Agrobacterium tumefaciens-Mediated Transformation of NHEJ Mutant Aspergillus nidulans Conidia: An Efficient Tool for Targeted Gene Recombination Using Selectable Nutritional Markers
Abstract
1. Introduction
2. Materials and Methods
2.1. Strains, Media, Growth Conditions, and Reagents
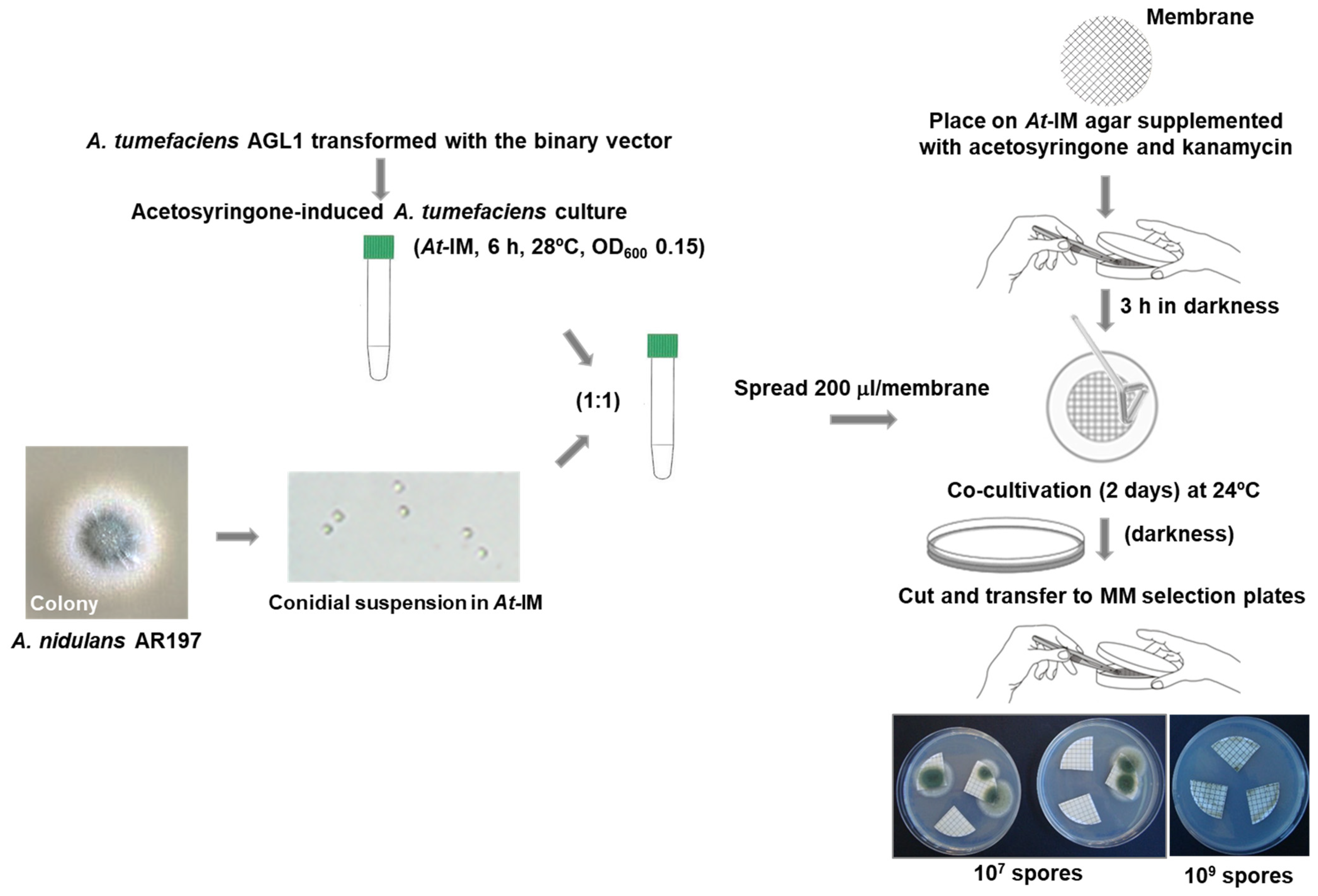
2.2. A. tumefaciens-Mediated Transformation
2.2.1. Preparation of A. tumefaciens Competent Cells
2.2.2. Transformation of A. tumefaciens by Electroporation
2.2.3. Transformation of A. nidulans Conidia by A. tumefaciens
2.3. Nucleic Acid Manipulations
2.3.1. General
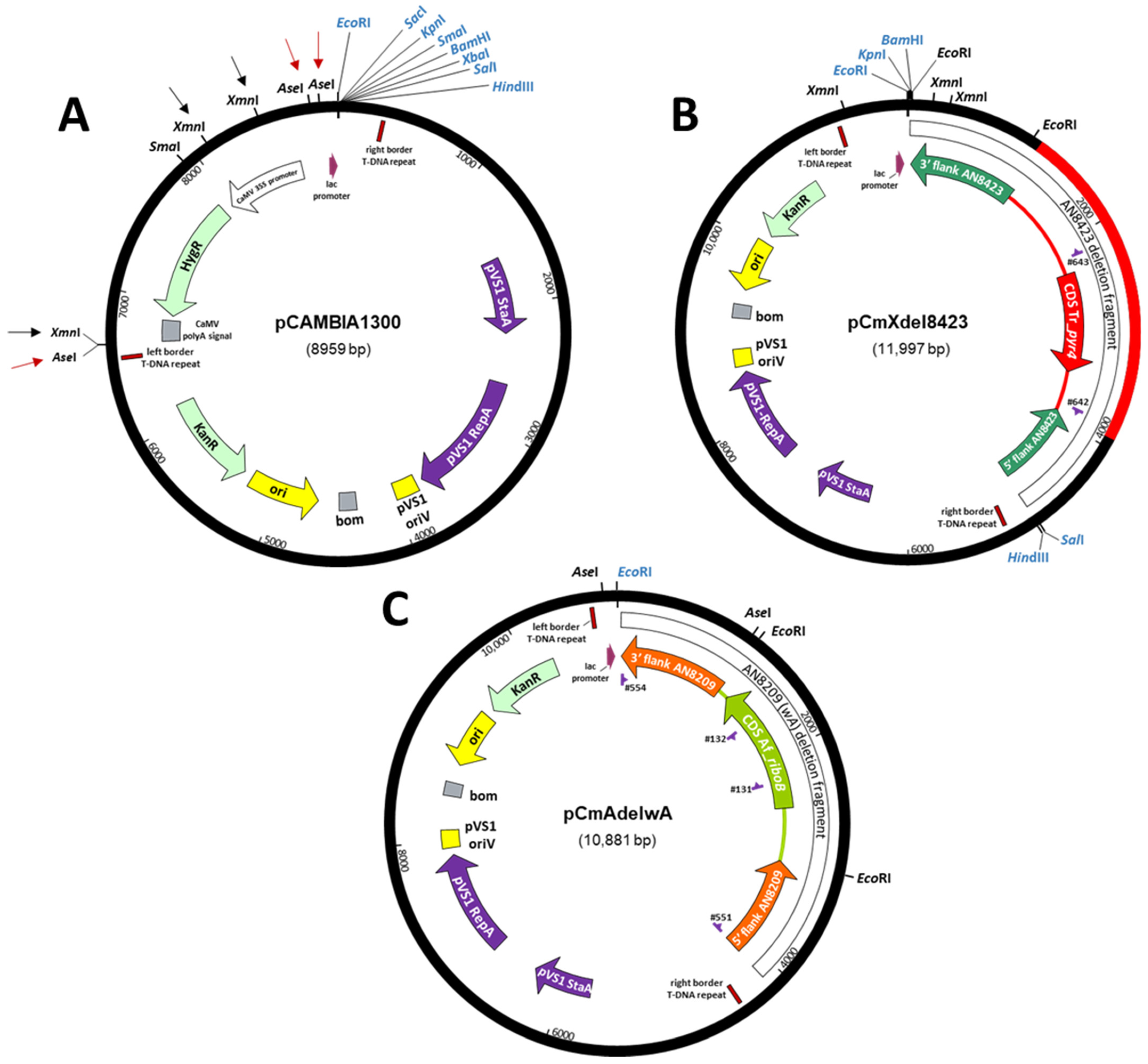
2.3.2. Construction of ATMT Binary Vectors
2.3.3. PCR of Conidia
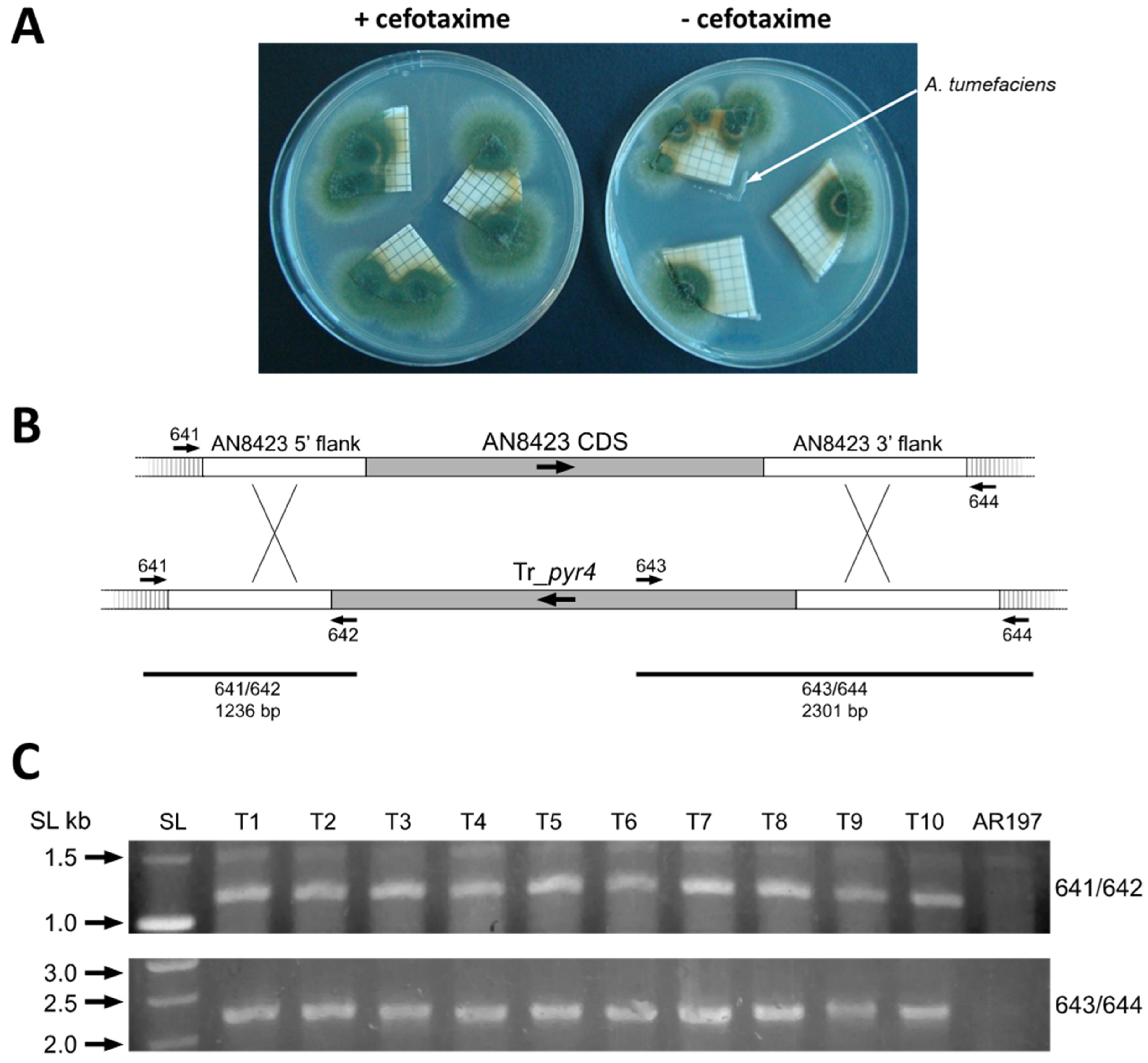
2.3.4. Deletion of AN8423
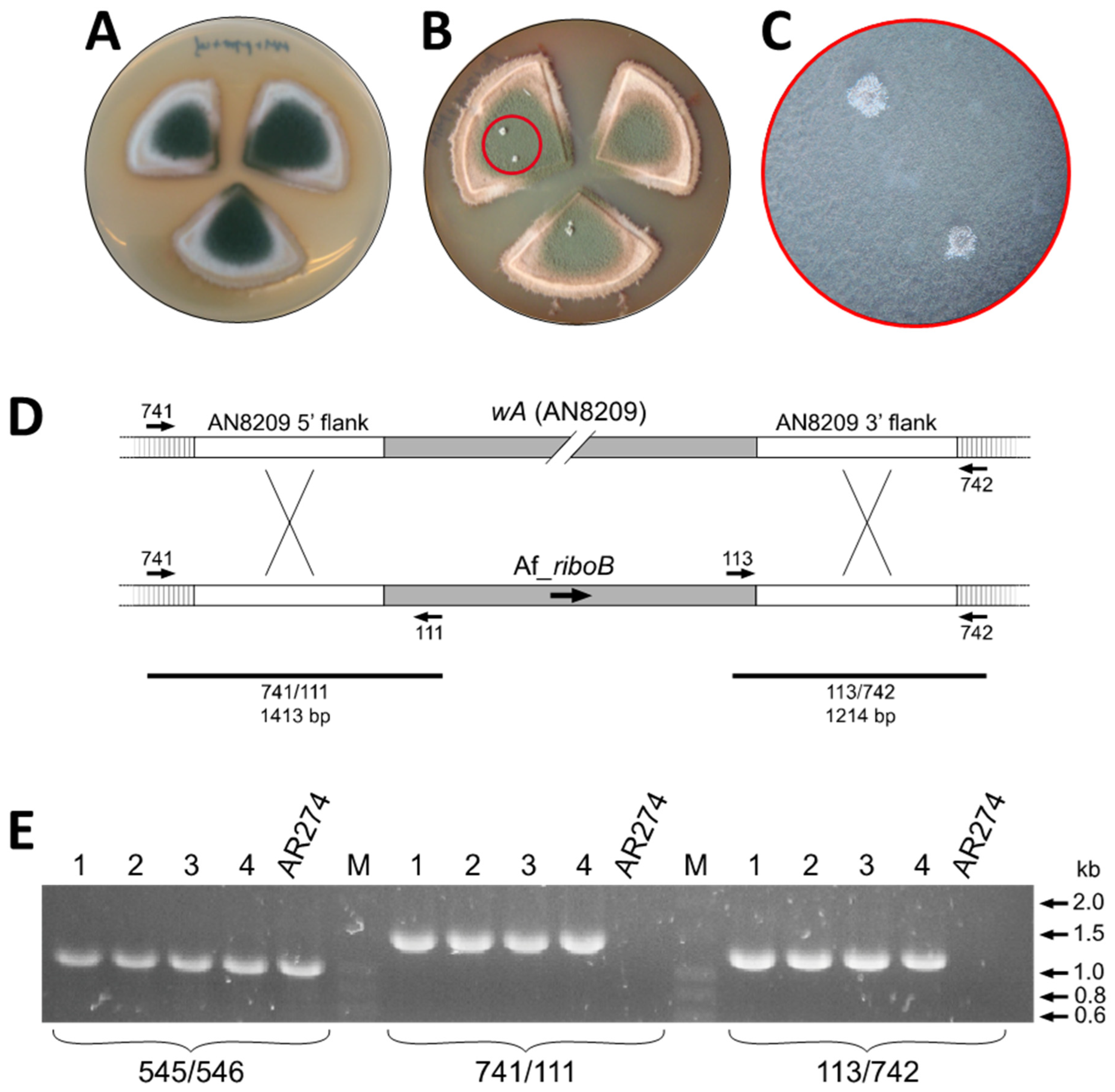
2.3.5. Integration at wA
3. Results
3.1. Adaptation of ATMT to A. nidulans and Deletion of AN8423 as a Prototype of a Targeted Gene
3.2. A Visual Reporter of Transformation
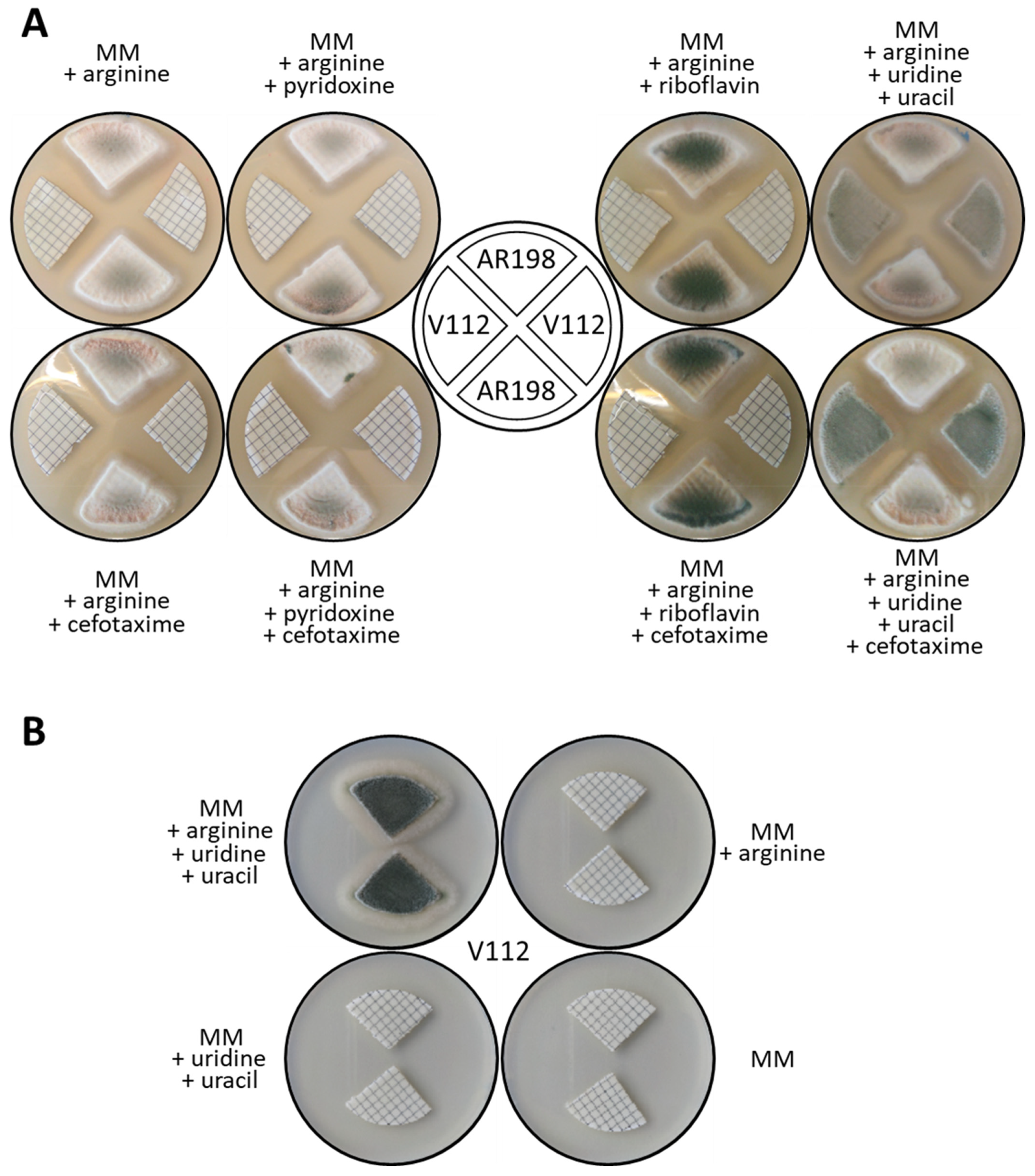
3.3. Nutrient Cross-Feeding Nullifies Certain Selection Systems
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data availability Statement
Acknowledgments
Conflicts of Interest
References
