P-Type ATPase Apt1 of the Fungal Pathogen Cryptococcus neoformans Is a Lipid Flippase of Broad Substrate Specificity
Abstract
:1. Introduction
2. Materials and Methods
3. Results
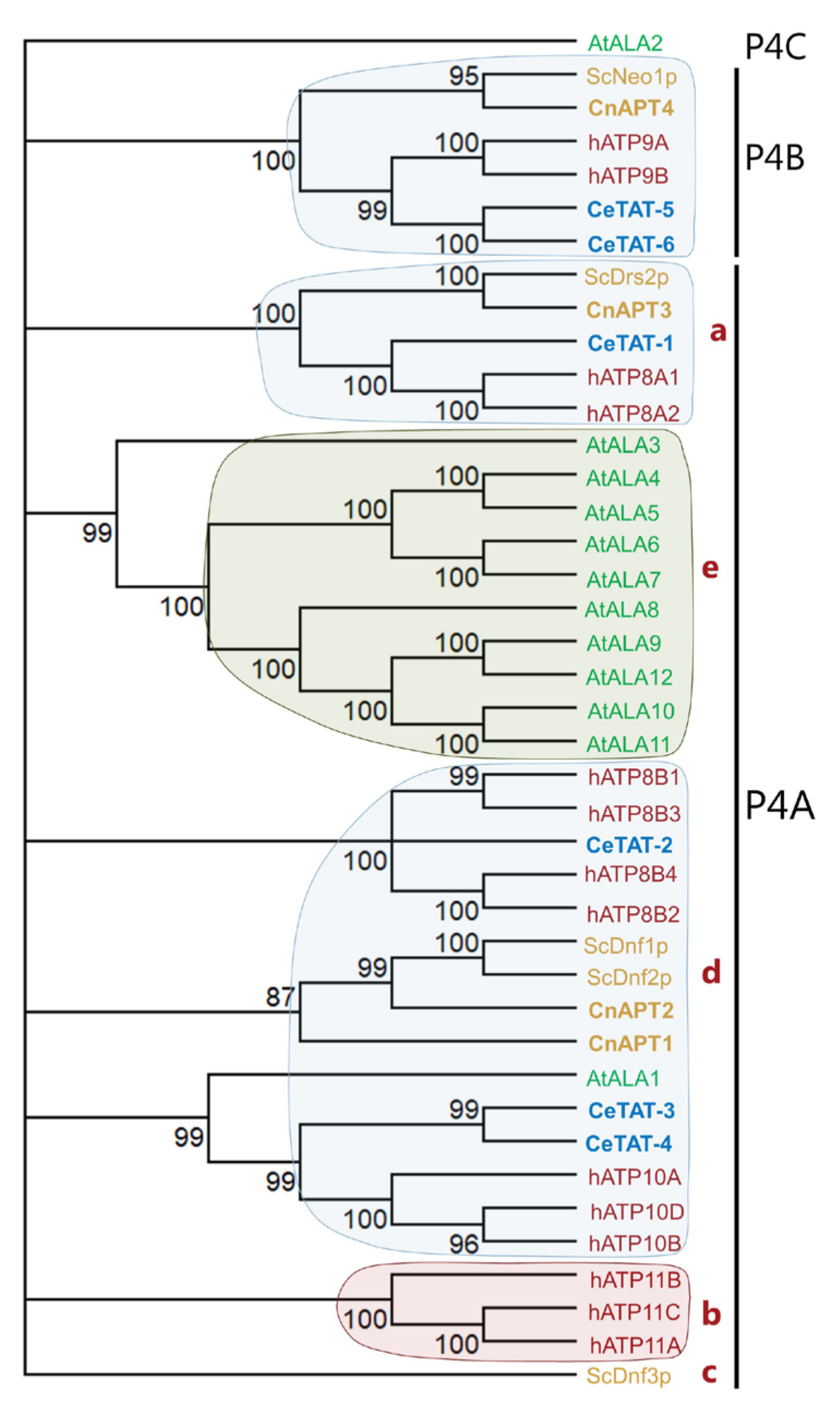
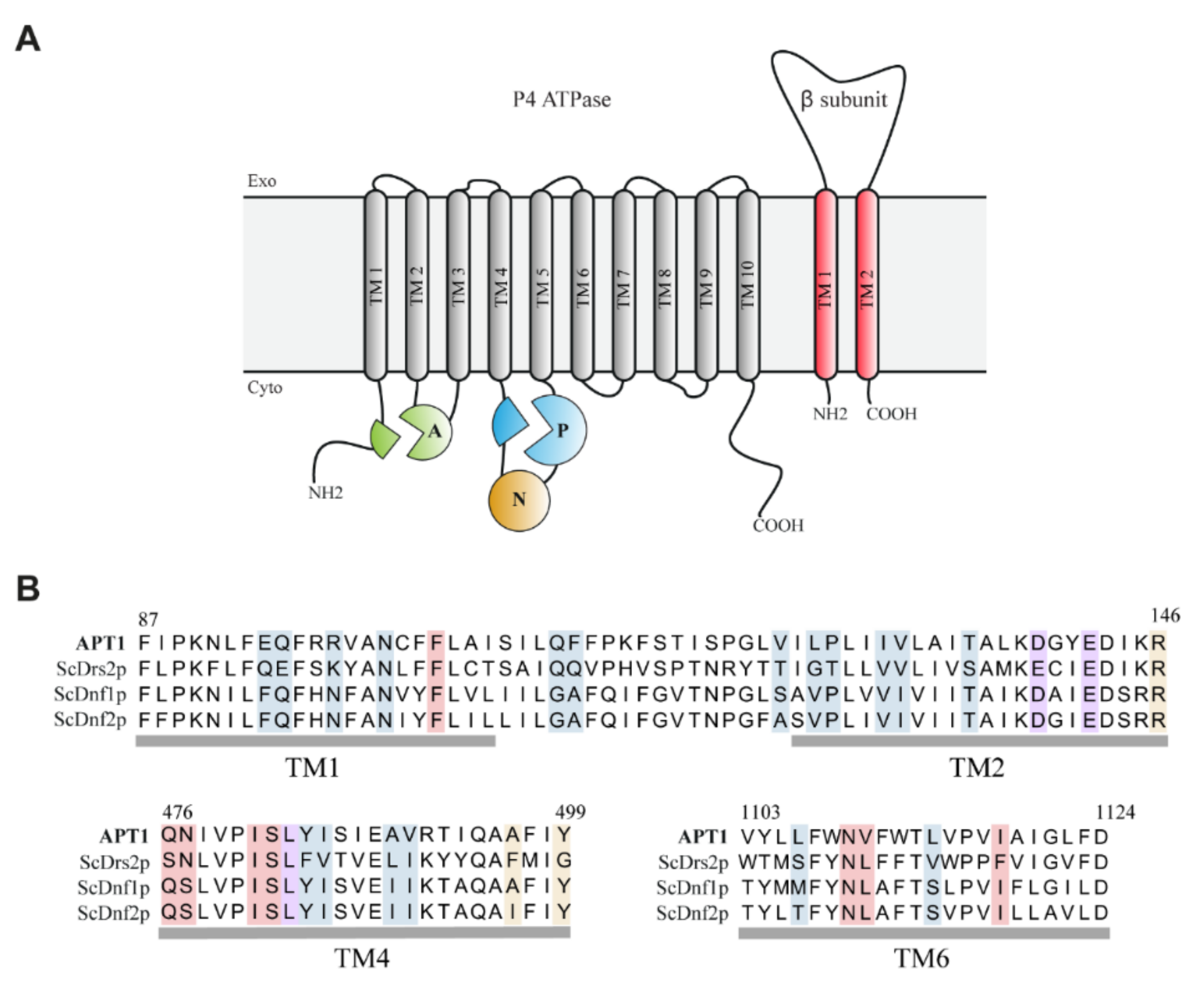
3.1. C. neoformans Genome Encodes Four Potential P4-ATPases
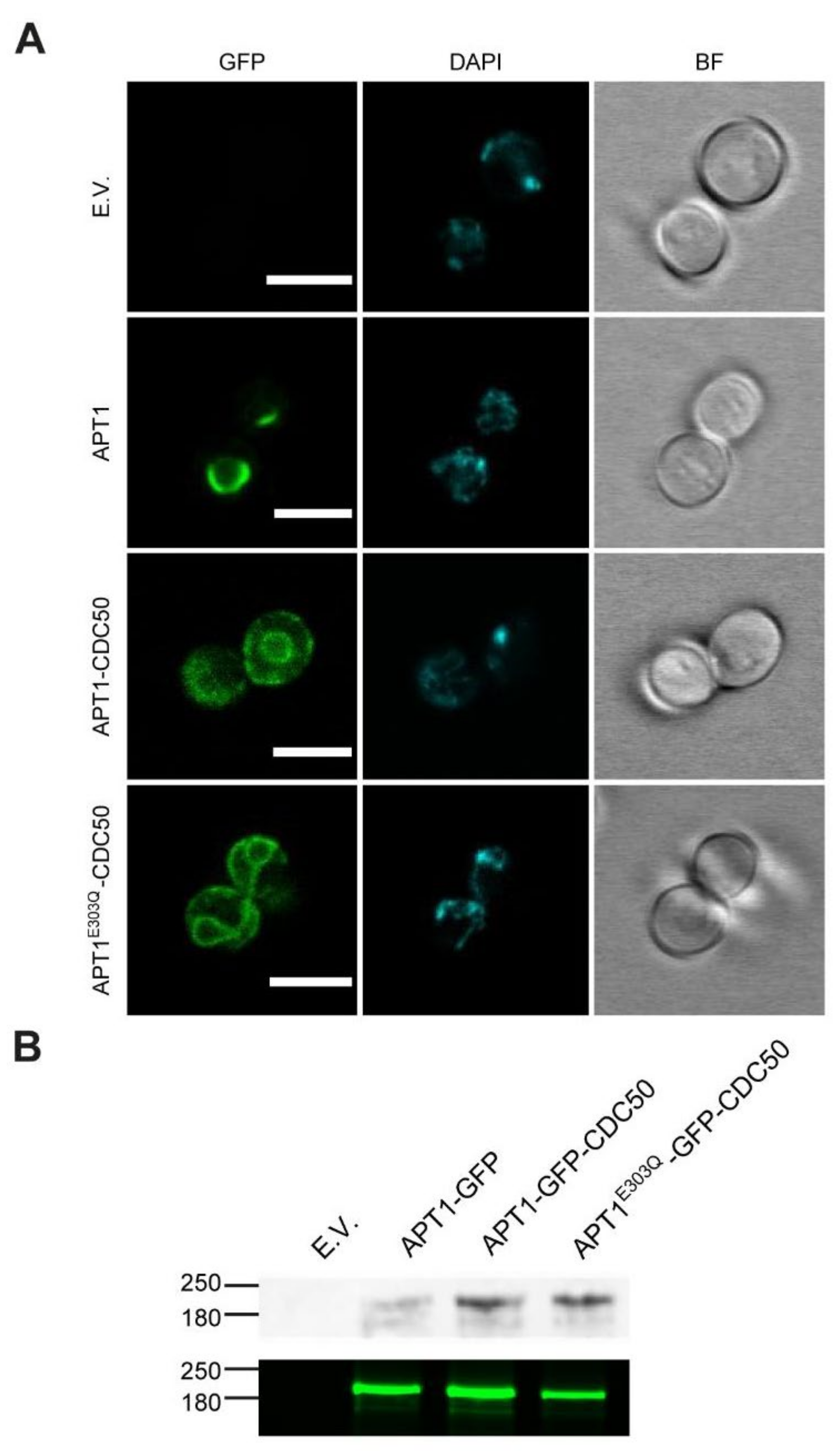
3.2. Heterologous Expression and Localization of Apt1 and Cdc50 Proteins in S. cerevisiae
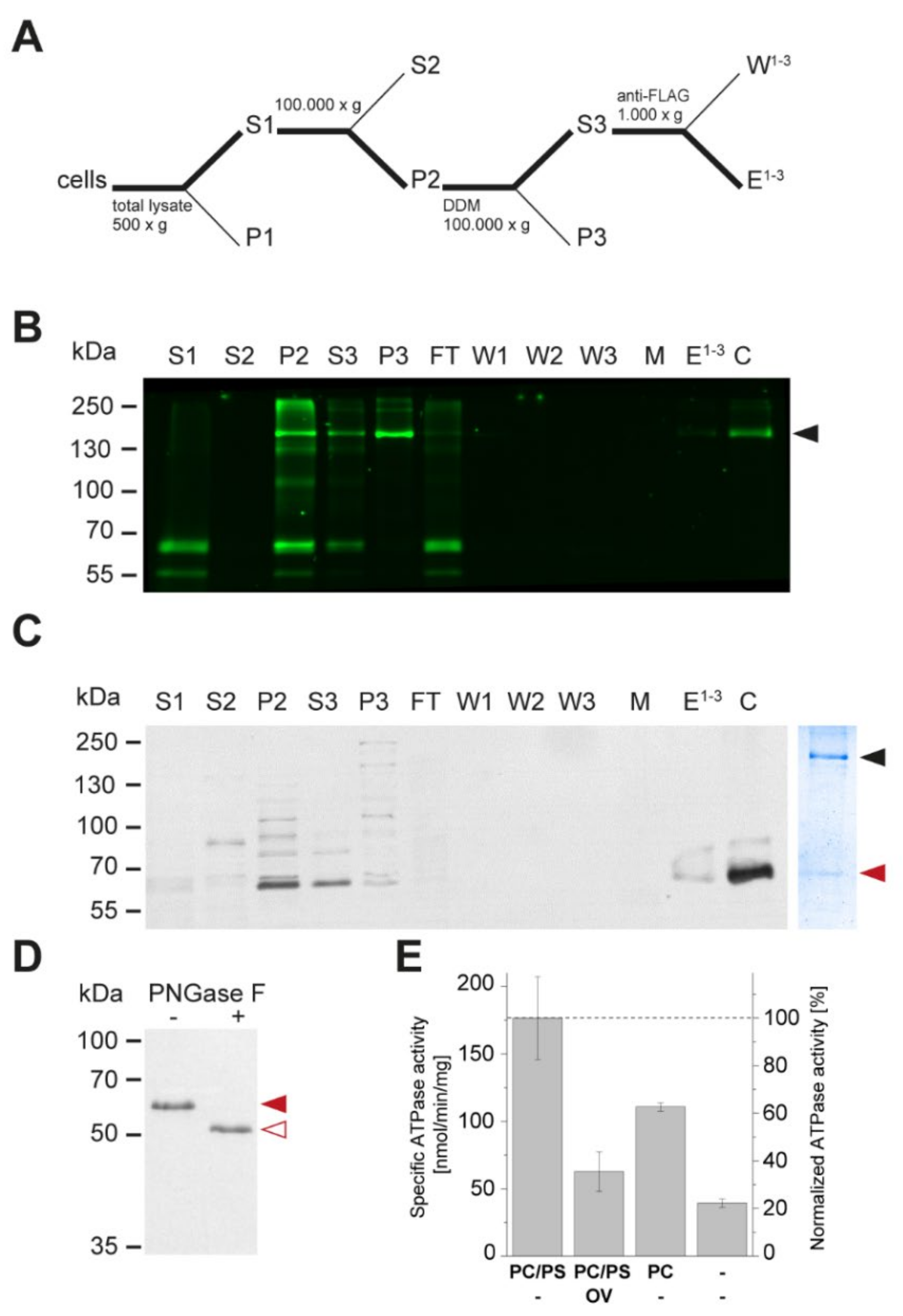
3.3. Apt1 and Cdc50 Proteins Form a Stable Complex Showing ATPase Activity
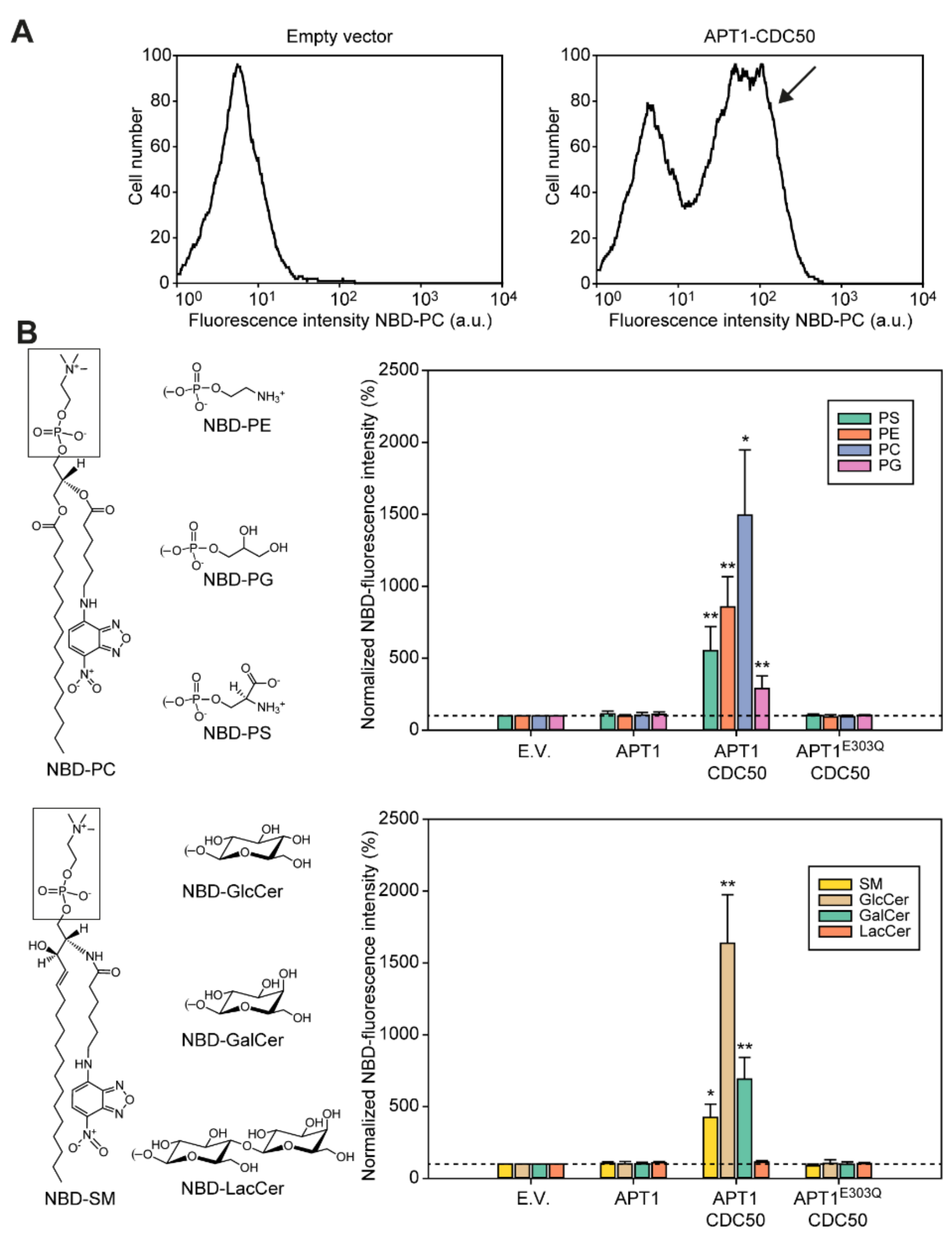
3.4. Apt1p in Complex with Cdc50p Is a Broad-Specificity Lipid Flippase
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Rodrigues, M.L.; Djordjevic, J.T. Unravelling Secretion in Cryptococcus neoformans: More than One Way to Skin a Cat. Mycopathologia 2012, 173, 407–418. [Google Scholar] [CrossRef] [PubMed]
- McClelland, E.E.; Bernhardt, P.; Casadevall, A. Estimating the relative contributions of virulence factors for pathogenic microbes. Infect. Immun. 2006, 74, 1500–1504. [Google Scholar] [CrossRef] [Green Version]
- Zaragoza, O.; Rodrigues, M.L.; De Jesus, M.; Frases, S.; Dadachova, E.; Casadevall, A. Chapter 4 The Capsule of the Fungal Pathogen Cryptococcus neoformans. Adv. Appl. Microbiol. 2009, 68, 133–216. [Google Scholar] [PubMed] [Green Version]
- Sebastian, T.T.; Baldridge, R.D.; Xu, P.; Graham, T.R. Phospholipid flippases: Building asymmetric membranes and transport vesicles. Biochim. Biophys. Acta Mol. Cell Biol. Lipids 2012, 1821, 1068–1077. [Google Scholar] [CrossRef] [Green Version]
- Andersen, J.P.; Vestergaard, A.L.; Mikkelsen, S.A.; Mogensen, L.S.; Chalat, M.; Molday, R.S. P4-ATPases as phospholipid flippases-structure, function, and enigmas. Front. Physiol. 2016, 7, 275. [Google Scholar] [CrossRef]
- Pomorski, T.; Lombardi, R.; Riezman, H.; Devaux, P.F.; van Meer, G.; Holthuis, J.C.M. Drs2p-related P-type ATPases Dnf1p and Dnf2p Are Required for Phospholipid Translocation across the Yeast Plasma Membrane and Serve a Role in Endocytosis. Mol. Biol. Cell 2003, 14, 1240–1254. [Google Scholar] [CrossRef] [Green Version]
- Chen, C.Y.; Ingram, M.F.; Rosal, P.H.; Graham, T.R. Role for Drs2p, a P-type ATPase and potential aminophospholipid translocase, in yeast late Golgi function. J. Cell Biol. 1999, 147, 1223–1236. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gall, W.E.; Geething, N.C.; Hua, Z.; Ingram, M.F.; Liu, K.; Chen, S.I.; Graham, T.R. Drs2p-dependent formation of exocytic clathrin-coated vesicles in vivo. Curr. Biol. 2002, 12, 1623–1627. [Google Scholar] [CrossRef] [Green Version]
