Gcorn fungi: A Web Tool for Detecting Biases between Gene Evolution and Speciation in Fungi
Abstract
1. Introduction
2. Materials and Methods
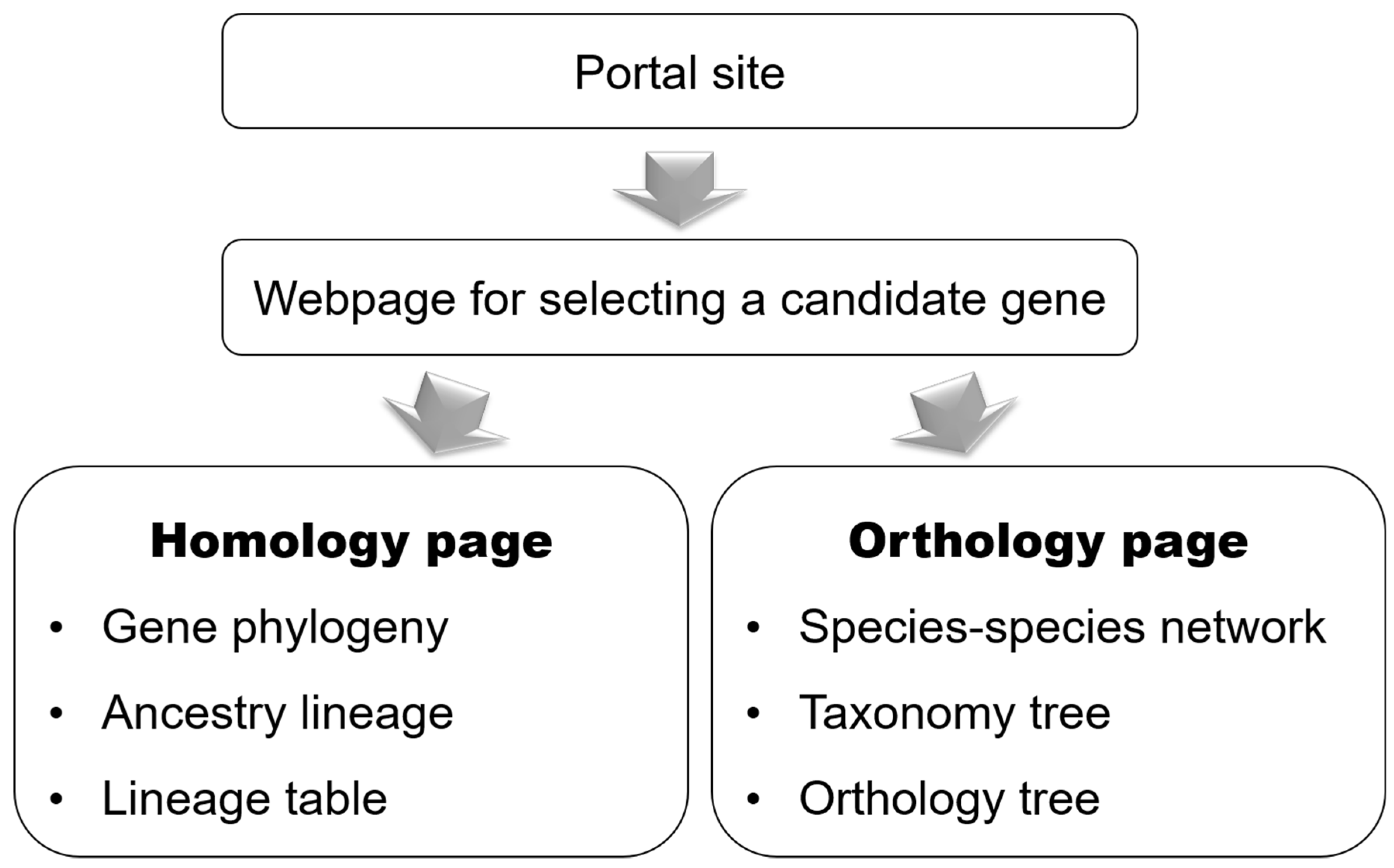
2.1. Schema
2.2. Data Source
2.3. Implementation
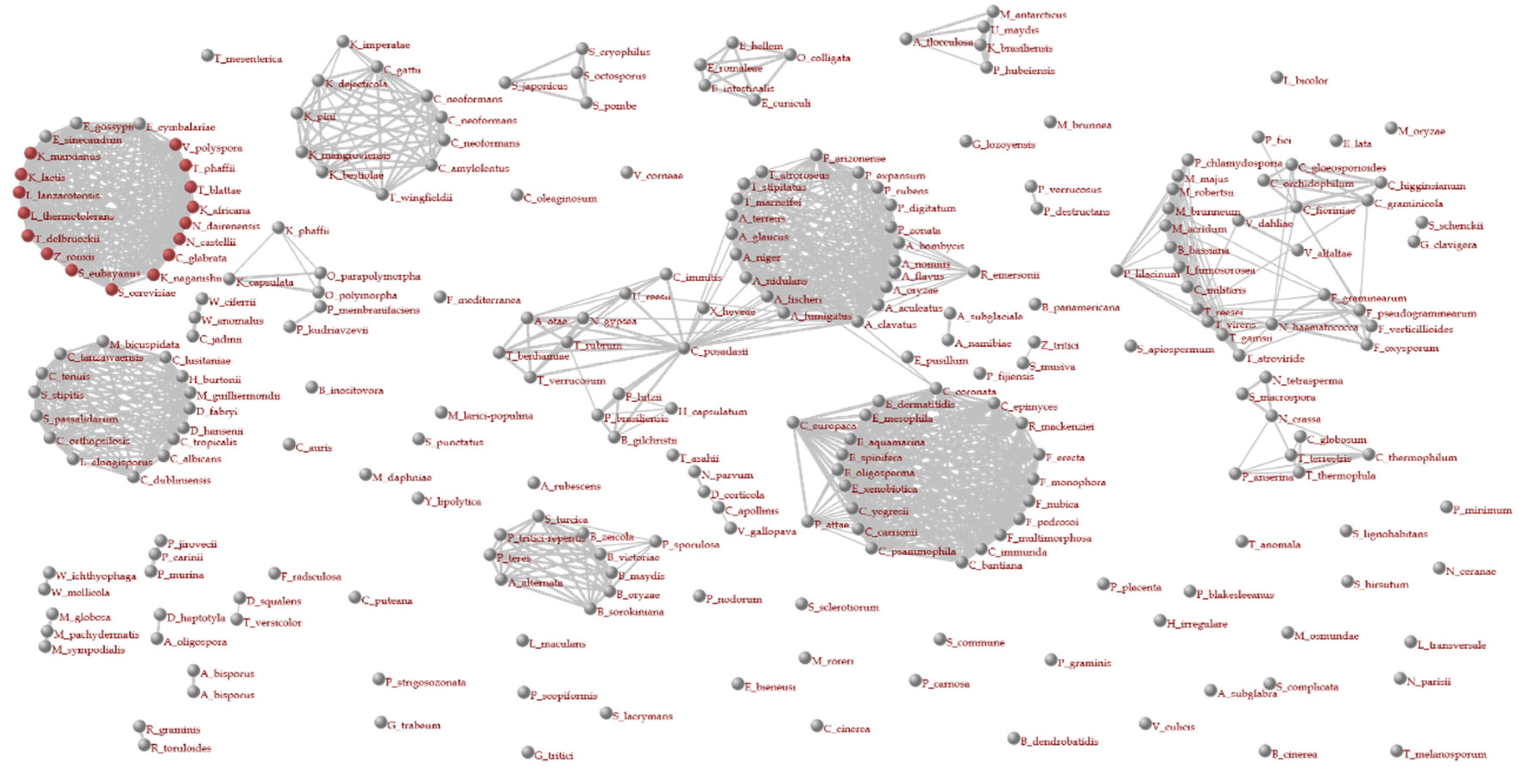
2.4. Detection of Homologous Gene Groups
2.5. Construction of a Phylogenetic Tree for Species (Taxonomy Tree)
2.6. Construction of a Phylogenetic Tree for Genes (Orthology Tree)
3. Results
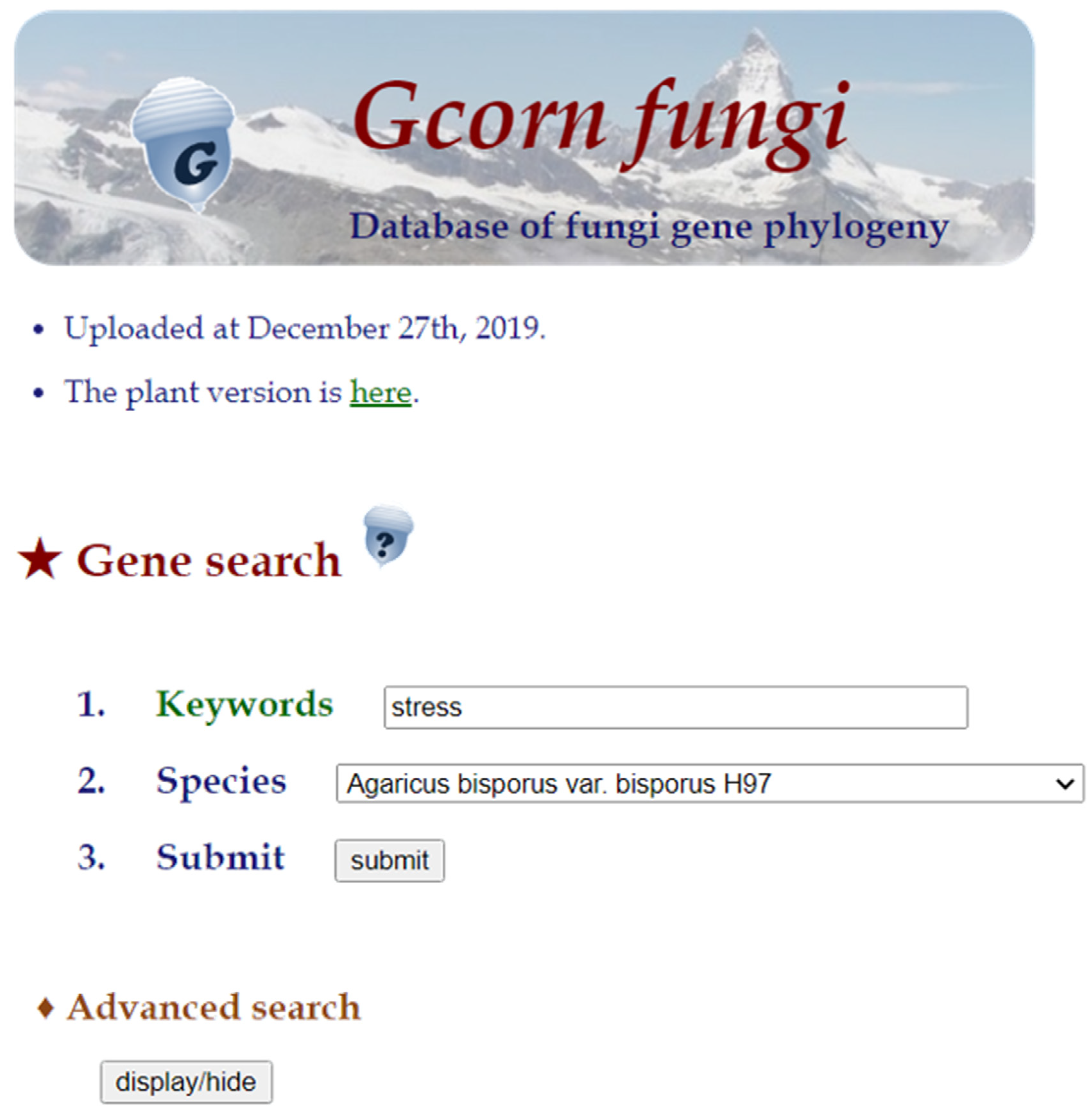
3.1. User Interface
3.2. Homology Page
3.3. Orthology Page
3.4. Other Downloadable Datasets
4. Discussion
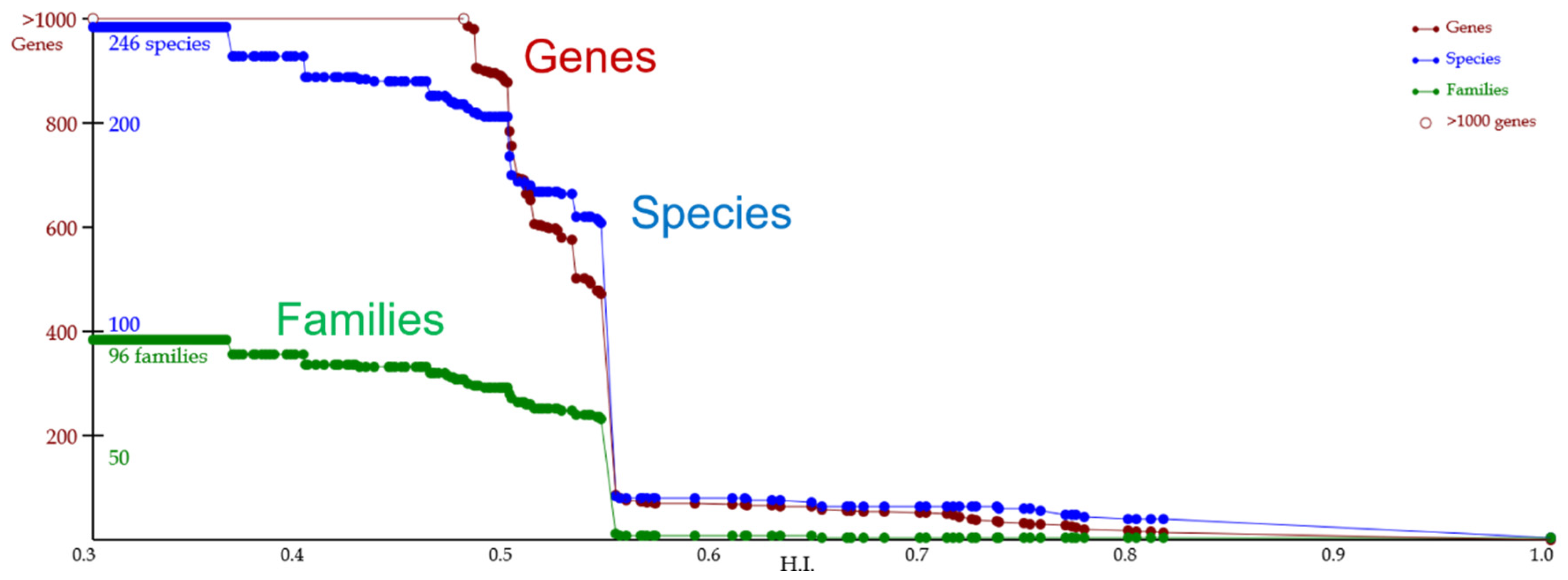
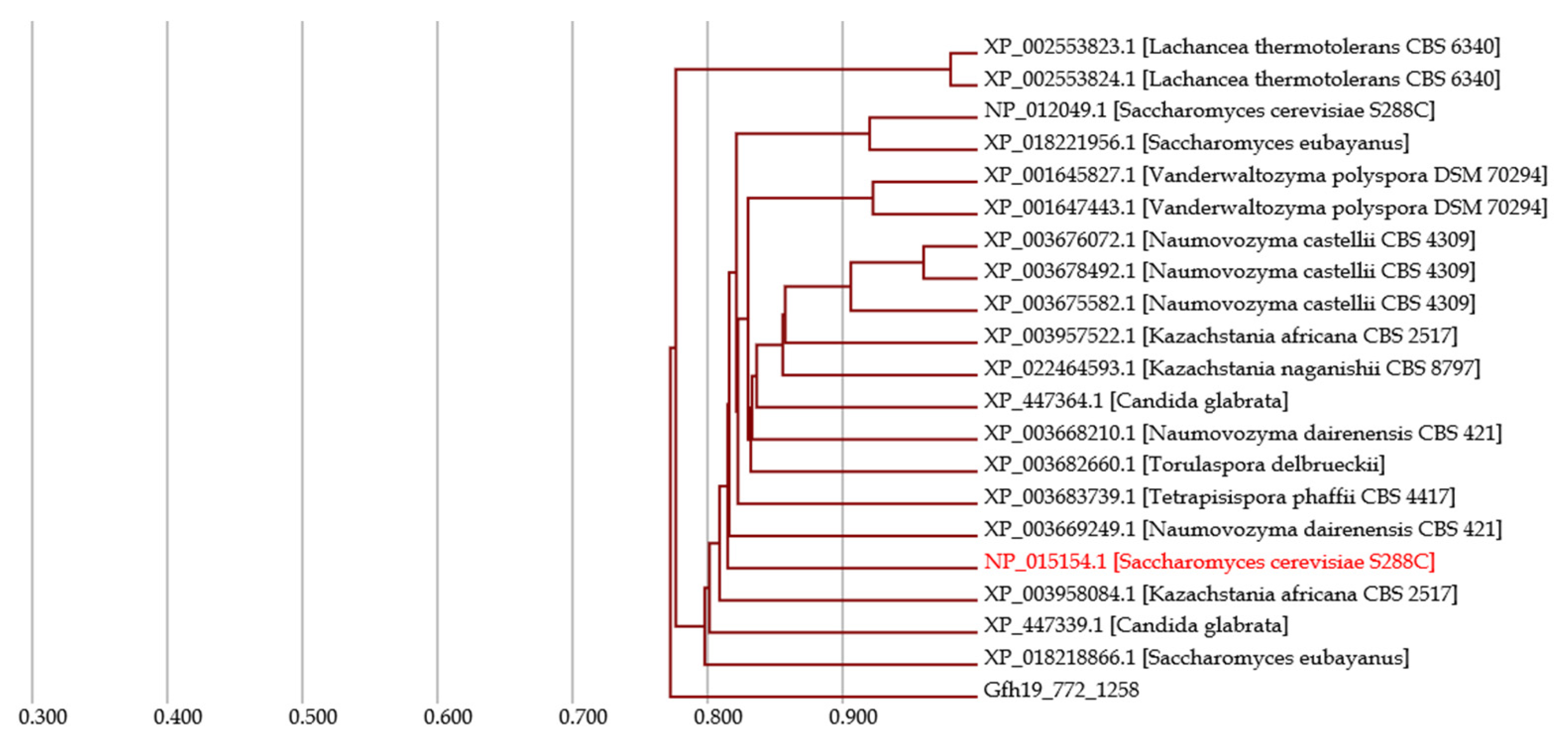
4.1. Case Study
- Homologous genes are detected throughout fungi.
- The gene sequence is well conserved in Saccharomycetaceae.
- Although there are some biases between the gene evolution and speciation of the gene of interest, evolution is generally consistent with speciation.
- The gene of interest has the potential to serve as a reference gene for DNA barcoding based on the above consistency.
4.2. Future Development
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Buckley, M. The Fungal Kingdom: Diverse and Essential Roles in Earth’s Ecosystem; American Society for Microbiology: Washington, DC, USA, 2008. [Google Scholar]
- Hawksworth, D.L.; Lücking, R. Fungal diversity revisited: 2.2 to 3.8 million species. Microbiol. Spectr. 2017, 5. [Google Scholar] [CrossRef]
