.; Vágvölgyi, C.; Papp, T. Expression of three isoprenoid biosynthesis genes and their effects on the carotenoid production of the zygomycete Mucor circinelloides. Fungal Genet. Biol. 2011, 48, 696–703. [Google Scholar] [CrossRef]
- Naz, T.; Yang, J.; Nosheen, S.; Sun, C.; Nazir, Y.; Mohamed, H.; Fazili, A.A.; Ullah, S.; Li, S.; Yang, W. Genetic modification of Mucor circinelloides for canthaxanthin production by heterologous expression of β-carotene ketolase gene. Front. Nutr. 2021, 718. [Google Scholar] [CrossRef]
- Jin, M.J.; Huang, H.; Xiao, A.H.; Gao, Z.; Liu, X.; Peng, C. Enhancing arachidonic acid production by Mortierella alpina ME-1 using improved mycelium aging technology. Bioprocess. Biosyst. Eng. 2009, 32, 117–122. [Google Scholar] [CrossRef]
- Huang, M.; Chen, H.; Tang, X.; Lu, H.; Zhao, J.; Zhang, H.; Chen, Y.; Chen, W. Two-stage pH control combined with oxygen-enriched air strategies for the highly efficient production of EPA by Mortierella alpina CCFM698 with fed-batch fermentation. Bioprocess. Biosyst. Eng. 2020, 43, 1725–1733. [Google Scholar] [CrossRef]
- Takeno, S.; Sakuradani, E.; Tomi, A.; Inohara-Ochiai, M.; Kawashima, H.; Ashikari, T.; Shimizu, S. Improvement of the fatty acid composition of an oil-producing filamentous fungus, Mortierella alpina 1S-4, through RNA interference with Δ12-desaturase gene expression. Appl. Environ. Microbiol. 2005, 71, 4992–4997. [Google Scholar] [CrossRef][Green Version]
- Gheinani, A.H.; Jahromi, N.H.; Feuk-Lagerstedt, E.; Taherzadeh, M.J. RNA silencing of lactate dehydrogenase gene in Rhizopus oryzae. J. RNAi Gene Silenc. 2011, 7, 443–448. [Google Scholar]
- Ibrahim, A.S.; Skory, C.D. Genetic Manipulation of Zygomycetes. In Medical Mycology: Cellular and Molecular Techniques; Wiley: New York, NY, USA, 2007; ISBN 9780470019238. [Google Scholar]
- Trieu, T.A.; Navarro-Mendoza, M.I.; Pérez-Arques, C.; Sanchis, M.; Capilla, J.; Navarro-Rodriguez, P.; Lopez-Fernandez, L.; Torres-Martínez, S.; Garre, V.; Ruiz-Vázquez, R.M.; et al. RNAi-Based Functional Genomics Identifies New Virulence Determinants in Mucormycosis. PLoS Pathog. 2017, 13, e1006150. [Google Scholar] [CrossRef]
- Calo, S.; Shertz-Wall, C.; Lee, S.C.; Bastidas, R.J.; Nicolás, F.E.; Granek, J.A.; Mieczkowski, P.; Torres-Martínez, S.; Ruiz-Vázquez, R.M.; Cardenas, M.E.; et al. Antifungal drug resistance evoked via RNAi-dependent epimutations. Nature 2014, 513, 555–558. [Google Scholar] [CrossRef] [PubMed]
- Murray, N.E. Type I Restriction Systems: Sophisticated Molecular Machines (a Legacy of Bertani and Weigle). Microbiol. Mol. Biol. Rev. 2000, 64, 412–434. [Google Scholar] [CrossRef] [PubMed]
