Isolation of Fungi from a Textile Industry Effluent and the Screening of Their Potential to Degrade Industrial Dyes
Abstract
:1. Introduction
2. Materials and Methods
2.1. Chemicals
2.2. Culture Conditions and Isolation of Fungi from a Textile Industry Effluent
2.3. Fungal Strains Identification
2.3.1. Genomic DNA Extraction and PCR Conditions
2.3.2. PCR Amplification
2.3.3. Restriction Analysis
2.3.4. RAPD Analysis
2.4. Fungal Characterization
2.4.1. Dye Decolorization on Agar Plate
2.4.2. Dye Decolorization in a Carbon-Limited Liquid System
2.4.3. Plate Screening for Lignin-Degrading Enzymes
2.5. Growth and Ligninolytic Enzyme Production of the Emmia Latemarginata (MAP03) Strain in Submerged Fermentation under Different Conditions
2.6. Ligninolytic Enzyme Assays
2.7. Statistical Analysis
3. Results
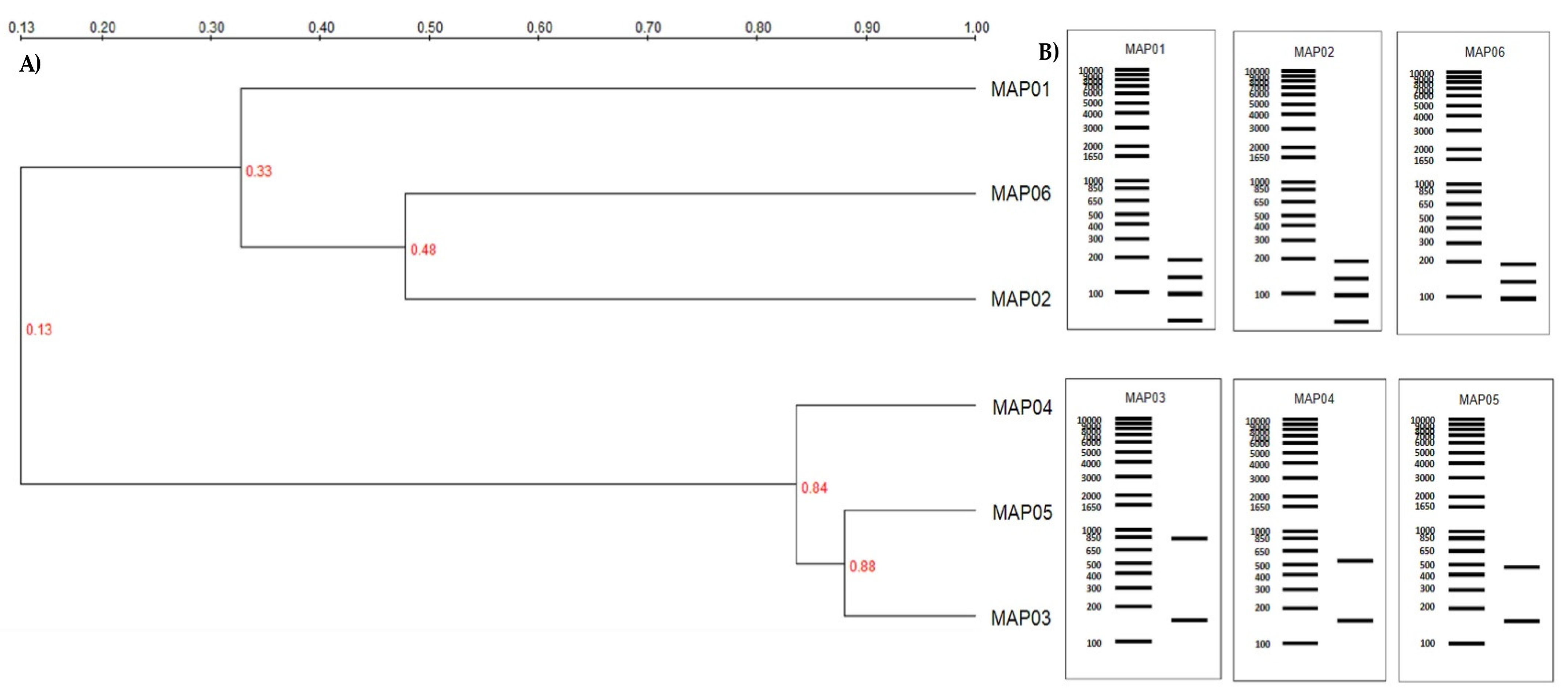
3.1. Strains Identification
3.2. Fungal Characterization
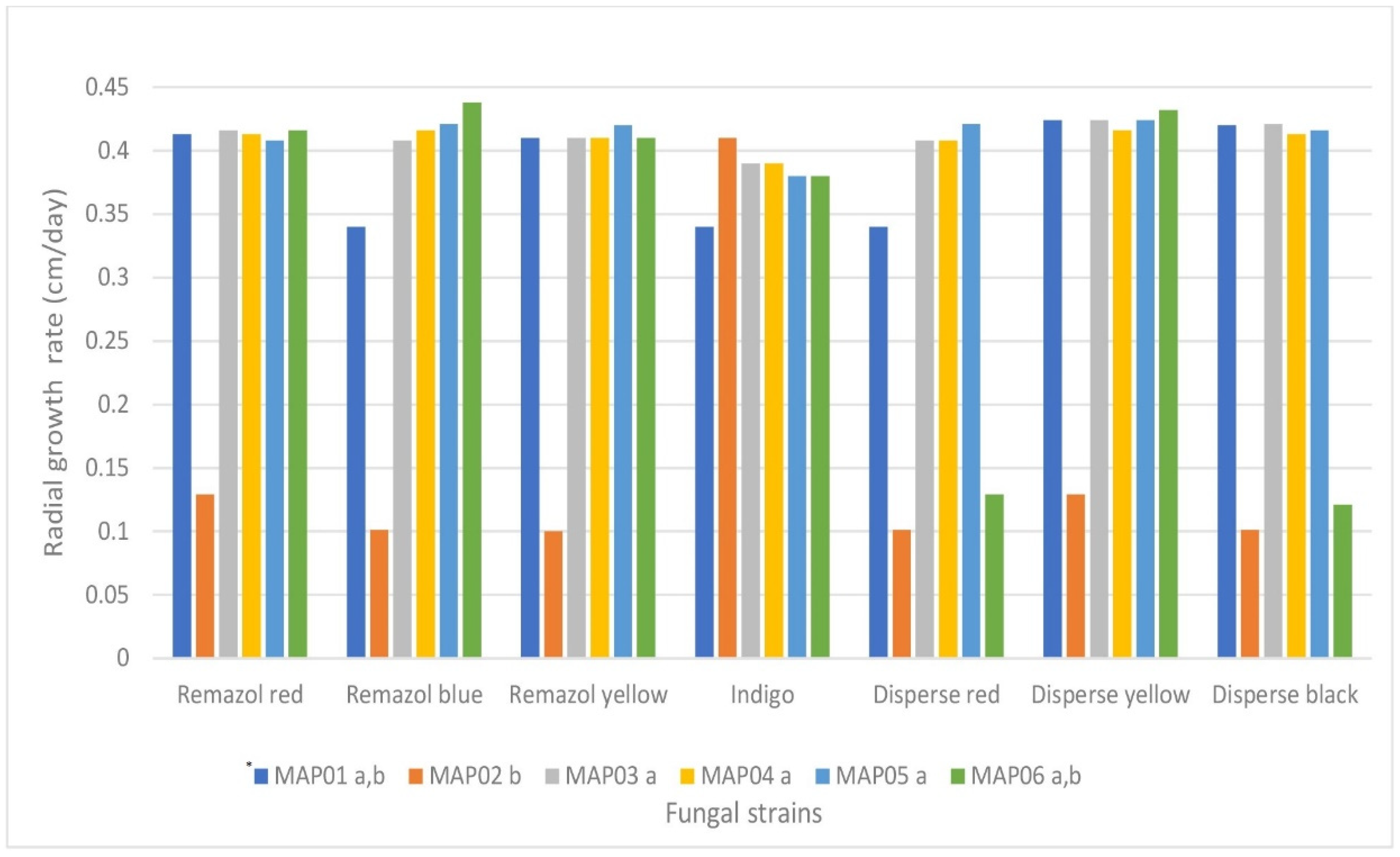
3.2.1. Agar-Plate Screening for the Decolorization of Different Dyes
3.2.2. Dye Decolorization in a Carbon-Limited Liquid System
3.2.3. Plate Screening of Lignin-Degrading Enzymes
3.3. Characterization of the Growth and Ligninolytic Enzyme Production of the MAP03 Strain in Submerged Fermentation under Different Conditions
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Maas, R.; Chaudhari, S. Adsorption and biological decolourization of azo dye Reactive Red 2 in semicontinuous anaerobic reactors. Process. Biochem. 2005, 40, 699–705. [Google Scholar] [CrossRef] [Green Version]
- Shindhal, T.; Rakholiya, P.; Varjani, S.; Pandey, A.; Ngo, H.H.; Guo, W.; Ng, H.Y.; Taherzadeh, M.J. A critical review on advances in the practices and perspectives for the treatment of dye industry wastewater. Bioengineered 2020, 12, 70–87. [Google Scholar] [CrossRef] [PubMed]
- Ali, N.; Hameed, A.; Ahmed, S.; Khan, A.G. Decolorization of structurally different textile dyes by Aspergillus niger SA1. World J. Microbiol. Biotechnol. 2007, 24, 1067–1072. [Google Scholar] [CrossRef]
- Ledakowicz, S.; Paździor, K. Recent Achievements in Dyes Removal Focused on Advanced Oxidation Processes Integrated with Biological Methods. Molecules 2021, 26, 870. [Google Scholar] [CrossRef] [PubMed]