- Giraud, T.; Refrégier, G.; Le Gac, M.; de Vienne, D.M.; Hood, M.E. Speciation in fungi. Fungal Genet. Biol. 2008, 45, 791–802. [Google Scholar] [CrossRef]
- Zhang, J. Evolution by gene duplication: An update. Trends Ecol. Evol. 2003, 18, 292–298. [Google Scholar] [CrossRef]
- Ochman, H.; Lawrence, J.G.; Groisman, E.A. Lateral gene transfer and the nature of bacterial innovation. Nature 2000, 405, 299–304. [Google Scholar] [CrossRef]
- Dunning Hotopp, J.C. Horizontal gene transfer between bacteria and animals. Trends Genet. 2011, 47, 157–163. [Google Scholar] [CrossRef]
- Robinson, K.M.; Sieber, K.B.; Dunning Hotopp, J.C. A review of bacteria-animal lateral gene transfer may inform our understanding of diseases like cancer. PLoS Genet. 2013, 9, e1003877. [Google Scholar] [CrossRef] [PubMed]
- Ambrosino, L.; Chiusano, M.L. Transcriptologs: A transcriptome-based approach to predict orthology relationships. Bioinfom. Biol. Insights 2017, 11, 1177932217600136. [Google Scholar] [CrossRef] [PubMed]
- Ullah, I.; Sjöstrand, J.; Andersson, P.; Sennblad, B.; Lagergren, J. Integrating sequence evolution into probabilistic orthology analysis. Syst. Biol. 2015, 64, 969–982. [Google Scholar] [CrossRef] [PubMed]
- The Angiosperm Phylogeny Group. An update of the Angiosperm Phylogeny Group classification for the orders and families of flowering plants: APG IV. Bot. J. Linn. Soc. 2016, 181, 1–20. [Google Scholar] [CrossRef]
