The Cell Wall Integrity Receptor Mtl1 Contributes to Articulate Autophagic Responses When Glucose Availability Is Compromised
Abstract
1. Introduction
2. Materials and Methods
2.1. Yeast Strains and Plasmids
2.2. Media, Growth Conditions and Reagents
2.3. Vacuole and Dihydroethidium Staining
2.4. Cell Survival and Chronological Life Span
2.5. Protein Extraction and Immunoblot Analyses
2.6. Autophagy Detection
2.7. Glucose Determination
2.8. Statistical Analysis
3. Results
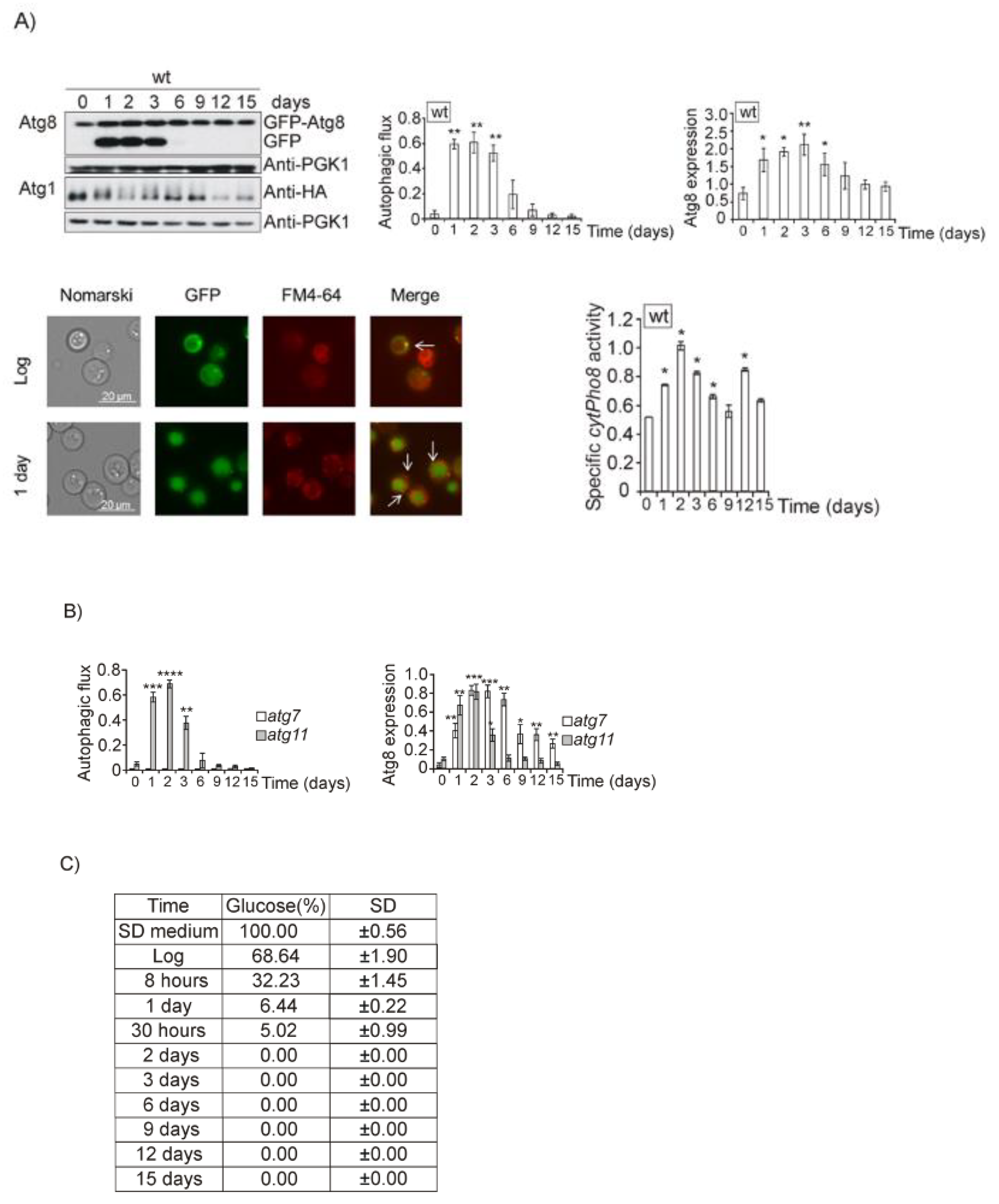
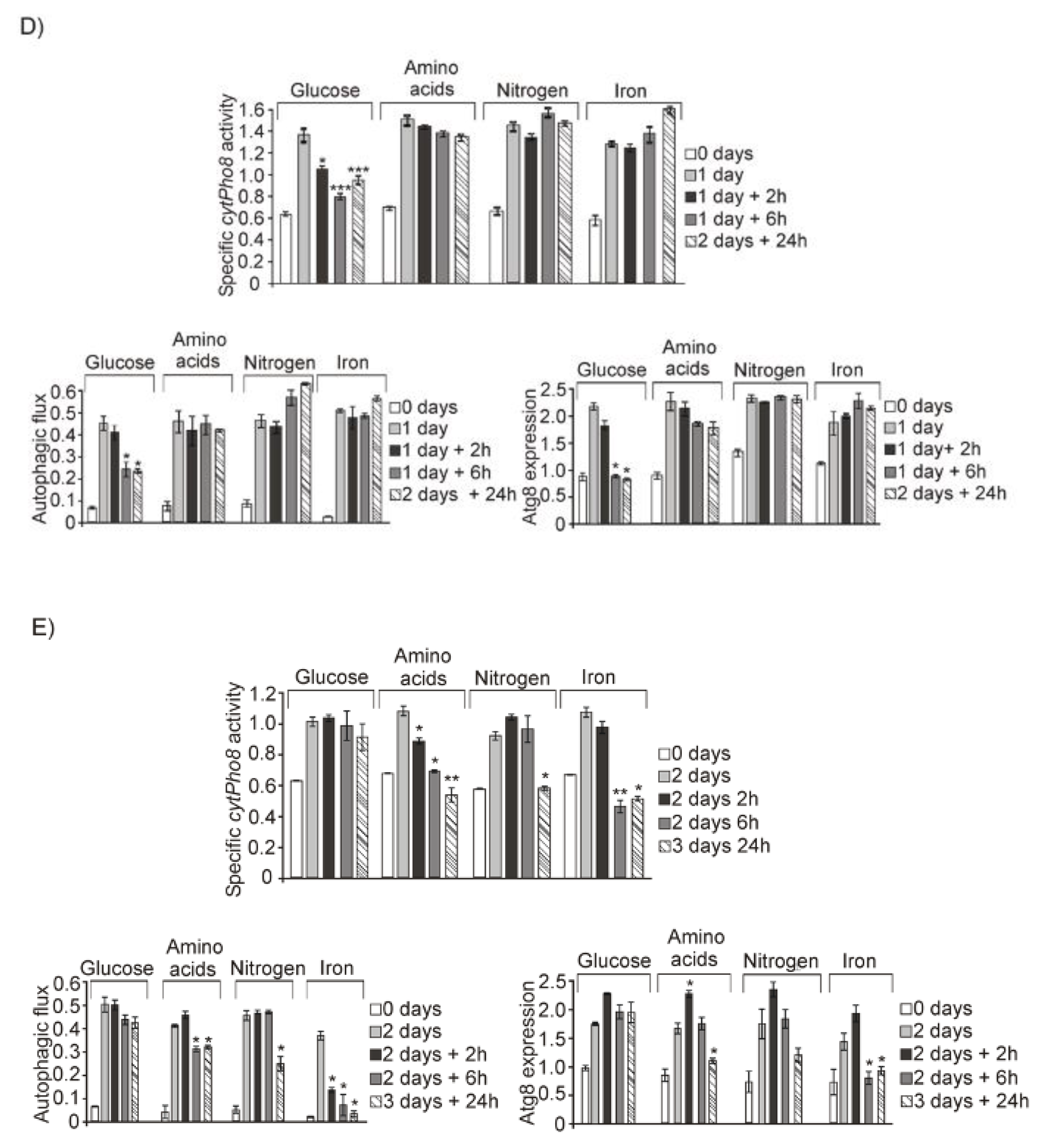
3.1. Glucose, Amino Acids, Nitrogen and Iron Deprivation Determine the Induction of Bulk Autophagy during Diauxic Transition
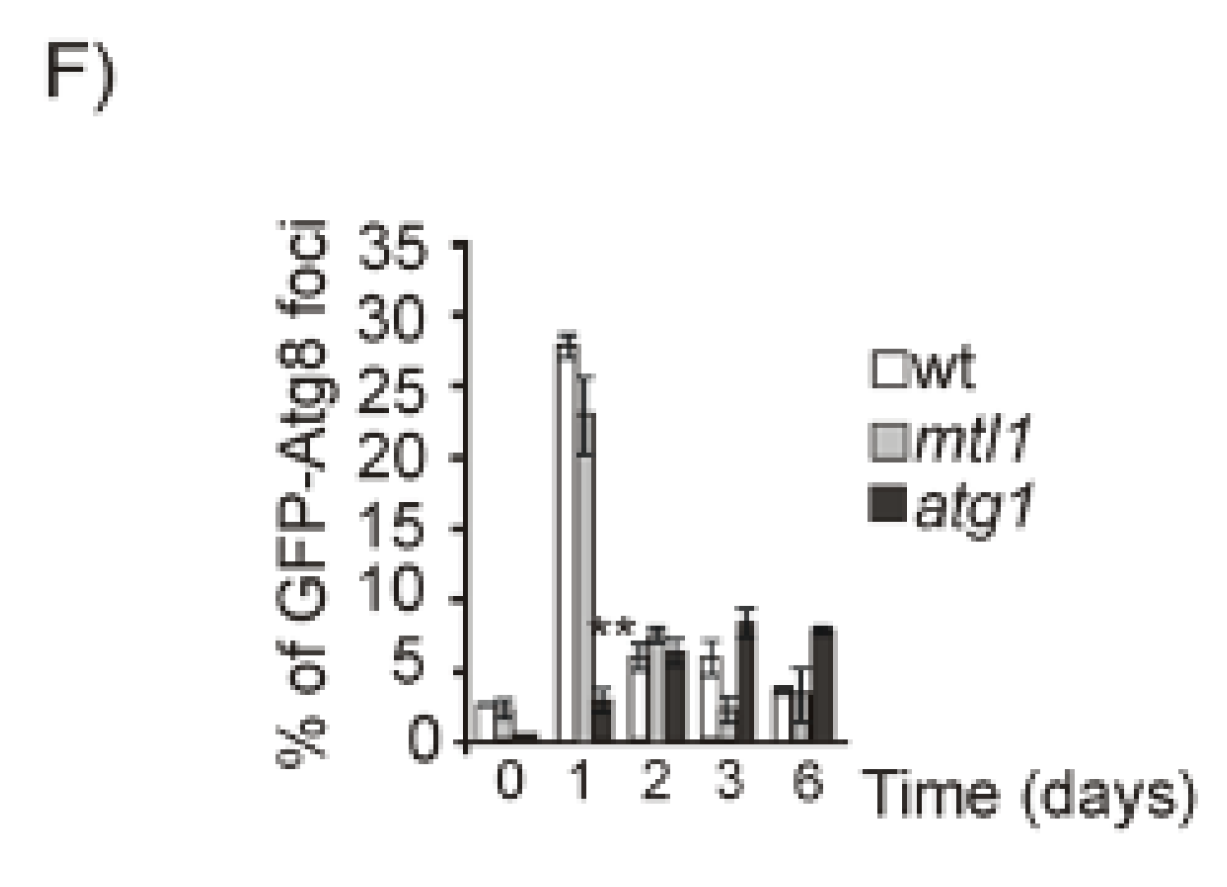
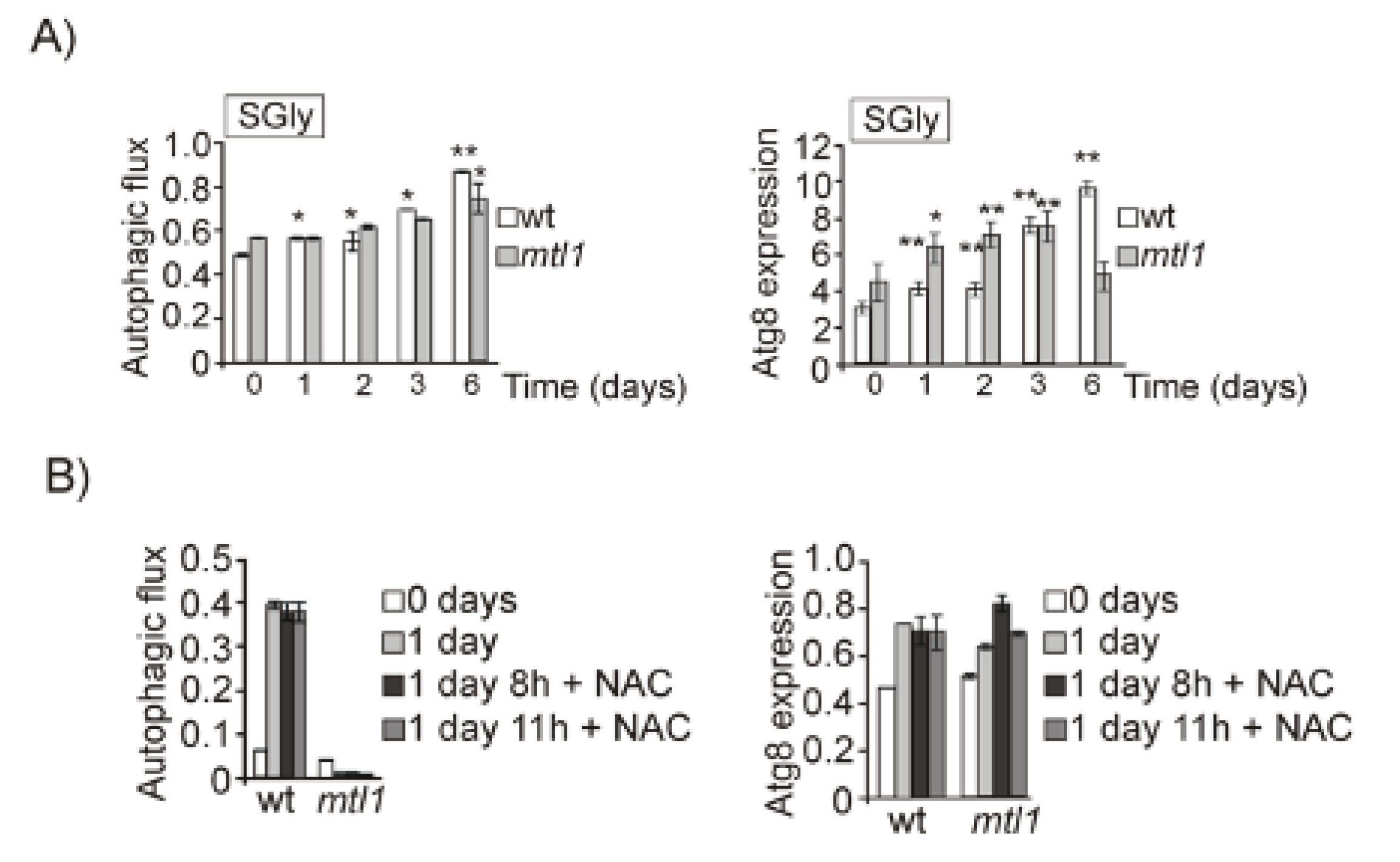
3.2. Both Mtl1 and Gcn2 Control Autophagy Induction during Diauxic Transition
3.3. Mtl1 CWI Cell Wall Receptor Signals Glucose Concentration to the Autophagy Machinery in a Manner Partly Dependent on Intracellular ATP Levels
3.4. Both Ras2 or Sch9 Suppress mtl1 Deficiency in Bulk Autophagy Activation in All Metabolic Conditions That Imply Reduced Glucose Levels
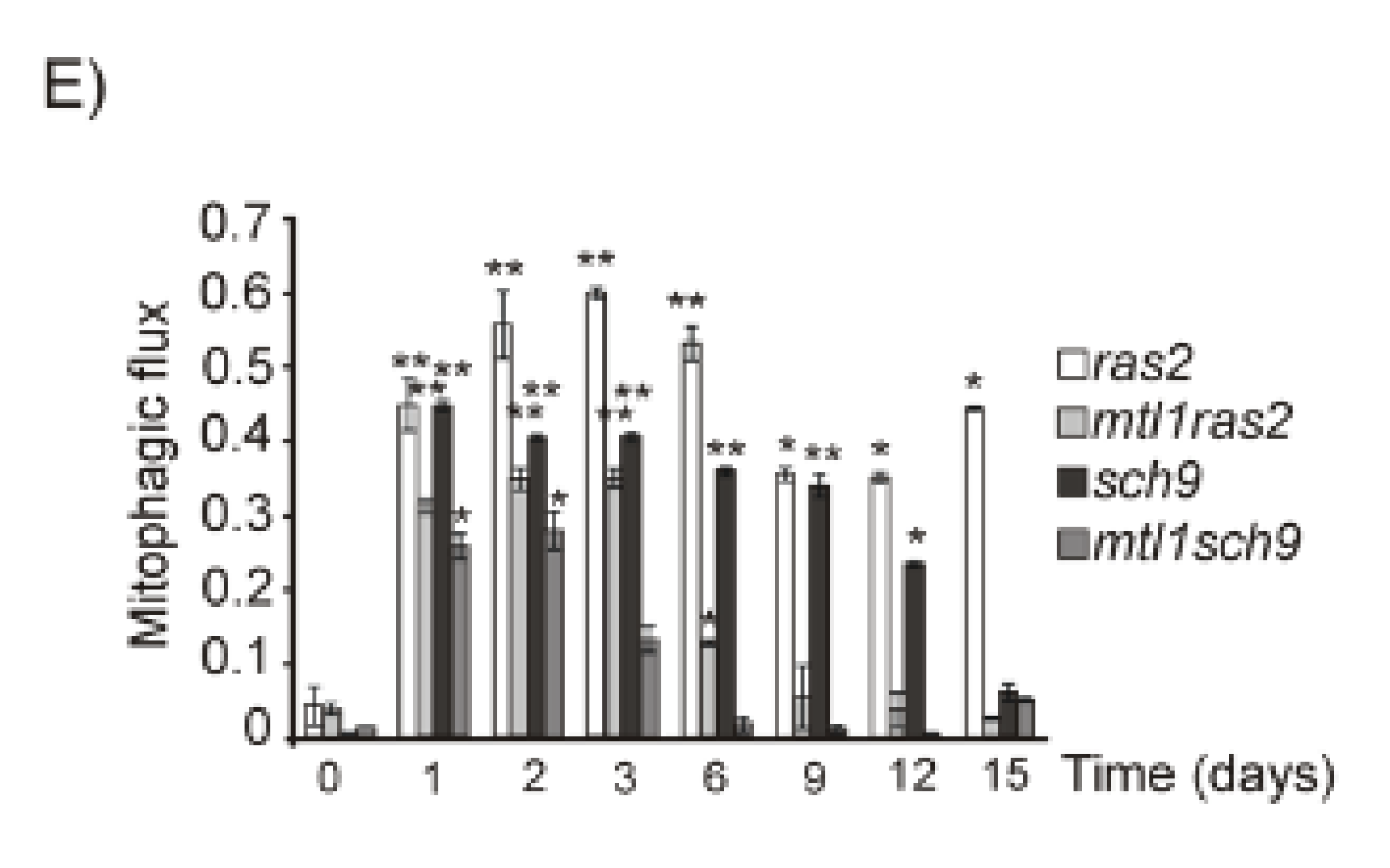
3.5. Mtl1 Is Required for Mitochondrial Degradation Dependent on Atg33 and Independent of Atg32 during Chronological Ageing
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- de Virgilio, C. The essence of yeast quiescence. FEMS Microbiol. Rev. 2012, 36, 306–339. [Google Scholar] [CrossRef]
- Hughes Hallett, J.E.; Luo, X.; Capaldi, A.P. State transitions in the TORC1 signaling pathway and information processing in Saccharomyces cerevisiae. Genetics 2014, 198, 773–786. [Google Scholar] [CrossRef] [PubMed]
- Picazo, C.; Orozco, H.; Matallana, E.; Aranda, A. Interplay among Gcn5, Sch9 and mitochondria during chronological aging of wine yeast is dependent on growth conditions. PLoS ONE 2015, 10, e0117267. [Google Scholar] [CrossRef] [PubMed]
- Santos, J.; Leão, C.; Sousa, M.J. Ammonium-dependent shortening of CLS in yeast cells starved for essential amino acids is determined by the specific amino acid deprived, through different signaling pathways. Oxid. Med. Cell. Longev. 2013, 2013, 161986. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Wu, J.; Huang, D. New arahypins isolated from fungal-challenged peanut seeds and their glucose uptake-stimulatory activity in 3T3-L1 adipocytes. Phytochem. Lett. 2013, 6, 123–127. [Google Scholar] [CrossRef]
- Fabrizio, P.; Pozza, F.; Pletcher, S.D.; Gendron, C.M.; Longo, V.D. Regulation of longevity and stress resistance by Sch9 in yeast. Science 2001, 292, 288–290. [Google Scholar] [CrossRef] [PubMed]
- Longo, V.D. The Ras and Sch9 pathways regulate stress resistance and longevity. Exp. Gerontol. 2003, 38, 807–811. [Google Scholar] [CrossRef]
- Kaeberlein, M.; Powers, R.W.; Steffen, K.K.; Westman, E.A.; Hu, D.; Dang, N.; Kerr, E.O.; Kirkland, K.T.; Fields, S.; Kennedy, B.K.; et al. Regulation of Yeast Replicative Life Span by TOR and Sch9 in Response to Nutrients. Science 2005, 310, 1193–1196. [Google Scholar] [CrossRef] [PubMed]