- Case, M.E.; Schweizer, M.; Kushner, S.R.; Giles, N.H. Efficient transformation of Neurospora crassa by utilizing hybrid plasmid DNA. Proc. Natl. Acad. Sci. USA 1979, 76, 5259–5263. [Google Scholar] [CrossRef] [PubMed]
- Li, D.; Tang, Y.; Lin, J.; Cai, W. Methods for genetic transformation of filamentous fungi. Microb. Cell Factories 2017, 16, 168. [Google Scholar] [CrossRef] [PubMed]
- Tilburn, J.; Scazzocchio, C.; Taylor, G.G.; Zabicky-Zissman, J.H.; Lockington, R.A.; Davies, R.W. Transformation by integration in Aspergillus nidulans. Gene 1983, 26, 205–221. [Google Scholar] [CrossRef]
- Jung, M.K.; Ovechkina, Y.; Prigozhina, N.; Oakley, C.E.; Oakley, B.R. The use of beta-D-glucanase as a substitute for Novozyme 234 in immunofluorescence and protoplasting. Fungal Genet. Rep. 2000, 47, 8. [Google Scholar] [CrossRef]
- Szewczyk, E.; Nayak, T.; Oakley, C.E.; Edgerton, H.; Xiong, Y.; Taheri-Talesh, N.; Osmani, S.A.; Oakley, B.R. Fusion PCR and gene targeting in Aspergillus nidulans. Nat. Protoc. 2006, 1, 3111–3120. [Google Scholar] [CrossRef] [PubMed]
- De Groot, M.J.A.; Bundock, P.; Hooykaas, P.J.J.; Beijersbergen, A.G.M. Agrobacterium tumefaciens-mediated transformation of filamentous fungi. Nat. Biotechnol. 1998, 16, 839–842. [Google Scholar] [CrossRef]