- Ogata, Y.; Kimura, N.; Sano, R. Gcorn Plant: A database for retrieving functional and evolutionary traits of plant genes. Plant Physiol. 2019, 180, 732–742. [Google Scholar] [CrossRef]
- Yates, A.D.; Achuthan, P.; Akanni, W.; Allen, J.; Allen, J.; Alvarez-Jarreta, J.; Amode, M.R.; Armean, I.M.; Azov, A.G.; Bennett, R.; et al. Ensembl 2020. Nucleic Acids Res. 2020, 48, D682–D688. [Google Scholar] [CrossRef] [PubMed]
- Kriventseva, E.V.; Kuznetsov, D.; Tegenfeldt, F.; Manni, M.; Dias, R.; Simão, F.A.; Zdobnov, E.M. OrthoDB v10: Sampling the diversity of animal, plant, fungal, protist, bacterial and viral genomes for evolutionary and functional annotations of orthologs. Nucleic Acids Res. 2019, 47, D807–D811. [Google Scholar] [CrossRef]
- Correia, K.; Yu, S.M.; Mahadevan, R. AYbRAH: A curated ortholog database for yeasts and fungi spanning 600 million years of evolution. Database 2019, 2919, baz022. [Google Scholar] [CrossRef]
- Basenko, E.Y.; Pulman, J.A.; Shanmugasundram, A.; Harb, O.S.; Crouch, K.; Starns, D.; Warrenfeltz, S.; Aurrecoechea, C.; Stoeckert, C.J., Jr.; Kissinger, J.C.; et al. FungiDB: An integrated bioinformatic resource for fungi and oomycetes. J. Fungi. 2018, 4, 39. [Google Scholar] [CrossRef] [PubMed]
- Altschul, S.F.; Madden, T.L.; Schäffer, A.A.; Zhang, J.; Xhang, Z.; Miller, W.; Lipman, D.J. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 1997, 25, 3389–3402. [Google Scholar] [CrossRef] [PubMed]