- Yan, G.; Lai, Y.; Jiang, Y. The TOR Complex 1 Is a Direct Target of Rho1 GTPase. Mol. Cell 2012, 45, 743–753. [Google Scholar] [CrossRef] [PubMed]
- Thomas, J.D.; Zhang, Y.J.; Wei, Y.H.; Cho, J.H.; Morris, L.E.; Wang, H.Y.; Zheng, X.S. Rab1A Is an mTORC1 Activator and a Colorectal Oncogene. Cancer Cell 2014, 26, 754–769. [Google Scholar] [CrossRef] [PubMed]
- Yuan, W.; Guo, S.; Gao, J.; Zhong, M.; Yan, G.; Wu, W.; Chao, Y.; Jiang, Y. General control nonderepressible 2 (GCN2) kinase inhibits target of rapamycin complex 1 in response to amino acid starvation in Saccharomyces cerevisiae. J. Biol. Chem. 2017, 292, 2660–2669. [Google Scholar] [CrossRef]
- Postnikoff, S.D.L.; Johnson, J.E.; Tyler, J.K. The integrated stress response in budding yeast lifespan extension. Microb. Cell 2017, 4, 368–375. [Google Scholar] [CrossRef] [PubMed]
- Stephan, J.S.; Yeh, Y.-Y.; Ramachandran, V.; Deminoff, S.J.; Herman, P.K. The Tor and PKA signaling pathways independently target the Atg1/Atg13 protein kinase complex to control autophagy. Proc. Natl. Acad. Sci. USA 2009, 106, 17049–17054. [Google Scholar] [CrossRef] [PubMed]
- Kamada, Y.; Funakoshi, T.; Shintani, T.; Nagano, K.; Ohsumi, M.; Ohsumi, Y. Tor-mediated induction of autophagy via an Apg1 protein kinase complex. J. Cell Biol. 2000, 150, 1507–1513. [Google Scholar] [CrossRef]
- Kim, Y.C.; Guan, K. mTOR: A pharmacologic target for autophagy regulation. J. Clin. Investig. 2015, 2, 25–32. [Google Scholar] [CrossRef] [PubMed]
- Budovskaya, Y.V.; Stephan, J.S.; Reggiori, F.; Klionsky, D.J.; Herman, P.K. The Ras/cAMP-dependent Protein Kinase Signaling Pathway Regulates an Early Step of the Autophagy Process in Saccharomyces cerevisiae. J. Biol. Chem. 2004, 279, 20663–20671. [Google Scholar] [CrossRef]
- Yorimitsu, T.; Zaman, S.; Broach, J.R.; Klionsky, D.J. Protein Kinase A and Sch9 Cooperatively Regulate Induction of Autophagy in Saccharomyces cerevisiae. Mol. Biol. Cell 2007, 18, 4180–4189. [Google Scholar] [CrossRef] [PubMed]
- Conrad, M.; Schothorst, J.; Kankipati, H.N.; Van Zeebroeck, G.; Rubio-Texeira, M.; Thevelein, J.M. Nutrient sensing and signaling in the yeast Saccharomyces cerevisiae. FEMS Microbiol. Rev. 2014, 38, 254–299. [Google Scholar] [CrossRef]
- Cazzanelli, G.; Pereira, F.; Alves, S.; Francisco, R.; Azevedo, L.; Dias Carvalho, P.; Almeida, A.; Côte-Real, M.; Oliveira, M.J.; Lucas, C.; et al. The Yeast Saccharomyces cerevisiae as a Model for Understanding RAS Proteins and their Role in Human Tumorigenesis. Cells 2018, 7, 14. [Google Scholar] [CrossRef]
- Celenza, J.L.; Carlson, M. A yeast gene that is essential for release from glucose repression encodes a protein kinase. Science 1986, 233, 1175–1180. [Google Scholar] [CrossRef] [PubMed]
- Shashkova, S.; Welkenhuysen, N.; Hohmann, S. Molecular communication: Crosstalk between the Snf1 and other signaling pathways. FEMS Yeast Res. 2015, 15, fov026. [Google Scholar] [CrossRef]
