Comment on Dimou et al. Profile of Membrane Cargo Trafficking Proteins and Transporters Expressed under N Source Derepressing Conditions in Aspergillus nidulans. J. Fungi 2021, 7, 560
Abstract
:1. Introduction
“Impressively (sic erat scriptum), all proteins essential for cargo biogenesis were indeed present in the proteome analyzed. The only exception is the product of AN11127, which was recently reported as the Sec12 protein of A. nidulans [52, this refers to our 2019 paper]. However, to our opinion, AN11127 might not be an orthologous protein to the Sec12 protein, which in yeast acts as an essential guanine nucleotide exchange factor (GEF) for activating Sar1p and initiation of COPII vesicle formation. Evidence against AN11127 being an isofunctional Sec12 orthologue comes from the observation that is has no amino acid similarity with Sec12 proteins, and importantly, is not present in several Aspergilli and most ascomycetes.”
“Evidence against AN11127 being an isofunctional Sec12 orthologue comes from the observation that is has no amino acid similarity with Sec12 proteins, and importantly, is not present in several Aspergilli and most ascomycetes” is incorrect.
2. Results
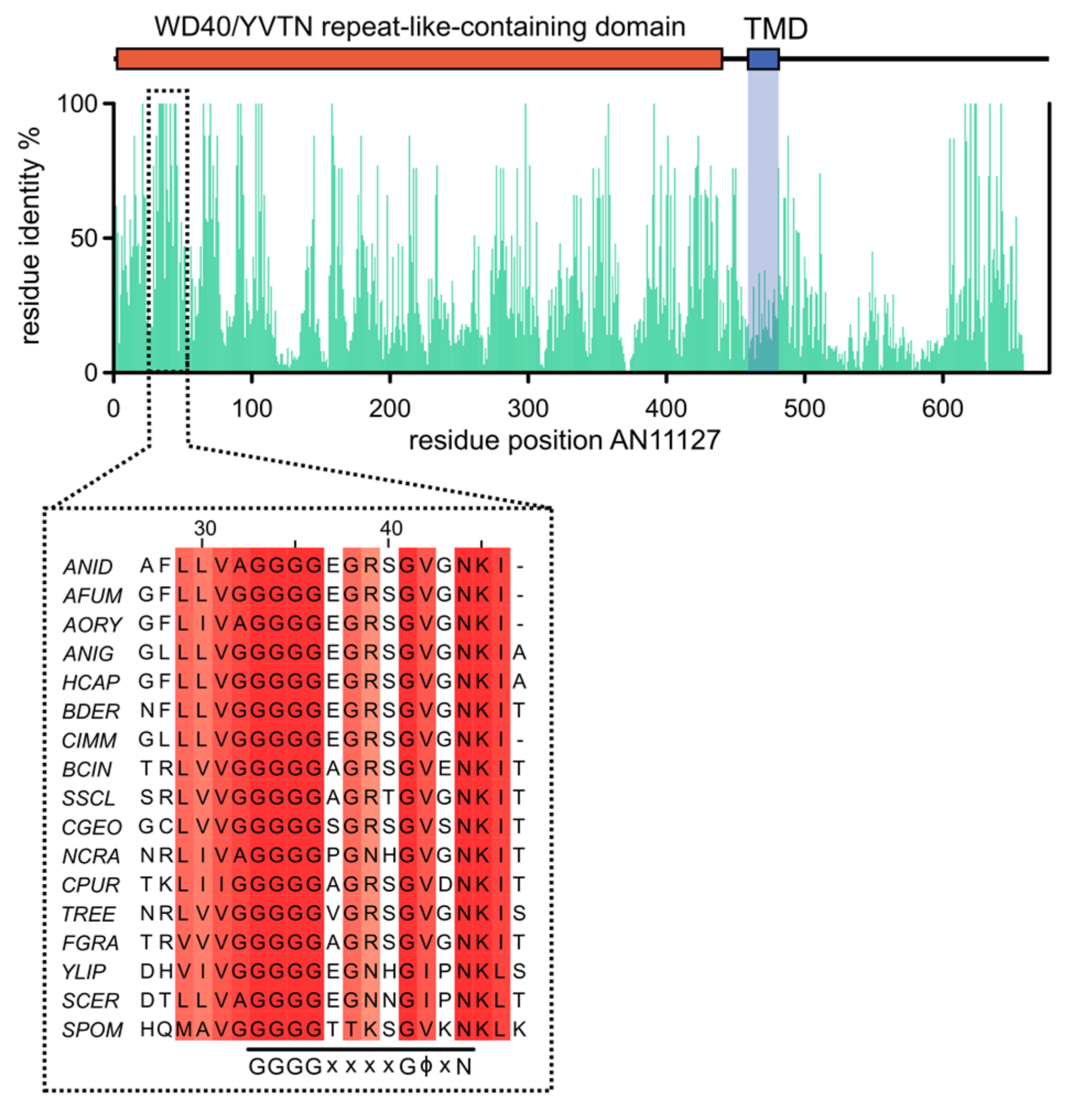
2.1. Compelling Evidence Previously Reported by Bravo-Plaza et al. Demonstrating That the AN11127 Product Has Convincing Sequence Similarity with Other Sec12 Proteins and That It Is Indeed an Isofunctional Sec12 Orthologue
“This (lack of sequence conservation) is also likely the case for Sec12; low sequence conservation and the presence of multiple WD40 repeats make it difficult to distinguish from other WD40 repeat containing proteins. This became apparent when trying to identify the S. cerevisiae Sec12 using the H. sapiens sequence; multiple rounds of psi-BLAST were required to show that they are indeed homologs, as BLASTp did not provide enough sensitivity to do so”.
2.2. Further Evidence Disproving the Statement by Dimou et al.
3. Conclusions
Supplementary Materials
Author Contributions
Funding
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Nakano, A.; Muramatsu, M. A novel GTP-binding protein, Sar1p, is involved in transport from the endoplasmic reticulum to the Golgi apparatus. J. Cell Biol. 1989, 109, 2677–2691. [Google Scholar] [CrossRef] [Green Version]
- Miller, E.A.; Schekman, R. COPII—A flexible vesicle formation system. Curr. Opin. Cell Biol. 2013, 25, 420–427. [Google Scholar] [CrossRef] [PubMed]