- Tuck, S. Extracellular Vesicles: Budding Regulated by a Phosphatidylethanolamine Translocase. Curr. Biol. 2011, 21, R988–R990. [Google Scholar] [CrossRef] [Green Version]
- Wehman, A.M.; Poggioli, C.; Schweinsberg, P.; Grant, B.D.; Nance, J. The P4-ATPase TAT-5 inhibits the budding of extracellular vesicles in C. elegans embryos. Curr. Biol. 2011, 21, 1951–1959. [Google Scholar] [CrossRef] [Green Version]
- Naik, J.; Hau, C.M.; ten Bloemendaal, L.; Mok, K.S.; Hajji, N.; Wehman, A.M.; Meisner, S.; Muncan, V.; Paauw, N.J.; de Vries, H.E.; et al. The P4-ATPase ATP9A is a novel determinant of exosome release. PLoS ONE 2019, 14, e0213069. [Google Scholar] [CrossRef]
- Paulusma, C.C.; Folmer, D.E.; Ho-Mok, K.S.; De Waart, D.R.; Hilarius, P.M.; Verhoeven, A.J.; Oude Elferink, R.P.J. ATP8B1 requires an accessory protein for endoplasmic reticulum exit and plasma membrane lipid flippase activity. Hepatology 2008, 47, 268–278. [Google Scholar] [CrossRef] [PubMed]
- Bryde, S.; Hennrich, H.; Verhulst, P.M.; Devaux, P.F.; Lenoir, G.; Holthuis, J.C.M. CDC50 proteins are critical components of the human class-1 P 4-ATPase transport machinery. J. Biol. Chem. 2010, 285, 40562–40572. [Google Scholar] [CrossRef] [Green Version]
- López-Marqués, R.L.; Poulsen, L.R.; Hanisch, S.; Meffert, K.; Buch-Pedersen, M.J.; Jakobsen, M.K.; Pomorski, T.G.; Palmgren, M.G. Intracellular targeting signals and lipid specificity determinants of the ALA/ALIS P4-ATPase complex reside in the catalytic ALA α-subunit. Mol. Biol. Cell 2010, 21, 791–801. [Google Scholar] [CrossRef] [Green Version]
- Timcenko, M.; Lyons, J.A.; Januliene, D.; Ulstrup, J.J.; Dieudonné, T.; Montigny, C.; Ash, M.R.; Karlsen, J.L.; Boesen, T.; Kühlbrandt, W.; et al. Structure and autoregulation of a P4-ATPase lipid flippase. Nature 2019, 571, 366–370. [Google Scholar] [CrossRef]
- Hiraizumi, M.; Yamashita, K.; Nishizawa, T.; Nureki, O. Cryo-EM structures capture the transport cycle of the P4-ATPase flippase. Science 2019, 365, 1149–1155. [Google Scholar] [CrossRef]
- Bai, L.; Kovach, A.; You, Q.; Hsu, H.C.; Zhao, G.; Li, H. Autoinhibition and activation mechanisms of the eukaryotic lipid flippase Drs2p-Cdc50p. Nat. Commun. 2019, 10, 4142. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- He, Y.; Xu, J.; Wu, X.; Li, L. Structures of a P4-ATPase lipid flippase in lipid bilayers. Protein Cell 2020, 11, 458–463. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nakanishi, H.; Irie, K.; Segawa, K.; Hasegawa, K.; Fujiyoshi, Y.; Nagata, S.; Abe, K. Crystal structure of a human plasma membrane phospholipid flippase. J. Biol. Chem. 2020, 295, 10180–10194. [Google Scholar] [CrossRef] [PubMed]