- Wang, Z.; Wilson, W.; Fujino, M.; Roach, P.J. Antagonistic controls of autophagy and glycogen accumulation by Snf1p, the yeast homolog of AMP-activated protein kinase, and the cyclin-dependent kinase Pho85p. Mol. Cell. Biol. 2001, 21, 5742–5752. [Google Scholar] [CrossRef] [PubMed]
- Montella-Manuel, S.; Pujol-Carrion, N.; Mechoud, M.A.; de la Torre-Ruiz, M.A. Bulk autophagy induction and life extension is achieved when iron is the only limited nutrient in Saccharomyces cerevisiae. Biochem. J. 2021, 478, 811–837. [Google Scholar] [CrossRef]
- Petkova, M.I.; Pujol-Carrion, N.; de la Torre-Ruiz, M.A. Mtl1 O-mannosylation mediated by both Pmt1 and Pmt2 is important for cell survival under oxidative conditions and TOR blockade. Fungal Genet. Biol. 2012, 49, 903–914. [Google Scholar] [CrossRef] [PubMed]
- Kock, C.; Dufrêne, Y.F.; Heinisch, J.J. Up against the wall: Is yeast cell wall integrity ensured by mechanosensing in plasma membrane microdomains? Appl. Environ. Microbiol. 2015, 81, 806–811. [Google Scholar] [CrossRef] [PubMed]
- Rajavel, M.; Philip, B.; Buehrer, B.M.; Errede, B.; Levin, D.E. Mid2 Is a Putative Sensor for Cell Integrity Signaling in. Mol. Cell. Biol. 1999, 19, 3969–3976. [Google Scholar] [CrossRef]
- Vilella, F.; Herrero, E.; Torres, J.; De La Torre-Ruiz, M.A. Pkc1 and the upstream elements of the cell integrity pathway in Saccharomyces cerevisiae, Rom2 and Mtl1, are required for cellular responses to oxidative stress. J. Biol. Chem. 2005, 280, 9149–9159. [Google Scholar] [CrossRef]
- Petkova, M.I.; Pujol-Carrion, N.; Arroyo, J.; García-Cantalejo, J.; De La Torre-Ruiz, M.A. Mtl1 is required to activate general stress response through TOR1 and RAS2 inhibition under conditions of glucose starvation and oxidative stress. J. Biol. Chem. 2010, 285, 19521–19531. [Google Scholar] [CrossRef] [PubMed]
- Jin, C.; Parshin, A.V.; Daly, I.; Strich, R.; Cooper, K.F. The cell wall sensors Mtl1, Wsc1, and Mid2 are required for stress-induced nuclear to cytoplasmic translocation of cyclin C and programmed cell death in yeast. Oxid. Med. Cell. Longev. 2013, 2013, 320823. [Google Scholar] [CrossRef] [PubMed]
- Sundaram, V.; Petkova, M.I.; Pujol-Carrion, N.; Boada, J.; de la Torre-Ruiz, M.A. Tor1, Sch9 and PKA downregulation in quiescence rely on Mtl1 to preserve mitochondrial integrity and cell survival. Mol. Microbiol. 2015, 97, 93–109. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Levin, D.E. Regulation of cell wall biogenesis in Saccharomyces cerevisiae: The cell wall integrity signaling pathway. Genetics 2011, 189, 1145–1175. [Google Scholar] [CrossRef] [PubMed]
- de la Torre-Ruiz, M.A.; Pujol, N.; Sundaram, V. Coping with oxidative stress. Curr. Drug Targets 2015, 16, 2–12. [Google Scholar] [CrossRef]
- Staleva, L.; Hall, A.; Orlow, S.J. Oxidative Stress Activates FUS1 and RLM1 Transcription in the Yeast Saccharomyces cerevisiae in an Oxidant-dependent Manner. Mol. Biol. Cell 2004, 15, 5574–5582. [Google Scholar] [CrossRef]