- Shankar, S.; Uddin, I.; Afzali, S.F. Restriction endonucleases. In Plant Biotechnology, Volume 1: Principles, Techniques, and Applications; CRC Press Book: Boca Raton, FL, USA, 2017; ISBN 9781351819510. [Google Scholar]
- Brown, D.H.; Slobodkin, I.V.; Kumamoto, C.A. Stable transformation and regulated expression of an inducible reporter construct in Candida albicans using restriction enzyme-mediated integration. Mol. Gen. Genet. 1996, 251, 75–80. [Google Scholar] [CrossRef] [PubMed]
- Brown, J.S.; Aufauvre-Brown, A.; Holden, D.W. Insertional mutagenesis of Aspergillus fumigatus. Mol. Gen. Genet. 1998, 259, 327–335. [Google Scholar] [CrossRef] [PubMed]
- Warrilow, A.G.S.; Parker, J.E.; Price, C.L.; Nes, W.D.; Kelly, S.L.; Kelly, D.E. In Vitro biochemical study of CYP51-mediated azole resistance in Aspergillus fumigatus. Antimicrob. Agents Chemother. 2015, 59, 7771–7778. [Google Scholar] [CrossRef]
- Jiang, C.; Dong, D.; Yu, B.; Cai, G.; Wang, X.; Ji, Y.; Peng, Y. Mechanisms of azole resistance in 52 clinical isolates of Candida tropicalis in China. J. Antimicrob. Chemother. 2013, 68, 778–785. [Google Scholar] [CrossRef]
- Hussain, S.A.; Hameed, A.; Khan, M.A.K.; Zhang, Y.; Zhang, H.; Garre, V.; Song, Y. Engineering of fatty acid synthases (FASs) to boost the production of medium-chain fatty acids (MCFAs) in Mucor circinelloides. Int. J. Mol. Sci. 2019, 20, 786. [Google Scholar] [CrossRef]
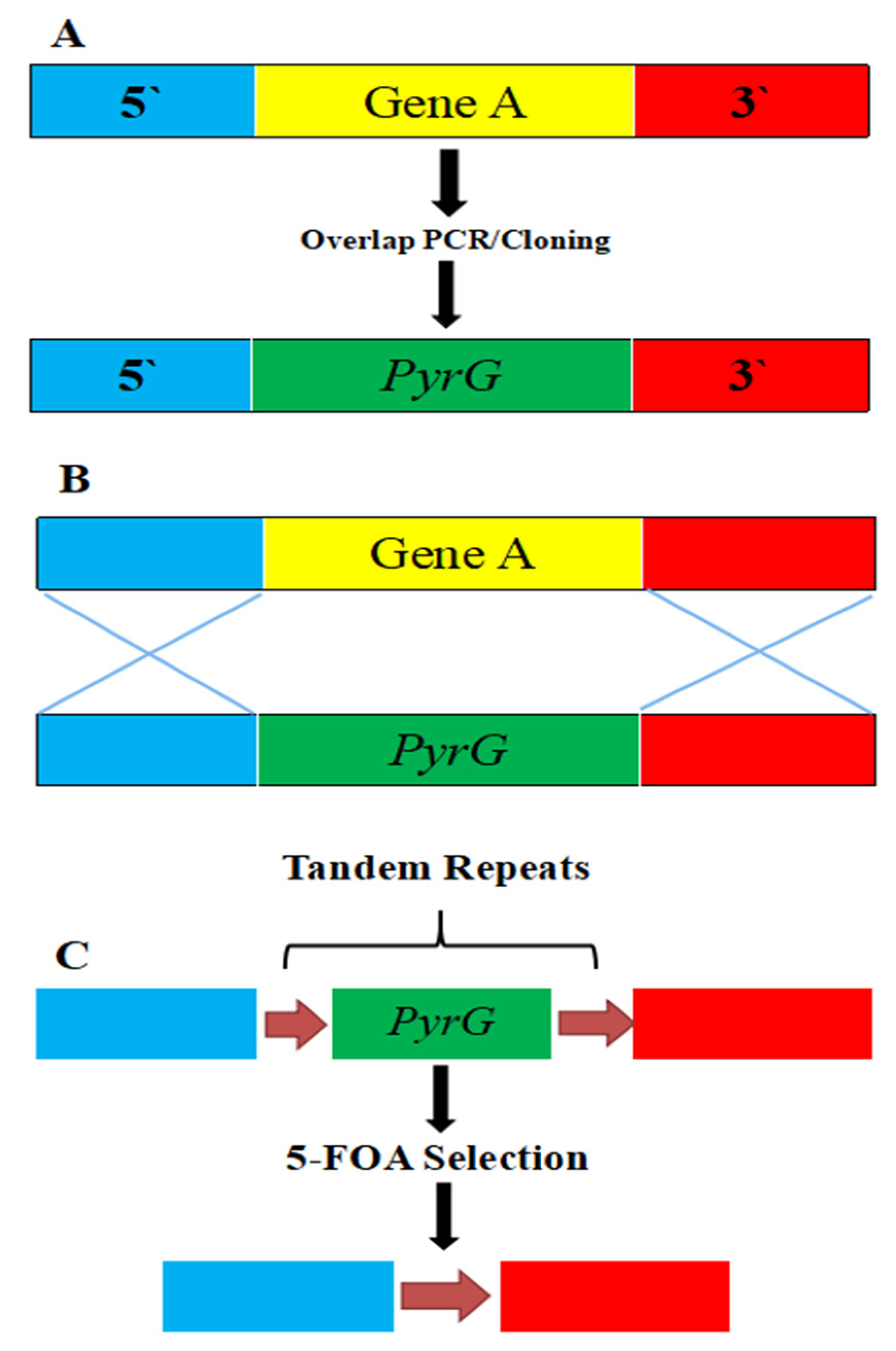
- Garcia, A.; Adedoyin, G.; Heitman, J.; Lee, S.C. Construction of a recyclable genetic marker and serial gene deletions in the human pathogenic mucorales Mucor circinelloides. G3 Genes Genomes Genet. 2017, 7, 2047–2054. [Google Scholar] [CrossRef]
- Roncero, M.I.; Jepsen, L.P.; Stroman, P.; van Heeswijck, R. Characterization of a leuA gene and an ARS element from Mucor circinelloides. Gene 1989, 84, 335–343. [Google Scholar] [CrossRef]
- Skory, C.D.; Ibrahim, A.S. Native and modified lactate dehydrogenase expression in a fumaric acid producing isolate Rhizopus oryzae 99-880. Curr. Genet. 2007, 52, 23–33. [Google Scholar] [CrossRef]
- Skory, C.D.; Freer, S.N.; Bothast, R.J. Production of L-lactic acid by Rhizopus oryzae under oxygen limiting conditions. Biotechnol. Lett. 1998, 20, 191–194. [Google Scholar] [CrossRef]
- Bai, D.M.; Zhao, X.M.; Li, X.G.; Xu, S.M. Strain improvement of Rhizopus oryzae for over-production of L(+)-lactic acid and metabolic flux analysis of mutants. Biochem. Eng. J. 2004, 18, 41–48. [Google Scholar] [CrossRef]
- Ge, C.M.; Gu, S.B.; Zhou, X.H.; Yao, J.M.; Pan, R.R.; Yu, Z.L. Breeding of L(+)-lactic acid producing strain by low-energy ion implantation. J. Microbiol. Biotechnol. 2004, 14, 363–366. [Google Scholar]
- Velayos, A.; López-Matas, M.A.; Ruiz-Hidalgo, M.J.; Eslava, A.P. Complementation analysis of carotenogenic mutants of Mucor circinelloides. Fungal Genet. Biol. 1997, 22, 19–27. [Google Scholar] [CrossRef]