- Nakanishi, H.; Nishizawa, T.; Segawa, K.; Nureki, O.; Fujiyoshi, Y.; Nagata, S.; Abe, K. Transport Cycle of Plasma Membrane Flippase ATP11C by Cryo-EM. Cell Rep. 2020, 32, 108208. [Google Scholar] [CrossRef]
- Hua, Z.; Fatheddin, P.; Graham, T.R. An essential subfamily of Drs2p-related P-type ATPases is required for protein trafficking between golgi complex and endosomal/vacuolar system. Mol. Biol. Cell 2002, 13, 3162–3177. [Google Scholar] [CrossRef] [Green Version]
- Wicky, S.; Schwarz, H.; Singer-Krüger, B. Molecular Interactions of Yeast Neo1p, an Essential Member of the Drs2 Family of Aminophospholipid Translocases, and Its Role in Membrane Trafficking within the Endomembrane System. Mol. Cell. Biol. 2004, 24, 7402–7418. [Google Scholar] [CrossRef] [Green Version]
- Natarajan, P.; Wang, J.; Hua, Z.; Graham, T.R. Drs2p-coupled aminophospholipid translocase activity in yeast Golgi membranes and relationship to in vivo function. Proc. Natl. Acad. Sci. USA 2004, 101, 10614–10619. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhou, X.; Graham, T.R. Reconstitution of phospholipid translocase activity with purified Drs2p, a type-IV P-type ATPase from budding yeast. Proc. Natl. Acad. Sci. USA 2009, 106, 16586–16591. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hachiro, T.; Yamamoto, T.; Nakano, K.; Tanaka, K. Phospholipid flippases Lem3p-Dnf1p and Lem3p-Dnf2p are involved in the sorting of the tryptophan permease Tat2p in yeast. J. Biol. Chem. 2013, 288, 3594–3608. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Alder-Baerens, N.; Lisman, Q.; Luong, L.; Pomorski, T.; Holthuis, J.C.M. Loss of P4 ATPases Drs2p and Dnf3p disrupts aminophospholipid transport and asymmetry in yeast post-Golgi secretory vesicles. Mol. Biol. Cell 2006, 17, 1632–1642. [Google Scholar] [CrossRef] [Green Version]
- Frøsig, M.M.; Costa, S.R.; Liesche, J.; Østerberg, J.T.; Hanisch, S.; Nintemann, S.; Sørensen, H.; Palmgren, M.; Pomorski, T.G.; López-Marqués, R.L. Pseudohyphal growth in Saccharomyces cerevisiae involves protein kinase-regulated lipid flippases. J. Cell Sci. 2020, 133, jcs235994. [Google Scholar] [CrossRef]
- Iwamoto, K.; Kobayashi, S.; Fukuda, R.; Umeda, M.; Kobayashi, T.; Ohta, A. Local exposure of phosphatidylethanolamine on the yeast plasma membrane is implicated in cell polarity. Genes Cells 2004, 9, 891–903. [Google Scholar] [CrossRef]