- Jendretzki, A.; Wittland, J.; Wilk, S.; Straede, A.; Heinisch, J.J. How do I begin? Sensing extracellular stress to maintain yeast cell wall integrity. Eur. J. Cell Biol. 2011, 90, 740–744. [Google Scholar] [CrossRef]
- Yang, Z.; Klionsky, D.J. An overview of the molecular mechanism of autophagy. Curr. Top. Microbiol. Immunol. 2009, 335, 1–32. [Google Scholar] [CrossRef]
- Kawamata, T.; Horie, T.; Matsunami, M.; Sasaki, M.; Ohsumi, Y. Zinc starvation induces autophagy in yeast. J. Biol. Chem. 2017, 292, 8520–8530. [Google Scholar] [CrossRef] [PubMed]
- Yokota, H.; Gomi, K.; Shintani, T. Induction of autophagy by phosphate starvation in an Atg11-dependent manner in Saccharomyces cerevisiae. Biochem. Biophys. Res. Commun. 2017, 483, 522–527. [Google Scholar] [CrossRef]
- Nakatogawa, H.; Suzuki, K.; Kamada, Y.; Ohsumi, Y. Dynamics and diversity in autophagy mechanisms: Lessons from yeast. Nat. Rev. Mol. Cell Biol. 2009, 10, 458–467. [Google Scholar] [CrossRef] [PubMed]
- Yorimitsu, T.; Ke Wang, C.H.; Klionsky, D.J. Tap42-associated protein phosphatase type 2A negatively regulates induction of autophagy. Autophagy 2009, 5, 616–624. [Google Scholar] [CrossRef] [PubMed]
- Fujioka, Y.; Suzuki, S.W.; Yamamoto, H.; Kondo-Kakuta, C.; Kimura, Y.; Hirano, H.; Akada, R.; Inagaki, F.; Ohsumi, Y.; Noda, N.N. Structural basis of starvation-induced assembly of the autophagy initiation complex. Nat. Struct. Mol. Biol. 2014, 21, 513–521. [Google Scholar] [CrossRef]
- Yamamoto, H.; Fujioka, Y.; Suzuki, S.W.; Noshiro, D.; Suzuki, H.; Kondo-Kakuta, C.; Kimura, Y.; Hirano, H.; Ando, T.; Noda, N.N.; et al. The Intrinsically Disordered Protein Atg13 Mediates Supramolecular Assembly of Autophagy Initiation Complexes. Dev. Cell 2016, 38, 86–99. [Google Scholar] [CrossRef] [PubMed]
- Ohsumi, Y. Historical landmarks of autophagy research. Cell Res. 2014, 24, 9–23. [Google Scholar] [CrossRef]
- Feng, Y.; He, D.; Yao, Z.; Klionsky, D.J. The machinery of macroautophagy. Cell Res. 2014, 24, 24–41. [Google Scholar] [CrossRef]
- Powers, R.W.; Kaeberlein, M.; Caldwell, S.D.; Kennedy, B.K.; Fields, S. Extension of chronological life span in yeast by decreased TOR pathway signaling. Genes Dev. 2006, 20, 174–184. [Google Scholar] [CrossRef]
- Fabrizio, P.; Hoon, S.; Shamalnasab, M.; Galbani, A.; Wei, M.; Giaever, G.; Nislow, C.; Longo, V.D. Genome-wide screen in Saccharomyces cerevisiae identifies vacuolar protein sorting, autophagy, biosynthetic, and tRNA methylation genes involved in life span regulation. PLoS Genet. 2010, 6, e1001024. [Google Scholar] [CrossRef]
- Gatica, D.; Lahiri, V.; Klionsky, D.J. Cargo Recognition and Degradation by Selective Autophagy. Nat. Cell Biomol. 2018, 20, 233–242. [Google Scholar] [CrossRef] [PubMed]
- Mamaev, D.V.; Zvyagilskaya, R.A. Mitophagy in Yeast. Biochemistry 2019, 84, 225–232. [Google Scholar] [CrossRef] [PubMed]
- Fukuda, T.; Kanki, T. Mechanisms and physiological roles of mitophagy in yeast. Mol. Cells 2018, 41, 35–44. [Google Scholar] [CrossRef] [PubMed]