- Gouka, R.J.; Gerk, C.; Hooykaas, P.J.; Bundock, P.; Musters, W.; Verrips, C.T.; de Groot, M.J. Transformation of Aspergillus awamori by Agrobacterium tumefaciens–mediated homologous recombination. Nat. Biotechnol. 1999, 17, 598–601. [Google Scholar] [CrossRef]
- Idnurm, A.; Bailey, A.M.; Cairns, T.C.; Elliott, C.E.; Foster, G.D.; Ianiri, G.; Jeon, J. A silver bullet in a golden age of functional genomics: The impact of Agrobacterium-mediated transformation of fungi. Fungal Biol. Biotechnol. 2017, 4, 6. [Google Scholar] [CrossRef]
- Frandsen, R.J.N. A guide to binary vectors and strategies for targeted genome modification in fungi using Agrobacterium tumefaciens-mediated transformation. J. Microbiol. Methods 2011, 87, 247–262. [Google Scholar] [CrossRef]
- Kalleda, N.; Naorem, A.; Manchikatla, R.V. Targeting fungal genes by diced siRNAs: A rapid tool to decipher gene function in Aspergillus nidulans. PLoS ONE 2013, 8, e75443. [Google Scholar] [CrossRef]
- Lazo, G.R.; Stein, P.A.; Ludwig, R.A. A DNA transformation–competent Arabidopsis genomic library in Agrobacterium. Bio/Technology 1991, 9, 963–967. [Google Scholar] [CrossRef]