- Baldridge, R.D.; Xu, P.; Graham, T.R. Type IV p-type ATPases distinguish mono-versus diacyl phosphatidylserine using a cytofacial exit gate in the membrane domain. J. Biol. Chem. 2013, 288, 19516–19527. [Google Scholar] [CrossRef] [Green Version]
- Roland, B.P.; Naito, T.; Best, J.T.; Arnaiz-Yépez, C.; Takatsu, H.; Yu, R.J.; Shin, H.W.; Graham, T.R. Yeast and human P4-ATPases transport glycosphingolipids using conserved structural motifs. J. Biol. Chem. 2019, 294, 1794–1806. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hanson, P.K.; Malone, L.; Birchmore, J.L.; Nichols, J.W. Lem3p is essential for the uptake and potency of alkylphosphocholine drugs, edelfosine and miltefosine. J. Biol. Chem. 2003, 278, 36041–36050. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kato, U.; Emoto, K.; Fredriksson, C.; Nakamura, H.; Ohta, A.; Kobayashi, T.; Murakami-Murofushi, K.; Kobayashi, T.; Umeda, M. A novel membrane protein, Ros3p, is required for phospholipid translocation across the plasma membrane in Saccharomyces cerevisiae. J. Biol. Chem. 2002, 277, 37855–37862. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Riekhof, W.R.; Voelker, D.R. Uptake and utilization of lyso-phosphatidylethanolamine by Saccharomyces cerevisiae. J. Biol. Chem. 2006, 281, 36588–36596. [Google Scholar] [CrossRef] [Green Version]
- Riekhof, W.R.; Wu, J.; Gijón, M.A.; Zarini, S.; Murphy, R.C.; Voelker, D.R. Lysophosphatidylcholine metabolism in Saccharomyces cerevisiae: The role of P-type ATPases in transport and a broad specificity acyltransferase in acylation. J. Biol. Chem. 2007, 282, 36853–36861. [Google Scholar] [CrossRef] [Green Version]
- Takar, M.; Wu, Y.; Graham, T.R. The essential Neo1 protein from budding yeast plays a role in establishing aminophospholipid asymmetry of the plasma membrane. J. Biol. Chem. 2016, 291, 15727–15739. [Google Scholar] [CrossRef] [Green Version]
- Rizzo, J.; Oliveira, D.L.; Joffe, L.S.; Hu, G.; Gazos-Lopes, F.; Fonseca, F.L.; Almeida, I.C.; Frases, S.; Kronstad, J.W.; Rodrigues, M.L. Role of the Apt1 protein in polysaccharide secretion by Cryptococcus neoformans. Eukaryot. Cell 2014, 13, 715–726. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hu, G.; Kronstad, J.W. A putative P-type ATPase, Apt1, is involved in stress tolerance and virulence in Cryptococcus neoformans. Eukaryot. Cell 2010, 9, 74–83. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rizzo, J.; Colombo, A.C.; Zamith-Miranda, D.; Silva, V.K.A.; Allegood, J.C.; Casadevall, A.; Del Poeta, M.; Nosanchuk, J.D.; Kronstad, J.W.; Rodrigues, M.L. The putative flippase Apt1 is required for intracellular membrane architecture and biosynthesis of polysaccharide and lipids in Cryptococcus neoformans. Biochim. Biophys. Acta Mol. Cell Res. 2018, 1865, 532–541. [Google Scholar] [CrossRef] [PubMed]