- Vellanki, S.; Navarro-Mendoza, M.I.; Garcia, A.; Murcia, L.; Perez-Arques, C.; Garre, V.; Nicolas, F.E.; Lee, S.C. Mucor circinelloides: Growth, Maintenance, and Genetic Manipulation. Curr. Protoc. Microbiol. 2018, 49, e53. [Google Scholar] [CrossRef]
- Navarro, E.; Lorca-Pascual, J.; Quiles-Rosillo, M.; Nicolás, F.; Garre, V.; Torres-Martínez, S.; Ruiz-Vázquez, R. A negative regulator of light-inducible carotenogenesis in Mucor circinelloides. Mol. Genet. Genom. 2001, 266, 463–470. [Google Scholar] [CrossRef]
- Velayos, A.; Blasco, J.L.; Alvarez, M.I.; Iturriaga, E.A.; Eslava, A.P. Blue-light regulation of phytoene dehydrogenase (carB) gene expression in Mucor circinelloides. Planta 2000, 210, 938–946. [Google Scholar] [CrossRef]
- Velayos, A.; Eslava, A.P.; Iturriaga, E.A. A bifunctional enzyme with lycopene cydase and phytoene synthase activities is encoded by the carRP gene of Mucor circinelloides. Eur. J. Biochem. 2000, 267, 5509–5519. [Google Scholar] [CrossRef]
- Lee, S.C.; Blake Billmyre, R.; Li, A.; Carson, S.; Sykes, S.M.; Huh, E.Y.; Mieczkowski, P.; Ko, D.C.; Cuomo, C.A.; Heitman, J. Analysis of a food-borne fungal pathogen outbreak: Virulence and genome of a Mucor circinelloides isolate from yogurt. MBio 2014, 5, e01390–e01414. [Google Scholar] [CrossRef]
- Perina, D.; Mikoč, A.; Ahel, J.; Ćetković, H.; Žaja, R.; Ahel, I. Distribution of protein poly(ADP-ribosyl)ation systems across all domains of life. DNA Repair 2014, 23, 4–16. [Google Scholar] [CrossRef]
- Skory, C.D. Homologous recombination and double-strand break repair in the transformation of Rhizopus oryzae. Mol. Genet. Genom. 2002, 268, 397–406. [Google Scholar] [CrossRef]
- Fire, A.; Xu, S.; Montgomery, M.K.; Kostas, S.A.; Driver, S.E.; Mello, C.C. Potent and specific genetic interference by double-stranded RNA in caenorhabditis elegans. Nature 1998, 391, 806–811. [Google Scholar] [CrossRef]
- Calo, S.; Nicolás, F.E.; Vila, A.; Torres-Martínez, S.; Ruiz-Vázquez, R.M. Two distinct RNA-dependent RNA polymerases are required for initiation and amplification of RNA silencing in the basal fungus Mucor circinelloides. Mol. Microbiol. 2012, 83, 379–394. [Google Scholar] [CrossRef]
- Carthew, R.W.; Sontheimer, E.J. Origins and Mechanisms of miRNAs and siRNAs. Cell 2009, 136, 642–655. [Google Scholar] [CrossRef]
- Nicolás, F.E.; Torres-Martínez, S.; Ruiz-Vázquez, R.M. Two classes of small antisense RNAs in fungal RNA silencing triggered by non-integrative transgenes. EMBO J. 2003, 22, 3983–3991. [Google Scholar] [CrossRef]
- Billmyre, R.B.; Calo, S.; Feretzaki, M.; Wang, X.; Heitman, J. RNAi function, diversity, and loss in the fungal kingdom. Chromosom. Res. 2013, 21, 561–572. [Google Scholar] [CrossRef]