- Sambrook, J.; Russell, D.W. Molecular Cloning: A Laboratory Manual, 3rd ed.; Cold Spring Harbor Laboratory Press: Cold Spring Harbor, NY, USA, 2001. [Google Scholar]
- Nayak, T.; Szewczyk, E.; Oakley, C.E.; Osmani, A.; Ukil, L.; Murray, S.L.; Hynes, M.J.; Osmani, S.A.; Oakley, B. A versatile and efficient gene-targeting system for Aspergillus nidulans. Genetics 2006, 172, 1557–1566. [Google Scholar] [CrossRef]
- Pardo, E.; Orejas, M. The Aspergillus nidulans Zn(II)2Cys6 transcription factor AN5673/RhaR mediates L-rhamnose utilization and the production of α-L-rhamnosidases. Microb. Cell Factories 2014, 13, 161. [Google Scholar] [CrossRef]
- Pontecorvo, G.; Roper, J.A.; Hemmons, L.M.; Macdonald, K.D.; Bufton, A.W.J. The genetics of Aspergillus nidulans. Adv. Genet. 1953, 5, 141–238. [Google Scholar] [CrossRef] [PubMed]
- Cove, D.J. The induction and repression of nitrate reductase in the fungus Aspergillus nidulans. Biochim. Biophys. Acta 1966, 113, 51–56. [Google Scholar] [CrossRef]
- Gruber, F.; Visser, J.; Kubicek, C.P.; de Graaff, L.H. Cloning of the Trichoderma reesei pyrG gene and its use as a homologous marker for a high-frequency transformation system. Curr. Genet. 1990, 18, 447–451. [Google Scholar] [CrossRef]