- Huang, W.; Liao, G.; Baker, G.M.; Wang, Y.; Lau, R.; Paderu, P.; Perlin, D.S.; Xue, C. Lipid flippase subunit Cdc50 mediates drug resistance and virulence in Cryptococcus neoformans. MBio 2016, 7, e00478-16. [Google Scholar] [CrossRef] [Green Version]
- Hu, G.; Caza, M.; Bakkeren, E.; Kretschmer, M.; Bairwa, G.; Reiner, E.; Kronstad, J. A P4-ATPase subunit of the Cdc50 family plays a role in iron acquisition and virulence in Cryptococcus neoformans. Cell. Microbiol. 2017, 19, e12718. [Google Scholar] [CrossRef] [Green Version]
- Gietz, R.D.; Woods, R.A. Transformation of yeast by lithium acetate/single-stranded carrier DNA/polyethylene glycol method. Methods Enzymol. 2002, 350, 87–96. [Google Scholar] [CrossRef]
- Jensen, M.S.; Costa, S.; Günther-Pomorski, T.; López-Marqués, R.L. Cell-Based Lipid Flippase Assay Employing Fluorescent Lipid Derivatives. Methods Mol. Biol. 2016, 1377, 371–382. [Google Scholar] [CrossRef]
- Stanchev, L.D.; Marek, M.; Xian, F.; Klöhn, M.; Silvestro, D.; Dittmar, G.; López-Marqués, R.L.; Günther Pomorski, T. Functional Significance of Conserved Cysteines in the Extracellular Loops of the ATP Binding Cassette Transporter Pdr11p. J. Fungi 2020, 7, 2. [Google Scholar] [CrossRef] [PubMed]
- Gorbulev, S.; Abele, R.; Tampe, R. Allosteric crosstalk between peptide-binding, transport, and ATP hydrolysis of the ABC transporter TAP. Proc. Natl. Acad. Sci. USA 2001, 98, 3732–3737. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Marek, M.; Milles, S.; Schreiber, G.; Daleke, D.L.; Dittmar, G.; Herrmann, A.; Müller, P.; Pomorski, T.G. The Yeast Plasma Membrane ATP Binding Cassette (ABC) Transporter Aus1. J. Biol. Chem. 2011, 286, 21835–21843. [Google Scholar] [CrossRef] [Green Version]
- Edgar, R.C. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004, 32, 1792–1797. [Google Scholar] [CrossRef] [Green Version]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef] [PubMed]
- Jones, D.T.; Taylor, W.R.; Thornton, J.M. The rapid generation of mutation data matrices from protein sequences. Bioinformatics 1992, 8, 275–282. [Google Scholar] [CrossRef] [PubMed]
- Palmgren, M.; Østerberg, J.T.; Nintemann, S.J.; Poulsen, L.R.; López-Marqués, R.L. Evolution and a revised nomenclature of P4 ATPases, a eukaryotic family of lipid flippases. Biochim. Biophys. Acta Biomembr. 2019, 1861, 1135–1151. [Google Scholar] [CrossRef]