- O’Leary, N.A.; Wright, M.W.; Brister, J.R.; Ciufo, S.; Haddad, D.; McVeigh, R.; Rajput, B.; Robbertse, B.; Smith-White, B.; Ako-Adjei, D.; et al. Reference sequence (RefSeq) database at NCBI: Current status, taxonomic expansion, and functional annotation. Nucleic Acids Res. 2016, 44, D733–D745. [Google Scholar] [CrossRef]
- Bansal, M.S.; Burleigh, J.G.; Eulenstein, O.; Fernández-Baca, D. Robinson-Foulds supertrees. Algorithms Mol. Biol. 2010, 5, 18. [Google Scholar] [CrossRef][Green Version]
- Federhen, S. The NCBI Taxonomy database. Nucleic Acids Res. 2012, 40, D136–D143. [Google Scholar] [CrossRef]
- Vohník, M.; Sadowsky, J.J.; Kohout, P.; Lhotáková, Z.; Nestby, R.; Kolařík, M. Novel root-fungus symbiosis in Ericaceae: Sheathed ericoid mycorrhiza formed by a hitherto undescribed basidiomycete with affinities to Trechisporales. PLoS ONE 2012, 7, e39524. [Google Scholar] [CrossRef] [PubMed]
- Woo, P.C.Y.; Ngan, A.H.Y.; Tsang, C.C.C.; Ling, I.W.H.; Chan, J.F.W.; Leung, S.Y.; Yuen, K.Y.; Lau, S.K.P. Clinical spectrum of exophiala infections and a novel Exophiala species, Exophiala hongkongensis. J. Clin. Microbiol. 2013, 51, 260–267. [Google Scholar] [CrossRef]
- Adhikary, S.; Cato, M.C.; McGary, K.L.; Rokas, A.; Eichman, B.F. Non-productive DNA damage binding by DNA glycosylase-like protein Mag2 from Schizosaccharomyces pombe. DNA Repair (Amst.) 2013, 12, 196–204. [Google Scholar] [CrossRef]
- Taverna, C.G.; Bosco-Borgeat, M.E.; Murisengo, O.A.; Davel, G.; Boité, M.C.; Cupolillo, E.; Canteros, C.E. Comparative analyses of classical phenotypic method and ribosomal RNA gene sequencing for identification of medically relevant Candida species. Mem. Inst. Oswaldo Cruz. 2013, 108, 178–185. [Google Scholar] [CrossRef]