- Lellis, B.; Fávaro-Polonio, C.Z.; Pamphile, J.A.; Polonio, J.C. Effects of textile dyes on health and the environment and bioremediation potential of living organisms. Biotechnol. Res. Innov. 2019, 3, 275–290. [Google Scholar] [CrossRef]
- Robinson, T.; Chandran, B.; Nigam, P. Studies on the production of enzymes by white-rot fungi for the decolourisation of textile dyes. Enzym. Microb. Technol. 2001, 29, 575–579. [Google Scholar] [CrossRef]
- Verma, P.; Baldrian, P.; Nerud, F. Decolorization of structurally different synthetic dyes using cobalt (II)/ascorbic acid/hydrogen peroxide system. Chemosphere 2002, 50, 975–979. [Google Scholar] [CrossRef]
- Baldrian, P.; Gabriel, J. Lignocellulose degradation byPleurotus ostreatusin the presence of cadmium. FEMS Microbiol. Lett. 2003, 220, 235–240. [Google Scholar] [CrossRef] [Green Version]
- Zhang, F.; Yediler, A.; Liang, X.; Kettrup, A. Effects of dye additives on the ozonation process and oxidation by-products: A comparative study using hydrolyzed C.I. Reactive Red 120. Dye. Pigment 2004, 60, 1–7. [Google Scholar] [CrossRef]
- Carmen, Z.; Daniel, S. Textile Organic Dyes—Characteristics, Polluting Effects and Separation/Elimination Procedures from Industrial Effluents—A Critical Overview; IntechOpen: Rijeka, Croatia, 2012. [Google Scholar] [CrossRef] [Green Version]
- Saxena, A.; Gupta, S. Bioefficacies of microbes for mitigation of Azo dyes in textile industry effluent: A review. BioResources 2020, 15, 9858–9881. [Google Scholar] [CrossRef]
- Pandey, A.K.; Gaur, V.K.; Udayan, A.; Varjani, S.; Kim, S.-H.; Wong, J.W. Biocatalytic remediation of industrial pollutants for environmental sustainability: Research needs and opportunities. Chemosphere 2021, 272, 129936. [Google Scholar] [CrossRef]
- Eichlerová, I.; Homolka, L.; Nerud, F. Evaluation of synthetic dye decolorization capacity in Ischnoderma resinosum. J. Ind. Microbiol. Biotechnol. 2006, 33, 759–766. [Google Scholar] [CrossRef] [PubMed]