- Cullen, D.; Leong, S.A.; Wilson, L.J.; Henner, D.J. Transformation of Aspergillus nidulans with the hygromycin-resistance gene, hph. Gene 1987, 57, 21–26. [Google Scholar] [CrossRef]
- Martinelli, S.D.; Zamir, A. Variable susceptibility of laboratory strains of Aspergillus nidulans to hygromycin B and other ribosomal antibiotics. Fungal Genet. Rep. 1987, 34, 10. [Google Scholar] [CrossRef][Green Version]
- Punt, P.J.; Oliver, R.P.; Dingemanse, M.A.; Pouwels, P.H.; van den Hondel, C.A.M.J.J. Transformation of Aspergillus based on the hygromycin B resistance marker from Escherichia coli. Gene 1987, 56, 117–124. [Google Scholar] [CrossRef]
- Meyer, V.; Mueller, D.; Strowig, T.; Stahl, U. Comparison of different transformation methods for Aspergillus giganteus. Curr. Genet. 2003, 43, 371–377. [Google Scholar] [CrossRef]
- Sugui, J.A.; Chang, Y.C.; Kwon-Chung, K.J. Agrobacterium tumefaciens-mediated transformation of Aspergillus fumigatus: An efficient tool for insertional mutagenesis and targeted gene disruption. Appl. Environ. Microbiol. 2005, 71, 1798–1802. [Google Scholar] [CrossRef] [PubMed]
- MacCabe, A.P.; Ninou, E.I.; Pardo, E.; Orejas, M. Catabolism of L-rhamnose in A. nidulans proceeds via the non-phosphorylated pathway and is glucose repressed by a CreA-independent mechanism. Microb. Cell Factories 2020, 19, 188. [Google Scholar] [CrossRef] [PubMed]