- Cervantes, M.; Vila, A.; Nicolás, F.E.; Moxon, S.; de Haro, J.P.; Dalmay, T.; Torres-Martínez, S.; Ruiz-Vázquez, R.M. A Single Argonaute Gene Participates in Exogenous and Endogenous RNAi and Controls Cellular Functions in the Basal Fungus Mucor circinelloides. PLoS ONE 2013, 8, e69283. [Google Scholar] [CrossRef]
- Nicolas, F.E.; Moxon, S.; de Haro, J.P.; Calo, S.; Grigoriev, I.V.; Torres-MartÍnez, S.; Moulton, V.; Ruiz-Vázquez, R.M.; Dalmay, T. Endogenous short RNAs generated by Dicer 2 and RNA-dependent RNA polymerase 1 regulate mRNAs in the basal fungus Mucor circinelloides. Nucleic Acids Res. 2010, 38, 5535–5541. [Google Scholar] [CrossRef]
- Nicolás, F.E.; Ruiz-Vázquez, R.M. Functional diversity of RNAi-associated sRNAs in fungi. Int. J. Mol. Sci. 2013, 14, 15348–15360. [Google Scholar] [CrossRef]
- De Haro, J.P.; Calo, S.; Cervantes, M.; Nicolás, F.E.; Torres-Martinez, S.; Ruiz-Vázquez, R.M. A single dicer gene is required for efficient gene silencing associated with two classes of small antisense RNAs in mucor circinelloides. Eukaryot. Cell 2009, 8, 1486–1497. [Google Scholar] [CrossRef]
- Nargesi, S.; Kaboli, S.; Thekkiniath, J.; Heidari, S.; Keramati, F.; Seyedmousavi, S.; Hedayati, M.T. Recent advances in genome editing tools in medical mycology research. J. Fungi 2021, 7, 257. [Google Scholar] [CrossRef]
- Pérez-Arques, C.; Navarro-Mendoza, M.I.; Murcia, L.; Navarro, E.; Garre, V.; Nicolás, F.E. A non-canonical RNAi pathway controls virulence and genome stability in Mucorales. PLoS Genet. 2020, 16, e1008611. [Google Scholar] [CrossRef] [PubMed]
- Alic, N.; Hoddinott, M.P.; Foley, A.; Slack, C.; Piper, M.D.W.; Partridge, L. Detrimental Effects of RNAi: A Cautionary Note on Its Use in Drosophila Ageing Studies. PLoS ONE 2012, 7, e45367. [Google Scholar] [CrossRef] [PubMed]
- Rath, D.; Amlinger, L.; Rath, A.; Lundgren, M. The CRISPR-Cas immune system: Biology, mechanisms and applications. Biochimie 2015, 117, 119–128. [Google Scholar] [CrossRef] [PubMed]
- Marraffini, L.A. The CRISPR-Cas System of Streptococcus Pyogenes: Function and Applications; University of Oklahoma Health Sciences Center: Oklahoma City, OK, USA, 2016. [Google Scholar]
- Bortesi, L.; Fischer, R. The CRISPR/Cas9 system for plant genome editing and beyond. Biotechnol. Adv. 2015, 33, 41–52. [Google Scholar] [CrossRef]
- Zheng, Y.M.; Lin, F.L.; Gao, H.; Zou, G.; Zhang, J.W.; Wang, G.Q.; Chen, G.D.; Zhou, Z.H.; Yao, X.S.; Hu, D. Development of a versatile and conventional technique for gene disruption in filamentous fungi based on CRISPR-Cas9 technology. Sci. Rep. 2017, 7, 9250. [Google Scholar] [CrossRef]