- López, M.; Guisado, G.; Vargas-García, M.; Suárez-Estrella, F.; Moreno, J. Decolorization of industrial dyes by ligninolytic microorganisms isolated from composting environment. Enzym. Microb. Technol. 2006, 40, 42–45. [Google Scholar] [CrossRef]
- Sen, S.K.; Raut, S.; Bandyopadhyay, P.; Raut, S. Fungal decolouration and degradation of azo dyes: A review. Fungal Biol. Rev. 2016, 30, 112–133. [Google Scholar] [CrossRef]
- Kumari, S.; Naraian, R. Decolorization of synthetic brilliant green carpet industry dye through fungal co-culture technology. J. Environ. Manag. 2016, 180, 172–179. [Google Scholar] [CrossRef] [PubMed]
- Khan, S.A.; Mehmood, S.; Iqbal, A.; Hamayun, M. Industrial polluted soil borne fungi decolorize the recalcitrant azo dyes Synozol red HF–6BN and Synozol black B. Ecotoxicol. Environ. Saf. 2020, 206, 111381. [Google Scholar] [CrossRef] [PubMed]
- Ben, A.B.; Amutha, E.; Pushpalaksmi, E.; Jenson, S.J.; Annadurai, G. Physicochemical Characteristics, Identification of Fungi and Optimization of Different Parameters for Degradation of Dye from Tannery Effluent. J. Appl. Sci. Environ. Manag. 2020, 24, 1203–1208. [Google Scholar] [CrossRef]
- Syafiuddin, A.; Fulazzaky, M.A. Decolorization kinetics and mass transfer mechanisms of Remazol Brilliant Blue R dye mediated by different fungi. Biotechnol. Rep. 2020, 29, e00573. [Google Scholar] [CrossRef] [PubMed]
- Borchert, M.; Libra, J. Decolorization of reactive dyes by the white rot fungusTrametes versicolor in sequencing batch reactors. Biotechnol. Bioeng. 2001, 75, 313–321. [Google Scholar] [CrossRef]
- Jarosz-Wilkołazka, A.; Kochmańska-Rdest, J.; Malarczyk, E.; Wardas, W.; Leonowicz, A. Fungi and their ability to decolourize azo and anthraquinonic dyes. Enzym. Microb. Technol. 2002, 30, 566–572. [Google Scholar] [CrossRef]
- Martins, L.O.; Soares, C.; Pereira, M.M.; Teixeira, M.; Costa, T.; Jones, G.H.; Henriques, A. Molecular and Biochemical Characterization of a Highly Stable Bacterial Laccase That Occurs as a Structural Component of the Bacillus subtilis Endospore Coat. J. Biol. Chem. 2002, 277, 18849–18859. [Google Scholar] [CrossRef] [Green Version]