- López-Marqués, R.L.; Gourdon, P.; Günther Pomorski, T.; Palmgren, M. The transport mechanism of P4 ATPase lipid flippases. Biochem. J. 2020, 477, 3769–3790. [Google Scholar] [CrossRef]
- Palmgren, M.G.; Nissen, P. P-Type ATPases. Annu. Rev. Biophys. 2011, 40, 243–266. [Google Scholar] [CrossRef] [PubMed]
- Vestergaard, A.L.; Coleman, J.A.; Lemmin, T.; Mikkelsen, S.A.; Molday, L.L.; Vilsen, B.; Molday, R.S.; Dal Peraro, M.; Andersen, J.P. Critical roles of isoleucine-364 and adjacent residues in a hydrophobic gate control of phospholipid transport by the mammalian P4-ATPase ATP8A2. Proc. Natl. Acad. Sci. USA 2014, 111, E1334-43. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Roland, B.P.; Graham, T.R. Directed evolution of a sphingomyelin flippase reveals mechanism of substrate backbone discrimination by a P4-ATPase. Proc. Natl. Acad. Sci. USA 2016, 113, E4460–E4466. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Baldridge, R.D.; Graham, T.R. Two-gate mechanism for phospholipid selection and transport by type IV P-type ATPases. Proc. Natl. Acad. Sci. USA 2013, 110, E358–E367. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huang, Y.; Takar, M.; Best, J.T.; Graham, T.R. Conserved mechanism of phospholipid substrate recognition by the P4-ATPase Neo1 from Saccharomyces cerevisiae. Biochim. Biophys. Acta Mol. Cell Biol. Lipids 2020, 1865, 158581. [Google Scholar] [CrossRef] [PubMed]
- Poulsen, L.R.; López-Marqués, R.L.; Pedas, P.R.; McDowell, S.C.; Brown, E.; Kunze, R.; Harper, J.F.; Pomorski, T.G.; Palmgren, M. A phospholipid uptake system in the model plant Arabidopsis thaliana. Nat. Commun. 2015, 6, 1–14. [Google Scholar] [CrossRef] [Green Version]
- Jensen, M.S.; Costa, S.R.; Duelli, A.S.; Andersen, P.A.; Poulsen, L.R.; Stanchev, L.D.; Gourdon, P.; Palmgren, M.; Günther Pomorski, T.; López-Marqués, R.L. Phospholipid flipping involves a central cavity in P4 ATPases. Sci. Rep. 2017, 7, 17621. [Google Scholar] [CrossRef] [Green Version]
- Rizzo, J.; Stanchev, L.D.; da Silva, V.K.A.; Nimrichter, L.; Pomorski, T.G.; Rodrigues, M.L. Role of lipid transporters in fungal physiology and pathogenicity. Comput. Struct. Biotechnol. J. 2019, 17, 1278–1289. [Google Scholar] [CrossRef]
- Chen, S.; Wang, J.; Muthusamy, B.-P.; Liu, K.; Zare, S.; Andersen, R.J.; Graham, T.R. Roles for the Drs2p-Cdc50p complex in protein transport and phosphatidylserine asymmetry of the yeast plasma membrane. Traffic 2006, 7, 1503–1517. [Google Scholar] [CrossRef]