- Fuller, K.K.; Chen, S.; Loros, J.J.; Dunlap, J.C. Development of the CRISPR/Cas9 system for targeted gene disruption in Aspergillus fumigatus. Eukaryot. Cell 2015, 14, 1073–1080. [Google Scholar] [CrossRef]
- Shi, T.Q.; Liu, G.N.; Ji, R.Y.; Shi, K.; Song, P.; Ren, L.J.; Huang, H.; Ji, X.J. CRISPR/Cas9-based genome editing of the filamentous fungi: The state of the art. Appl. Microbiol. Biotechnol. 2017, 101, 7435–7443. [Google Scholar] [CrossRef]
- Ran, F.A.; Hsu, P.D.; Wright, J.; Agarwala, V.; Scott, D.A.; Zhang, F. Genome engineering using the CRISPR-Cas9 system. Nat. Protoc. 2013, 8, 2281–2308. [Google Scholar] [CrossRef]
- Ryan, D.E.; Taussig, D.; Steinfeld, I.; Phadnis, S.M.; Lunstad, B.D.; Singh, M.; Vuong, X.; Okochi, K.D.; McCaffrey, R.; Olesiak, M.; et al. Improving CRISPR-Cas specificity with chemical modifications in single-guide RNAs. Nucleic Acids Res. 2018, 46, 792–803. [Google Scholar] [CrossRef]
- Katayama, T.; Tanaka, Y.; Okabe, T.; Nakamura, H.; Fujii, W.; Kitamoto, K.; Maruyama, J. ichi Development of a genome editing technique using the CRISPR/Cas9 system in the industrial filamentous fungus Aspergillus oryzae. Biotechnol. Lett. 2016, 38, 637–642. [Google Scholar] [CrossRef]
- Matsu-ura, T.; Baek, M.; Kwon, J.; Hong, C. Efficient gene editing in Neurospora crassa with CRISPR technology. Fungal Biol. Biotechnol. 2015, 2, 1–7. [Google Scholar] [CrossRef]
- Liu, R.; Chen, L.; Jiang, Y.; Zhou, Z.; Zou, G. Efficient genome editing in filamentous fungus Trichoderma reesei using the CRISPR/Cas9 system. Cell Discov. 2015, 1, 15007. [Google Scholar] [CrossRef]
- Nagy, G.; Szebenyi, C.; Csernetics, Á.; Vaz, A.G.; Tóth, E.J.; Vágvölgyi, C.; Papp, T. Development of a plasmid free CRISPR-Cas9 system for the genetic modification of Mucor circinelloides. Sci. Rep. 2017, 7, 16800. [Google Scholar] [CrossRef]
- Baldin, C.; Soliman, S.; Jeon, H.; Skory, C.; Edwards, J.; Ibrahim, A. Optimization of the CRISPR/Cas9 System to Manipulate Gene Function in Rhizopus delemar. Open Forum Infect. Dis. 2017, 4, S116. [Google Scholar] [CrossRef]
- Langfelder, K.; Jahn, B.; Gehringer, H.; Schmidt, A.; Wanner, G.; Brakhage, A.A. Identification of a polyketide synthase gene (pksP) of Aspergillus fumigatus involved in conidial pigment biosynthesis and virulence. Med. Microbiol. Immunol. 1998, 187, 79–89. [Google Scholar] [CrossRef]
- Yang, G.; Rose, M.S.; Turgeon, B.G.; Yoder, O.C. A polyketide synthase is required for fungal virulence and production of the polyketide T-toxin. Plant Cell 1996, 8, 2139–2150. [Google Scholar] [CrossRef]
- Umeyama, T.; Hayashi, Y.; Shimosaka, H.; Inukai, T.; Yamagoe, S.; Takatsuka, S.; Hoshino, Y.; Nagi, M.; Nakamura, S.; Kamei, K.; et al. CRISPR/Cas9 genome editing to demonstrate the contribution of Cyp51A Gly138Ser to azole resistance in aspergillus fumigatus. Antimicrob. Agents Chemother. 2018, 62, e00894–e00918. [Google Scholar] [CrossRef]