- MacCabe, A.; Sanmartín, G.; Orejas, M. Identification of the genes encoding the catalytic steps corresponding to LRA4 (L-2-keto-3-deoxyrhamnonate aldolase) and L-lactaldehyde dehydrogenase in Aspergillus nidulans: Evidence for involvement of the loci AN9425/lraD and AN0544/aldA in the L-rhamnose catabolic pathway. Environ. Microbiol. 2021, 23, 2420–2432. [Google Scholar] [CrossRef] [PubMed]
- Greenstein, S.; Shadkchan, Y.; Jadoun, J.; Sharon, C.; Markovich, S.; Osherov, N. Analysis of the Aspergillus nidulans thaumatin-like cetA gene and evidence for transcriptional repression of pyr4 expression in the cetA-disrupted strain. Fungal Genet. Biol. 2006, 43, 42–53. [Google Scholar] [CrossRef]
- Mayorga, M.E.; Timberlake, W.E. Isolation and molecular characterization of the Aspergillus nidulans wA gene. Genetics 1990, 126, 73–79. [Google Scholar] [CrossRef]
- Mayorga, M.E.; Timberlake, W.E. The developmentally regulated Aspergillus nidulans wA gene encodes a polypeptide homologous to polyketide and fatty acid synthases. Mol. Gen. Genet. 1992, 235, 205–212. [Google Scholar] [CrossRef]