- Saito, K.; Fujimura-Kamada, K.; Furuta, N.; Kato, U.; Umeda, M.; Tanaka, K. Cdc50p, a protein required for polarized growth, associates with the Drs2p P-type ATPase implicated in phospholipid translocation in Saccharomyces cerevisiae. Mol. Biol. Cell 2004, 15, 3418–3432. [Google Scholar] [CrossRef] [Green Version]
- Furuta, N.; Fujimura-Kamada, K.; Saito, K.; Yamamoto, T.; Tanaka, K. Endocytic recycling in yeast is regulated by putative phospholipid translocases and the Ypt31p/32p-Rcy1p pathway. Mol. Biol. Cell 2007, 18, 295–312. [Google Scholar] [CrossRef] [PubMed]
- van der Velden, L.M.; Wichers, C.G.K.; van Breevoort, A.E.D.; Coleman, J.A.; Molday, R.S.; Berger, R.; Klomp, L.W.J.; van de Graaf, S.F.J. Heteromeric interactions required for abundance and subcellular localization of human CDC50 proteins and class 1 P4-ATPases. J. Biol. Chem. 2010, 285, 40088–40096. [Google Scholar] [CrossRef] [Green Version]
- López-Marqués, R.L.; Poulsen, L.R.; Palmgren, M.G. A putative plant aminophospholipid flippase, the Arabidopsis P4 ATPase ALA1, localizes to the plasma membrane following association with a β-subunit. PLoS ONE 2012, 7, e33042. [Google Scholar] [CrossRef] [Green Version]
- Costa, S.R.; Marek, M.; Axelsen, K.B.; Theorin, L.; Pomorski, T.G.; López-Marqués, R.L. Role of post-translational modifications at the β-subunit ectodomain in complex association with a promiscuous plant P4-ATPase. Biochem. J. 2016, 473, 1605–1615. [Google Scholar] [CrossRef] [Green Version]
- Takatsu, H.; Baba, K.; Shima, T.; Umino, H.; Kato, U.; Umeda, M.; Nakayama, K.; Shin, H.W. ATP9B, a P4-ATPase (a putative aminophospholipid translocase), localizes to the trans-Golgi network in a CDC50 protein-independent manner. J. Biol. Chem. 2011, 286, 38159–38167. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- López-Marqués, R.L.; Davis, J.A.; Harper, J.F.; Palmgren, M. Dynamic membranes: The multiple roles of P4 and P5 ATPases. Plant. Physiol. 2021, 185, 619–631. [Google Scholar] [CrossRef] [PubMed]
- Rittershaus, P.C.; Kechichian, T.B.; Allegood, J.C.; Merrill, A.H.; Hennig, M.; Luberto, C.; Del Poeta, M. Glucosylceramide synthase is an essential regulator of pathogenicity of Cryptococcus neoformans. J. Clin. Invest. 2006, 116, 1651–1659. [Google Scholar] [CrossRef] [Green Version]
- Rodrigues, M.L.; Travassos, L.R.; Miranda, K.R.; Franzen, A.J.; Rozental, S.; de Souza, W.; Alviano, C.S.; Barreto-Bergter, E. Human antibodies against a purified glucosylceramide from Cryptococcus neoformans inhibit cell budding and fungal growth. Infect. Immun. 2000, 68, 7049–7060. [Google Scholar] [CrossRef] [Green Version]