- Ariyawansa, H.A.; Phookamsak, R.; Tibpromma, S.; Kang, J.C.; Hyde, K.D. A molecular and morphological reassessment of Diademaceae. Sci. World J. 2014, 2014, 675348. [Google Scholar] [CrossRef]
- Mardanov, A.V.; Beletsky, A.V.; Kadnikov, V.V.; Ignatov, A.N.; Ravin, N.V. The 203 kbp mitochondrial genome of the phytopathogenic fungus Sclerotinia borealis reveals multiple invasions of introns and genomic duplications. PLoS ONE 2014, 9, e107536. [Google Scholar] [CrossRef]
- Yilmaz, N.; Visagie, C.M.; Houbraken, J.; Frisvad, J.C.; Samson, R.A. Polyphasic taxonomy of the genus Talaromyces. Stud. Mycol. 2014, 78, 175–341. [Google Scholar] [CrossRef]
- Manamgoda, D.S.; Rossman, A.Y.; Castlebury, L.A.; Crous, P.W.; Madrid, H.; Chukeatirote, E.; Hyde, K.D. The genus Bipolaris. Stud. Mycol. 2014, 79, 221–288. [Google Scholar] [CrossRef] [PubMed]
- Réblová, M.; Fournier, J.; Štěpánek, V. Pisorisporiales, a new order of aquatic and terrestrial fungi for Achroceratosphaeria and Pisorisporium gen. nov. in the Sordariomycetes. Persoonia 2015, 34, 40–49. [Google Scholar] [CrossRef]
- Wang, L.; Groenewald, M.; Wang, Q.M.; Boekhout, T. Reclassification of Saccharomycodes sinensis, Proposal of Yueomyces sinensis gen. nov., comb. nov. within Saccharomycetaceae (Saccharomycetales, Saccharomycotina). PLoS ONE 2015, 10, e0136987. [Google Scholar] [CrossRef]