- Watanabe, A.; Fujii, I.; Sankawa, U.; Mayorga, M.E.; Timberlake, W.E.; Ebizuka, Y. Re-identification of Aspergillus nidulans wA gene to code for a polyketide synthase of naphthopyrone. Tetrahedron Lett. 1999, 40, 91–94. [Google Scholar] [CrossRef]
- Osmani, A.H.; Oakley, B.R.; Osmani, S.A. Identification and analysis of essential Aspergillus nidulans genes using the heterokaryon rescue technique. Nat. Protoc. 2006, 1, 2517–2526. [Google Scholar] [CrossRef]
- Schuster, M.; Kahmann, R. CRISPR-Cas9 genome editing approaches in filamentous fungi and oomycetes. Fungal Genet. Biol. 2019, 130, 43–53. [Google Scholar] [CrossRef]
- Jiang, C.; Lv, G.; Tu, Y.; Cheng, X.; Duan, Y.; Zeng, B.; He, B. Applications of CRISPR/Cas9 in the synthesis of secondary metabolites in filamentous fungi. Front. Microbiol. 2021, 12, 638096. [Google Scholar] [CrossRef]
| Strain Code | Name | Genotype 1 | References |
|---|---|---|---|
| AR197 | TN02A3 | ΔnkuA::argB, argB2, pyrG89, pyroA4 | FGSC A1149 |
| AR198 | TN02A21 | ΔnkuA::argB, argB2, riboB2, pyroA4 | [13] |
| AR271 | ΔnkuA::argB, argB2, ΔriboB2::Af_riboB, pyroA4 | [14] | |
| AR274 | FGSC A4 | [15] | |