- Connerly, P.L.; Esaki, M.; Montegna, E.A.; Strongin, D.E.; Levi, S.; Soderholm, J.; Glick, B.S. Sec16 is a determinant of transitional ER organization. Curr. Biol. 2005, 15, 1439–1447. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- d’Enfert, C.; Barlowe, C.; Nishikawa, S.; Nakano, A.; Schekman, R. Structural and functional dissection of a membrane glycoprotein required for vesicle budding from the endoplasmic reticulum. Mol. Cell Biol. 1991, 11, 5727–5734. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nakano, A.; Brada, D.; Schekman, R. A membrane glycoprotein, Sec12p, required for protein transport from the endoplasmic reticulum to the Golgi apparatus in yeast. J. Cell Biol. 1988, 107, 851–863. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Barlowe, C.; Schekman, R. SEC12 encodes a guanine-nucleotide-exchange factor essential for transport vesicle budding from the ER. Nature 1993, 365, 347–349. [Google Scholar] [CrossRef] [PubMed]
- Schekman, R.; Novick, P. 23 genes, 23 years later. Cell 2004, 116, S13–S15. [Google Scholar] [CrossRef] [Green Version]
- Bravo-Plaza, I.; Hernandez-Gonzalez, M.; Pinar, M.; Diaz, J.F.; Penalva, M.A. Identification of the guanine nucleotide exchange factor for SAR1 in the filamentous fungal model Aspergillus nidulans. Biochim. Biophys. Acta Mol. Cell Res. 2019, 1866, 118551. [Google Scholar] [CrossRef]
- Dimou, S.; Georgiou, X.; Sarantidi, E.; Diallinas, G.; Anagnostopoulos, A.K. Profile of Membrane Cargo Trafficking Proteins and Transporters Expressed under N Source Derepressing Conditions in Aspergillus nidulans. J. Fungi 2021, 7, 560. [Google Scholar] [CrossRef] [PubMed]
- Joiner, A.M.N.; Fromme, J.C. Structural basis for the initiation of COPII vesicle biogenesis. Structure 2021, 29, 859–872.e6. [Google Scholar] [CrossRef] [PubMed]
- McMahon, C.; Studer, S.M.; Clendinen, C.; Dann, G.P.; Jeffrey, P.D.; Hughson, F.M. The structure of Sec12 implicates potassium ion coordination in Sar1 activation. J. Biol. Chem. 2012, 287, 43599–43606. [Google Scholar] [CrossRef] [Green Version]
- Schlacht, A.; Dacks, J.B. Unexpected ancient paralogs and an evolutionary model for the COPII coat complex. Genome Biol. Evol. 2015, 7, 1098–1109. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hernández-González, M.; Peñalva, M.A.; Pantazopoulou, A. Conditional inactivation of Aspergillus nidulans sarA uncovers the morphogenetic potential of regulating endoplasmic reticulum (ER) exit. Mol. Microbiol. 2014, 95, 491–508. [Google Scholar] [CrossRef] [PubMed]