- Verma, S.; Gazara, R.K.; Nizam, S.; Parween, S.; Chattopadhyay, D.; Verma, P.K. Draft genome sequencing and secretome analysis of fungal phytopathogen Ascochyta rabiei provides insight into the necrotrophic effector repertoire. Sci. Rep. 2016, 6, 24638. [Google Scholar] [CrossRef] [PubMed]
- Riley, R.; Haridas, S.; Wolfe, K.H.; Lopes, M.R.; Hittinger, C.T.; Göker, M.; Salamov, A.A.; Wisecaver, J.H.; Long, T.M.; Calvey, C.H.; et al. Comparative genomics of biotechnologically important yeasts. Proc. Natl. Acad. Sci. USA 2016, 113, 9882–9887. [Google Scholar] [CrossRef] [PubMed]
- de Hoog, G.S.; Dukik, K.; Monod, M.; Packeu, A.; Stubbe, D.; Hendrickx, M.; Kupsch, C.; Stielow, J.B.; Freeke, J.; Göker, M.; et al. Toward a novel multilocus phylogenetic taxonomy for the Dermatophytes. Mycopathologia 2017, 182, 5–31. [Google Scholar] [CrossRef]
- Moreno, L.F.; Feng, P.; Weiss, V.A.; Vicente, V.A.; Stielow, J.B.; de Hoog, S. Phylogenomic analyses reveal the diversity of laccase-coding genes in Fonsecaea genomes. PLoS ONE 2017, 12, e0171291. [Google Scholar] [CrossRef]
- Tett, A.; Pasolli, E.; Farina, S.; Truong, D.T.; Asnicar, F.; Zolfo, M.; Beghini, F.; Armanini, F.; Jousson, O.; De Sanctis, V.; et al. Unexplored diversity and strain-level structure of the skin microbiome associated with psoriasis. NPJ Biofilms Microbiomes 2017, 3, 14. [Google Scholar] [CrossRef] [PubMed]
- Réblová, M.; Miller, A.N.; Réblová, K.; Štěpánek, V. Phylogenetic classification and generic delineation of Calyptosphaeria gen. nov., Lentomitella, Spadicoides and Torrentispora (Sordariomycetes). Stud. Mycol. 2018, 89, 1–62. [Google Scholar] [CrossRef]