- Gu, Y.; Gao, J.; Cao, M.; Dong, C.; Lian, J.; Huang, L.; Cai, J.; Xu, Z. Construction of a series of episomal plasmids and their application in the development of an efficient CRISPR/Cas9 system in Pichia pastoris. World J. Microbiol. Biotechnol. 2019, 35, 1–10. [Google Scholar] [CrossRef]
- Dong, L.; Lin, X.; Yu, D.; Huang, L.; Wang, B.; Pan, L. High-level expression of highly active and thermostable trehalase from Myceliophthora thermophila in Aspergillus niger by using the CRISPR/Cas9 tool and its application in ethanol fermentation. J. Ind. Microbiol. Biotechnol. 2020, 47, 133–144. [Google Scholar] [CrossRef]
- Rojas-Sánchez, U.; López-Calleja, A.C.; Millán-Chiu, B.E.; Fernández, F.; Loske, A.M.; Gómez-Lim, M.A. Enhancing the yield of human erythropoietin in Aspergillus niger by introns and CRISPR-Cas9. Protein Expr. Purif. 2020, 168, 105570. [Google Scholar] [CrossRef]
- Zalatan, J.G.; Lee, M.E.; Almeida, R.; Gilbert, L.A.; Whitehead, E.H.; La Russa, M.; Tsai, J.C.; Weissman, J.S.; Dueber, J.E.; Qi, L.S.; et al. Engineering complex synthetic transcriptional programs with CRISPR RNA scaffolds. Cell 2015, 160, 339–350. [Google Scholar] [CrossRef]
- Zhang, J.L.; Peng, Y.Z.; Liu, D.; Liu, H.; Cao, Y.X.; Li, B.Z.; Li, C.; Yuan, Y.J. Gene repression via multiplex gRNA strategy in Y. lipolytica. Microb. Cell Fact. 2018, 17, 1–13. [Google Scholar] [CrossRef]
- Ronda, C.; Maury, J.; Jakočiunas, T.; Baallal Jacobsen, S.A.; Germann, S.M.; Harrison, S.J.; Borodina, I.; Keasling, J.D.; Jensen, M.K.; Nielsen, A.T. CrEdit: CRISPR mediated multi-loci gene integration in Saccharomyces cerevisiae. Microb. Cell Fact. 2015, 14, 1–11. [Google Scholar] [CrossRef]
- Nielsen, J.C.; Nielsen, J. Development of fungal cell factories for the production of secondary metabolites: Linking genomics and metabolism. Synth. Syst. Biotechnol. 2017, 2, 5–12. [Google Scholar] [CrossRef]
- Calderone, R.; Sun, N.; Gay-Andrieu, F.; Groutas, W.; Weerawarna, P.; Prasad, S.; Alex, D.; Li, D. Antifungal drug discovery: The process and outcomes. Future Microbiol. 2014, 9, 791–805. [Google Scholar] [CrossRef]
- Hara, S.; Jin, F.J.; Takahashi, T.; Koyama, Y. A further study on chromosome minimization by protoplast fusion in Aspergillus oryzae. Mol. Genet. Genom. 2012, 287, 177–187. [Google Scholar] [CrossRef]
- Roemer, T.; Boone, C. Systems-level antimicrobial drug and drug synergy discovery. Nat. Chem. Biol. 2013, 9, 222–231. [Google Scholar] [CrossRef]
| Organism | Gene Name | Strategy Employed | Product Obtained | References |
|---|---|---|---|---|
| M. circinelloides CBS 277.49 | Delta 12 and delta 6 desaturase | Overexpression | GLA | [90] |