- Rodrigues, M.L.; Nimrichter, L.; Oliveira, D.L.; Frases, S.; Miranda, K.; Zaragoza, O.; Alvarez, M.; Nakouzi, A.; Feldmesser, M.; Casadevall, A. Vesicular polysaccharide export in Cryptococcus neoformans is a eukaryotic solution to the problem of fungal trans-cell wall transport. Eukaryot. Cell 2007, 6, 48–59. [Google Scholar] [CrossRef] [Green Version]
- Kobayashi, T.; Menon, A.K. Transbilayer lipid asymmetry. Curr. Biol. 2018, 28, R386–R391. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jiang, Y.; Wang, W.; Xie, Q.; Liu, N.; Liu, L.; Wang, D.; Zhang, X.; Yang, C.; Chen, X.; Tang, D.; et al. Plants transfer lipids to sustain colonization by mutualistic mycorrhizal and parasitic fungi. Science 2017, 356, 1172–1175. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mor, V.; Rella, A.; Farnoud, A.M.; Singh, A.; Munshi, M.; Bryan, A.; Naseem, S.; Konopka, J.B.; Ojima, I.; Bullesbach, E.; et al. Identification of a New Class of Antifungals Targeting the Synthesis of Fungal Sphingolipids. MBio 2015, 6, e00647. [Google Scholar] [CrossRef] [PubMed] [Green Version]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Stanchev, L.D.; Rizzo, J.; Peschel, R.; Pazurek, L.A.; Bredegaard, L.; Veit, S.; Laerbusch, S.; Rodrigues, M.L.; López-Marqués, R.L.; Günther Pomorski, T. P-Type ATPase Apt1 of the Fungal Pathogen Cryptococcus neoformans Is a Lipid Flippase of Broad Substrate Specificity. J. Fungi 2021, 7, 843. https://doi.org/10.3390/jof7100843
Stanchev LD, Rizzo J, Peschel R, Pazurek LA, Bredegaard L, Veit S, Laerbusch S, Rodrigues ML, López-Marqués RL, Günther Pomorski T. P-Type ATPase Apt1 of the Fungal Pathogen Cryptococcus neoformans Is a Lipid Flippase of Broad Substrate Specificity. Journal of Fungi. 2021; 7(10):843. https://doi.org/10.3390/jof7100843
Chicago/Turabian StyleStanchev, Lyubomir Dimitrov, Juliana Rizzo, Rebecca Peschel, Lilli A. Pazurek, Lasse Bredegaard, Sarina Veit, Sabine Laerbusch, Marcio L. Rodrigues, Rosa L. López-Marqués, and Thomas Günther Pomorski. 2021. "P-Type ATPase Apt1 of the Fungal Pathogen Cryptococcus neoformans Is a Lipid Flippase of Broad Substrate Specificity" Journal of Fungi 7, no. 10: 843. https://doi.org/10.3390/jof7100843
APA StyleStanchev, L. D., Rizzo, J., Peschel, R., Pazurek, L. A., Bredegaard, L., Veit, S., Laerbusch, S., Rodrigues, M. L., López-Marqués, R. L., & Günther Pomorski, T. (2021). P-Type ATPase Apt1 of the Fungal Pathogen Cryptococcus neoformans Is a Lipid Flippase of Broad Substrate Specificity. Journal of Fungi, 7(10), 843. https://doi.org/10.3390/jof7100843