- Persinoti, G.F.; Martinez, D.A.; Li, W.; Döğen, A.; Billmyre, R.B.; Averette, A.; Goldberg, J.M.; Shea, T.; Young, S.; Zeng, Q.; et al. Whole-Genome Analysis illustrates global clonal population structure of the ubiquitous dermatophyte pathogen Trichophyton Rubrum. Genetics 2018, 208, 1657–1669. [Google Scholar] [CrossRef] [PubMed]
- Liang, X.; Wang, B.; Dong, Q.; Li, L.; Rollins, J.A.; Zhang, R.; Sun, G. Pathogenic adaptations of Colletotrichum fungi revealed by genome wide gene family evolutionary analyses. PLoS ONE 2018, 13, e0196303. [Google Scholar] [CrossRef] [PubMed]
- Ma, L.; Cissé, O.H.; Kovacs, J.A. A Molecular Window into the Biology and Epidemiology of Pneumocystis spp. Clin. Microbiol. Rev. 2018, 31, e00009-18. [Google Scholar] [CrossRef] [PubMed]
- Tsang, C.C.; Tang, J.Y.M.; Lau, S.K.P.; Woo, P.C.Y. Taxonomy and evolution of Aspergillus, Penicillium and Talaromyces in the omics era—Past, present and future. Comput. Struct. Biotechnol. J. 2018, 16, 197–210. [Google Scholar] [CrossRef]
- Coelho, R.A.; Brito-Santos, F.; Figueiredo-Carvalho, M.H.G.; Silva, J.V.D.S.; Gutierrez-Galhardo, M.C.; do Valle, A.C.F.; Zancopé-Oliveira, R.M.; Trilles, L.; Meyer, W.; Freitas, D.F.S.; et al. Molecular identification and antifungal susceptibility profiles of clinical strains of Fonsecaea spp. isolated from patients with chromoblastomycosis in Rio de Janeiro, Brazil. PLoS Negl. Trop. Dis. 2018, 12, e0006675. [Google Scholar] [CrossRef]