- Gill, P.K.; Arora, D.S.; Chander, M. Biodecolourization of azo and triphenylmethane dyes by Dichomitus squalens and Phlebiaitalic spp. J. Ind. Microbiol. Biotechnol. 2002, 28, 201–203. [Google Scholar] [CrossRef]
- Liu, W.; Chao, Y.; Yang, X.; Bao, H.; Qian, S. Biodecolorization of azo, anthraquinonic and triphenylmethane dyes by white-rot fungi and a laccase-secreting engineered strain. J. Ind. Microbiol. Biotechnol. 2004, 31, 127–132. [Google Scholar] [CrossRef]
- Bezalel, L.; Hadar, Y.; Fu, P.P.; Freeman, J.P.; Cerniglia, C.E. Initial Oxidation Products in the Metabolism of Pyrene, Anthracene, Fluorene, and Dibenzothiophene by the White Rot Fungus Pleurotus ostreatus. Appl. Environ. Microbiol. 1996, 62, 2554–2559. [Google Scholar] [CrossRef] [Green Version]
- Buckley, K.F.; Dobson, A.D. Extracellular ligninolytic enzyme production and polymeric dye decolourization in immobilized cultures of Chrysosporium lignorum CL1. Biotechnol. Lett. 1998, 20, 301–306. [Google Scholar] [CrossRef]
- Pointing, S. Feasibility of bioremediation by white-rot fungi. Appl. Microbiol. Biotechnol. 2001, 57, 20–33. [Google Scholar] [CrossRef]
- Hatvani, N.; Mécs, I. Effect of the nutrient composition on dye decolorisation and extracellular enzyme production by Lentinus edodes on solid medium. Enzym. Microb. Technol. 2002, 30, 381–386. [Google Scholar] [CrossRef]
- White, T.J.; Bruns, T.; Lee, S.; Taylor, J. Amplification and Direct Sequencing of Fungal Ribosomal RNA Genes for Phylogenetics in Book PCR Protocols; Academic Press: Cambridge, MA, USA, 1990; pp. 315–322. [Google Scholar]
- Gardes, M.; Bruns, T.D. ITS primers with enhanced specificity for basidiomycetes—Application to the identification of mycorrhizae and rusts. Mol. Ecol. 1993, 2, 113–118. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K.; Battistuzzi, F.U. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef] [PubMed]
- Janshekar, H.; Fiechter, A. Cultivation of Phanerochaete chrysosporium and production of lignin peroxidases in submerged stirred tank reactors. J. Biotechnol. 1988, 8, 97–112. [Google Scholar] [CrossRef]
- Kiiskinen, L.-L.; Ratto, M.; Kruus, K. Screening for novel laccase-producing microbes. J. Appl. Microbiol. 2004, 97, 640–646. [Google Scholar] [CrossRef] [PubMed]