- Welter, E.; Montino, M.; Reinhold, R.; Schlotterhose, P.; Krick, R.; Dudek, J.; Rehling, P.; Thumm, M. Uth1 is a mitochondrial inner membrane protein dispensable for post-log-phase and rapamycin-induced mitophagy. FEBS J. 2013, 280, 4970–4982. [Google Scholar] [CrossRef]
- Wach, A.; Brachat, A.; Pöhlmann, R.; Philippsen, P. New heterologous modules for classical or PCR-based gene disruptions in Saccharomyces cerevisiae. Yeast 1994, 10, 1793–1808. [Google Scholar] [CrossRef]
- Pujol-Carrion, N.; Belli, G.; Herrero, E.; Nogues, A.; de la Torre-Ruiz, M.A. Glutaredoxins Grx3 and Grx4 regulate nuclear localisation of Aft1 and the oxidative stress response in Saccharomyces cerevisiae. J. Cell Sci. 2006, 119, 4554–4564. [Google Scholar] [CrossRef] [PubMed]
- Gallego, C.; Eloi, G.; Colomina, N.; Enrique, H.; Aldea, M. The Cln3 cyclin is down-regulated by translational repression and degradation during the G1 arrest caused by nitrogen deprivation in budding yeast. EMBO J. 1997, 16, 7196–7206. [Google Scholar] [CrossRef] [PubMed]
- Petkova, M.I.; Pujol-Carrion, N.; de la Torre-Ruiz, M.A. Signal flow between CWI/TOR and CWI/RAS in budding yeast under conditions of oxidative stress and glucose starvation. Commun. Integr. Biol. 2010, 3, 555–557. [Google Scholar] [CrossRef] [PubMed]
- Guedes, A.; Ludovico, P.; Sampaio-Marques, B. Caloric restriction alleviates alpha-synuclein toxicity in aged yeast cells by controlling the opposite roles of Tor1 and Sir2 on autophagy. Mech. Ageing Dev. 2016, 161, 270–276. [Google Scholar] [CrossRef] [PubMed]
- Ecker, N.; Mor, A.; Journo, D.; Abeliovich, H.; Ecker, N.; Mor, A. Deprivation is distinct from nitrogen starvation- induced macroautophagy Induction of autophagic flux by amino acid deprivation is distinct from nitrogen starvation-induced macroautophagy. Autophagy 2010, 6, 870–890. [Google Scholar] [CrossRef] [PubMed]
- Görner, W.; Durchschlag, E.; Martinez-Pastor, M.T.; Estruch, F.; Ammerer, G.; Hamilton, B.; Ruis, H.; Schüller, C. Nuclear localization of the C2H2 zinc finger protein Msn2p is regulated by stress and protein kinase A activity. Genes Dev. 1998, 12, 586–597. [Google Scholar] [CrossRef]
- Komeili, A.; Wedaman, K.P.; O’Shea, E.K.; Powers, T. Mechanism of metabolic control: Target of rapamycin signaling links nitrogen quality to the activity of the Rtg1 and Rtg3 transcription factors. J. Cell Biol. 2000, 151, 863–878. [Google Scholar] [CrossRef] [PubMed]
- Hagai, A.; Zareic, M.; Kristoffer, T.G.R.; Richard, J.Y.; Joern, D. Involvement of mitochondrial dynamics in the segregation of mitochondrial matrix proteins during stationary phase mitophagy. Physiol. Behav. 2017, 176, 100–106. [Google Scholar] [CrossRef]
- Kaiser, C.; Michaelis, S.; Mitchell, A. Methods in Yeast Genetics; Cold Spring Harbour Laboratory Press Ltd.: New York, NY, USA, 1994; pp. 107–121. ISBN 0-87969-451-3. [Google Scholar]
- Pujol-Carrion, N.; Petkova, M.I.; Serrano, L.; de la Torre-Ruiz, M.A. The MAP kinase Slt2 is involved in vacuolar function and actin remodeling in Saccharomyces cerevisiae mutants affected by endogenous oxidative stress. Appl. Environ. Microbiol. 2013, 79, 6459–6471. [Google Scholar] [CrossRef] [PubMed]