- Xie, T.; Wang, Y.; Yu, D.; Zhang, Q.; Zhang, T.; Wang, Z.; Huang, B. MrSVP, a secreted virulence-associated protein, contributes to thermotolerance and virulence of the entomopathogenic fungus Metarhizium robertsii. BMC Microbiol. 2019, 19, 25. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef] [PubMed]
- Larkin, M.A.; Blackshields, G.; Brown, N.P.; Chenna, R.; McGettigan, P.A.; McWilliam, H.; Valentin, F.; Wallace, I.M.; Wilm, A.; Lopez, R.; et al. Clustal W and Clustal X version 2.0. Bioinformatics 2007, 23, 2947–2948. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kawachi, T.; Inuki, Y.; Ogata, Y. Gcorn fungi: A Web Tool for Detecting Biases between Gene Evolution and Speciation in Fungi. J. Fungi 2021, 7, 959. https://doi.org/10.3390/jof7110959
Kawachi T, Inuki Y, Ogata Y. Gcorn fungi: A Web Tool for Detecting Biases between Gene Evolution and Speciation in Fungi. Journal of Fungi. 2021; 7(11):959. https://doi.org/10.3390/jof7110959
Chicago/Turabian StyleKawachi, Taiga, Yuta Inuki, and Yoshiyuki Ogata. 2021. "Gcorn fungi: A Web Tool for Detecting Biases between Gene Evolution and Speciation in Fungi" Journal of Fungi 7, no. 11: 959. https://doi.org/10.3390/jof7110959
APA StyleKawachi, T., Inuki, Y., & Ogata, Y. (2021). Gcorn fungi: A Web Tool for Detecting Biases between Gene Evolution and Speciation in Fungi. Journal of Fungi, 7(11), 959. https://doi.org/10.3390/jof7110959