| M. circinelloides | Delta-6 elongase | Overexpression | DGLA | [46] |
| M. circinelloides | Citrate transporter | Overexpression | 44% lipid content elevation | [91] |
| M. circinelloides | Snf-β | Overexpression and knockout | 32% lipid content elevation | [92] |
| M. circinelloides | crgA | Mutagenesis and deletion of crgA gene | 4 mg/g β-carotene 268–527 µg/g β-carotene | [23,93] |
| M. circinelloides | crtS and crtR | Overexpression | 190 µg/g canthaxanthin | [48] |
| M. circinelloides | crtW and crtZ | Co-expression | 145–443 µg/g canthaxanthin, 35 µg/g astaxanthin | [47] |
| M. circinelloides | bkt | Overexpression | 576 µg/g canthaxanthin | [94] |
| M. alpina | gamma linolenic acid elongase | Overexpression | Twofold greater production of ARA | [69] |
| M. alpina | GK and ME1 | Co-overexpression | Fatty acid accumulation by 81% | [70] |
| M. alpina | N/A | UV mutagenesis (optimized mycelia ageing technique) | 19 g/L of ARA production | [95] |
| M. alpina | ω-3 fatty acid desaturase | Heterologous expression | EPA | [96] |
| M. alpina | G6PD2 | Overexpression | 1.7-fold rise in total FA production | [60] |
| malE2 | 1.5-fold increase in ARA content | |||
| M. alpina | VHb | Heterologous overexpression | eight times more total lipid and ARA | [71] |
| M. alpina | 12-desaturase gene | RNA interference | 19.02 g/L of ARA | [97] |
| M. alpina | MaLeuB | Homologous overexpression | 20.2% higher total FA | [54] |
| R.arrhizus | ldhA and ldhB | Small interfering RNA | 15.4% increment in ethanol yield | [98] |
| C. echinulata | - | Grown on tomato waste and potato starch | 1018 and 540 mg/L of GLA, respectively | [81,82] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mohamed, H.; Naz, T.; Yang, J.; Shah, A.M.; Nazir, Y.; Song, Y. Recent Molecular Tools for the Genetic Manipulation of Highly Industrially Important Mucoromycota Fungi. J. Fungi 2021, 7, 1061. https://doi.org/10.3390/jof7121061
Mohamed H, Naz T, Yang J, Shah AM, Nazir Y, Song Y. Recent Molecular Tools for the Genetic Manipulation of Highly Industrially Important Mucoromycota Fungi. Journal of Fungi. 2021; 7(12):1061. https://doi.org/10.3390/jof7121061
Chicago/Turabian StyleMohamed, Hassan, Tahira Naz, Junhuan Yang, Aabid Manzoor Shah, Yusuf Nazir, and Yuanda Song. 2021. "Recent Molecular Tools for the Genetic Manipulation of Highly Industrially Important Mucoromycota Fungi" Journal of Fungi 7, no. 12: 1061. https://doi.org/10.3390/jof7121061
APA StyleMohamed, H., Naz, T., Yang, J., Shah, A. M., Nazir, Y., & Song, Y. (2021). Recent Molecular Tools for the Genetic Manipulation of Highly Industrially Important Mucoromycota Fungi. Journal of Fungi, 7(12), 1061. https://doi.org/10.3390/jof7121061