- Mechoud, M.A.; Pujol-Carrion, N.; Montella-Manuel, S.; De La Torre-Ruiz, M.A. Interactions of GMP with Human Glrx3 and with Saccharomyces cerevisiae Grx3 and Grx4 Converge in the Regulation of the Gcn2 Pathway. Am. Soc. Microbiol. 2020, 86, e00221-20. [Google Scholar] [CrossRef]
- Shintani, T.; Klionsky, D.J. Cargo Proteins Facilitate the Formation of Transport Vesicles in the Cytoplasm to Vacuole Targeting Pathway. J. Biol. Chem. 2004, 279, 29889–29894. [Google Scholar] [CrossRef] [PubMed]
- Noda, T.; Klionsky, D.J. The quantitative Pho8Delta60 assay of nonspecific autophagy. Methods Enzymol. 2008, 6879, 03203–03205. [Google Scholar] [CrossRef]
- Hernandez-Lopez, M.; Prieto, J.; Randez-Gil, F. Osmotolerance and leavening ability in sweet dough. Comparative analysis between Torulaspora delbrueckii and Saccharomyces cerevisiae baker’s yeast strains. Antonie Leeuwenhoek 2003, 84, 125–134. [Google Scholar] [CrossRef] [PubMed]
- Dever, T.E.; Feng, L.; Wek, R.C.; Cigan, A.M.; Donahue, T.F.; Hinnebusch, A.G. Phosphorylation of initiation factor 2α by protein kinase GCN2 mediates gene-specific translational control of GCN4 in yeast. Cell 1992, 68, 585–596. [Google Scholar] [CrossRef]
- Suzuki, K.; Kirisako, T.; Kamada, Y.; Mizushima, N.; Noda, T.; Ohsumi, Y. The pre-autophagosomal structure organized by concerted functions of APG genes is essential for autophagosome formation. EMBO J. 2001, 20, 5971–5981. [Google Scholar] [CrossRef] [PubMed]
- Adachi, A.; Koizumi, M.; Ohsumi, Y. Autophagy induction under carbon starvation conditions is negatively regulated by carbon catabolite repression. J. Biol. Chem. 2017, 292, 19905–19918. [Google Scholar] [CrossRef]
- Tomotake, K.; Daniel, J.K. Mitophagy in yeast occurs through a selective mechanism. J. Biol. Chem. 2008, 283, 32386–32393. [Google Scholar] [CrossRef]
- Kolitsida, P.; Abeliovich, H. Methods for Studying Mitophagy in Yeast. Methods Mol. Biol. 2019, 1880, 669–678. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Okamoto, K. Regulatory mechanisms of mitophagy in yeast. Biochim. Biophys. Acta-Gen. Subj. 2021, 1865, 129858. [Google Scholar] [CrossRef] [PubMed]
- Zientara-Rytter, K.; Subramani, S. Mechanistic insights into the role of Atg11 in selecetive autophagy. J. Mol. Biol. 2020, 432, 104–122. [Google Scholar] [CrossRef]
- Grisan, F.; Iannucci, L.F.; Surdo, N.C.; Gerbino, A.; Zanin, S.; Di Benedetto, G.; Pozzan, T.; Lefkimmiatis, K. PKA compartmentalization links cAMP signaling and autophagy. Cell Death Differ. 2021, 28, 2436–2449. [Google Scholar] [CrossRef] [PubMed]
- Coccetti, P.; Nicastro, R.; Tripodi, F. Conventional and emerging roles of the energy sensor snf1/AMPK in Saccharomyces cerevisiae. Microb. Cell 2018, 5, 482–494. [Google Scholar] [CrossRef] [PubMed]
- Hedbacker, K. SNF1/AMPK pathways in yeast. Front. Biosci. 2008, 13, 2408. [Google Scholar] [CrossRef] [PubMed]