| V112 | pyrG89, argB2 | Lab collection |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Casado-del Castillo, V.; MacCabe, A.P.; Orejas, M. Agrobacterium tumefaciens-Mediated Transformation of NHEJ Mutant Aspergillus nidulans Conidia: An Efficient Tool for Targeted Gene Recombination Using Selectable Nutritional Markers. J. Fungi 2021, 7, 961. https://doi.org/10.3390/jof7110961
Casado-del Castillo V, MacCabe AP, Orejas M. Agrobacterium tumefaciens-Mediated Transformation of NHEJ Mutant Aspergillus nidulans Conidia: An Efficient Tool for Targeted Gene Recombination Using Selectable Nutritional Markers. Journal of Fungi. 2021; 7(11):961. https://doi.org/10.3390/jof7110961
Chicago/Turabian StyleCasado-del Castillo, Virginia, Andrew P. MacCabe, and Margarita Orejas. 2021. "Agrobacterium tumefaciens-Mediated Transformation of NHEJ Mutant Aspergillus nidulans Conidia: An Efficient Tool for Targeted Gene Recombination Using Selectable Nutritional Markers" Journal of Fungi 7, no. 11: 961. https://doi.org/10.3390/jof7110961
APA StyleCasado-del Castillo, V., MacCabe, A. P., & Orejas, M. (2021). Agrobacterium tumefaciens-Mediated Transformation of NHEJ Mutant Aspergillus nidulans Conidia: An Efficient Tool for Targeted Gene Recombination Using Selectable Nutritional Markers. Journal of Fungi, 7(11), 961. https://doi.org/10.3390/jof7110961