- Rep, M.; Duyvesteijn, R.G.; Gale, L.; Usgaard, T.; Cornelissen, B.J.; Ma, L.J.; Ward, T.J. The presence of GC-AG introns in Neurospora crassa and other euascomycetes determined from analyses of complete genomes: Implications for automated gene prediction. Genomics 2006, 87, 338–347. [Google Scholar] [CrossRef] [Green Version]
| Sequence ID | Description | Score | E-Value |
|---|---|---|---|
| KV878136 | Aspergillus versicolor CBS 583.65 | 856 | 0 |
| DF126470 | Aspergillus luchuensis IFO 4308 | 572 | 3.0 × 10−178 |
| KV878249 | Aspergillus luchuensis CBS 106.47 | 565 | 4.0 × 10−176 |
| KV878690 | Aspergillus brasiliensis CBS 101740 | 565 | 6.0 × 10−176 |
| ACJE01000012.1 | Aspergillus niger ATCC 1015 | 545 | 4.0 × 10−169 |
| KZ851914 | Aspergillus niger ATCC 13496 | 545 | 4.0 × 10−169 |
| OGUI01000010.1 | Aspergillus niger strain N402 (ATCC64974) | 545 | 4.0 × 10−169 |
| VTFN01000009 | Aspergillus niger strain LDM3 | 543 | 2.0 × 10−168 |
| KV878203 | Aspergillus tubingensis CBS 134.48 | 530 | 8.0 × 10−164 |
| An17_A_niger_CBS_513_88 | Aspergillus niger CBS 513.88 | 283 | 3.0 × 10−79 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bravo-Plaza, I.; Hernández-González, M.; Peñalva, M.Á. Comment on Dimou et al. Profile of Membrane Cargo Trafficking Proteins and Transporters Expressed under N Source Derepressing Conditions in Aspergillus nidulans. J. Fungi 2021, 7, 560. J. Fungi 2021, 7, 1037. https://doi.org/10.3390/jof7121037
Bravo-Plaza I, Hernández-González M, Peñalva MÁ. Comment on Dimou et al. Profile of Membrane Cargo Trafficking Proteins and Transporters Expressed under N Source Derepressing Conditions in Aspergillus nidulans. J. Fungi 2021, 7, 560. Journal of Fungi. 2021; 7(12):1037. https://doi.org/10.3390/jof7121037
Chicago/Turabian StyleBravo-Plaza, Ignacio, Miguel Hernández-González, and Miguel Á. Peñalva. 2021. "Comment on Dimou et al. Profile of Membrane Cargo Trafficking Proteins and Transporters Expressed under N Source Derepressing Conditions in Aspergillus nidulans. J. Fungi 2021, 7, 560" Journal of Fungi 7, no. 12: 1037. https://doi.org/10.3390/jof7121037
APA StyleBravo-Plaza, I., Hernández-González, M., & Peñalva, M. Á. (2021). Comment on Dimou et al. Profile of Membrane Cargo Trafficking Proteins and Transporters Expressed under N Source Derepressing Conditions in Aspergillus nidulans. J. Fungi 2021, 7, 560. Journal of Fungi, 7(12), 1037. https://doi.org/10.3390/jof7121037