- Mayer, F.V.; Heath, R.; Underwood, E.; Sanders, M.J.; Carmena, D.; McCartney, R.R.; Leiper, F.C.; Xiao, B.; Jing, C.; Walker, P.A.; et al. ADP regulates SNF1, the Saccharomyces cerevisiae homolog of AMP-activated protein kinase. Cell Metab. 2011, 14, 707–714. [Google Scholar] [CrossRef]
- Hlavatá, L.; Nyström, T. Ras Proteins Control Mitochondrial Biogenesis and Function in Saccharomyces cerevisiae. Folia Microbiol. 2003, 48, 725–730. [Google Scholar] [CrossRef] [PubMed]
- Cebollero, E.; Reggiori, F. Regulation of autophagy in yeast Saccharomyces cerevisiae. Biochim. Biophys. Acta Mol. Cell Res. 2009, 1793, 1413–1421. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Klionsky, D.J. The regulation of autophagy—Unanswered questions. J. Cell Sci. 2011, 124, 161–170. [Google Scholar] [CrossRef]
- Pedruzzi, I.; Bürckert, N.; Egger, P.; De Virgilio, C. Saccharomyces cerevisiae Ras/cAMP pathway controls post-diauxic shift element-dependent transcription through the zinc finger protein Gis1. EMBO J. 2000, 19, 2569–2579. [Google Scholar] [CrossRef]
- Pedruzzi, I.; Dubouloz, F.; Cameroni, E.; Wanke, V.; Roosen, J.; Winderickx, J.; De Virgilio, C. TOR and PKA Signaling Pathways Converge on the Protein Kinase Rim15 to Control Entry into G0. Mol. Cell 2003, 12, 1607–1613. [Google Scholar] [CrossRef]
- Schmelzle, T.; Beck, T.; Martin, D.E.; Hall, M.N. Activation of the RAS/Cyclic AMP Pathway Suppresses a TOR Deficiency in Yeast. Mol. Cell. Biol. 2004, 24, 338–351. [Google Scholar] [CrossRef] [PubMed]
- Urban, J.; Soulard, A.; Huber, A.; Lippman, S.; Mukhopadhyay, D.; Deloche, O.; Wanke, V.; Anrather, D.; Ammerer, G.; Riezman, H.; et al. Sch9 Is a Major Target of TORC1 in Saccharomyces cerevisiae. Mol. Cell 2007, 26, 663–674. [Google Scholar] [CrossRef] [PubMed]
- Deprez, M.A.; Eskes, E.; Winderickx, J.; Wilms, T. The TORC1-Sch9 pathway as a crucial mediator of chronological lifespan in the yeast Saccharomyces cerevisiae. FEMS Yeast Res. 2018, 18, foy048. [Google Scholar] [CrossRef]
- Lakhani, R.; Vogel, K.R.; Till, A.; Liu, J.; Burnett, S.F.; Gibson, K.M.; Subramani, S. Defects in GABA metabolism affect selective autophagy pathways and are alleviated by mTOR inhibition. EMBO Mol. Med. 2014, 6, 551–566. [Google Scholar] [CrossRef] [PubMed]
- Waliullah, T.M.; Yeasmin, A.M.; Kaneko, A.; Koike, N.; Terasawa, M.; Totsuka, T.; Ushimaru, T. Rim15 and Sch9 kinases are involved in induction of autophagic degradation of ribosomes in budding yeast. Biosci. Biotechnol. Biochem. 2017, 81, 307–310. [Google Scholar] [CrossRef]








| Strain | Genotype | Source |
|---|---|---|
| CML128 | MATa leu2D3,112, ura3D52, trp1D0, his4D0 | [52] |
| GSL011 | MATa mtl1::NatMx4 | [53] |
| GSL053 | MATa ras2::Leu2MX5 | [53] |
| GSL054 | MATa mtl1::KanMx4 ras2::LEU2Mx5 | [53] |
| GSL197 | MATa leu2D3,112, ura3D52, trp1D0, his4D0 URA3::GFP-ATG8 | [23] |
| GSL198 | MATa mtl1::KanMx4 URA3::GFP-ATG8 | This work |
| GSL199 | MATa tor1::KanMx4 URA3::GFP-ATG8 | [23] |
| GSL200 | MATa mtl1::KanMx4 tor1::LEU2 URA3::GFP-ATG8 | This work |
| GSL201 | MATa ras2::Leu2MX5 URA3::GFP-ATG8 | [23] |
| GSL202 | MATa mtl1::KanMx4 ras2::LEU2 URA3::GFP-ATG8 | This work |
| GSL205 | MATa sch9::NatMx4 | [30] |
| GSL206 | MATa sch9::NatMx4 mtl1::KanMx4 | [30] |
| GSL218 | MATa atg7::NatMx4 | [23] |
| GSL226 | MATa atg7::NatMx4 URA3::GFP-ATG8 | [23] |
| GSL265 | MATa slt2::NatMx4 URA3::GFP-ATG8 | This work |
| GSL279 | MATa sch9::NatMx4 URA3::GFP-ATG8 | This work |
| GSL293 | MATa atg11::NatMx4 | [23] |
| GSL296 | MATa atg33::NatMX4 | This work |
| GSL297 | MATa atg11::NatMx4 URA3::GFP-ATG8 | [23] |
| GSL324 | MATa atg1::NatMx4 | [23] |
| GSL352 | MATa gcn2::KanMx4 URA3::GFP-ATG8 | [23] |
| GSL364 | MATa atg32::KanMx4 | [23] |
| GSL370 | MATa rho0 URA3::GFP-ATG8 | [23] |
| GSL372 | MATa leu2D3,112, uras3D52, trp1D0, his4D0 ATG1-HA::LEU2 | [23] |
| GSL382 | MATa snf1:KanMx4 URA3::GFP-ATG8 | [23] |
| GSL414 | MATa mtl1::KanMX4 sch9::NatMx4 URA3::GFP-ATG8 | This work |
| GSL415 | MATa mtl1::KanMx4 slt2::NatMx4 URA3::GFP-ATG8 | This work |
| GSL416 | MATa mtl1:NatMx4 ATG1-HA::LEU2 | This work |
| BY4741 pho8Δ | MATa pho8 his3D1, leu2D0, met15D0, ura3D0 | [54] |
| Plasmid | Restriction Sites to Clone the ORF | Marker | Promoter | Epitope | Source |
|---|---|---|---|---|---|
| pGFP-Atg8 | EcoRI, XhoI | URA3 | ATG8 | GFP | [55] |
| pSfp1-GFP | SalI, SmaI | URA3 | MET25 | GFP | [23] |
| pAdh1-Msn2-GFP | KspI, SalI | LEU2 | ADH1 | GFP | [56] |
| pRtg1-GFP | XhoI, EcoRI | URA3 | RTG1 | GFP | [57] |
| pIdp1-GFP | HindIII, XhoI | URA3 | IDP1 | GFP | [58] |
| pAtg13-HA | NotI, PstI | URA3 | ADH1 | HA | [23] |
| pAtg1-HA | NotI, PstI | LEU2 | ADH1 | HA | [23] |
| pMM351 | PstI, HindIII | LEU2 | ADH1 | HA | [51] |
| pBcy1HA | SmaI, XhoI | HIS3 | ADH1 | HA | [30] |
| pPkc1* | PmeI, NotI | LEU2 | ADH1 | HA | This work |
| pBCK1-20 | HindIII, PstI | TRP1 | LAC | [28] | |
| pYX242-cytPho8 | AvrII, MluI | LEU2 | PHO8 | [54] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Montella-Manuel, S.; Pujol-Carrion, N.; de la Torre-Ruiz, M.A. The Cell Wall Integrity Receptor Mtl1 Contributes to Articulate Autophagic Responses When Glucose Availability Is Compromised. J. Fungi 2021, 7, 903. https://doi.org/10.3390/jof7110903
Montella-Manuel S, Pujol-Carrion N, de la Torre-Ruiz MA. The Cell Wall Integrity Receptor Mtl1 Contributes to Articulate Autophagic Responses When Glucose Availability Is Compromised. Journal of Fungi. 2021; 7(11):903. https://doi.org/10.3390/jof7110903
Chicago/Turabian StyleMontella-Manuel, Sandra, Nuria Pujol-Carrion, and Maria Angeles de la Torre-Ruiz. 2021. "The Cell Wall Integrity Receptor Mtl1 Contributes to Articulate Autophagic Responses When Glucose Availability Is Compromised" Journal of Fungi 7, no. 11: 903. https://doi.org/10.3390/jof7110903
APA StyleMontella-Manuel, S., Pujol-Carrion, N., & de la Torre-Ruiz, M. A. (2021). The Cell Wall Integrity Receptor Mtl1 Contributes to Articulate Autophagic Responses When Glucose Availability Is Compromised. Journal of Fungi, 7(11), 903. https://doi.org/10.3390/jof7110903