- Téllez-Téllez, M.; Fernandez, F.; Montiel-González, A.M.; Sánchez, C.; Díaz-Godínez, G. Growth and laccase production by Pleurotus ostreatus in submerged and solid-state fermentation. Appl. Microbiol. Biotechnol. 2008, 81, 675–679. [Google Scholar] [CrossRef]
- Grandes-Blanco, A.I.; Díaz-Godínez, G.; Téllez-Téllez, M.; Delgado-Macuil, R.J.; Rojas-López, M.; Bibbins-Martínez, M.D. Ligninolytic activity patterns of Pleurotus ostreatus obtained by submerged fermentation in presence of 2,6-dimethoxyphenol and remazol brilliant blue r dye. Prep. Biochem. Biotechnol. 2013, 43, 468–480. [Google Scholar] [CrossRef] [PubMed]
- Have, R.T.; Hartmans, S.; Teunissen, P.J.; Field, J.A. Purification and characterization of two lignin peroxidase isozymes produced by Bjerkandera sp. strain BOS55. FEBS Lett. 1998, 422, 391–394. [Google Scholar] [CrossRef] [Green Version]
- Giardina, P.; Palmieri, G.; Fontanella, B.; Rivieccio, V.; Sannia, G. Manganese Peroxidase Isoenzymes Produced by Pleurotus ostreatus Grown on Wood Sawdust. Arch. Biochem. Biophys. 2000, 376, 171–179. [Google Scholar] [CrossRef]
- Salvachua, D.; Prieto, A.; Martinez, T.; Martinez, M.J. Characterization of a Novel Dye-Decolorizing Peroxidase (DyP)-Type Enzyme from Irpex lacteus and Its Application in Enzymatic Hydrolysis of Wheat Straw. Appl. Environ. Microbiol. 2013, 79, 4316–4324. [Google Scholar] [CrossRef] [Green Version]
- Pérez-Boada, M.; Ruiz-Dueñas, F.J.; Pogni, R.; Basosi, R.; Choinowski, T.; Martínez, M.J.; Piontek, K.; Martínez, A.T. Versatile Peroxidase Oxidation of High Redox Potential Aromatic Compounds: Site-directed Mutagenesis, Spectroscopic and Crystallographic Investigation of Three Long-Range Electron Transfer Pathways. J. Mol. Biol. 2005, 354, 385–402. [Google Scholar] [CrossRef] [PubMed]
- Morgulis, A.; Coulouris, G.; Raytselis, Y.; Madden, T.L.; Agarwala, R.; Schäffer, A.A. Database indexing for production MegaBLAST searches. Bioinformatics 2008, 24, 1757–1764. [Google Scholar] [CrossRef] [PubMed]
- Harms, H.; Schlosser, D.; Wick, L.Y. Untapped potential: Exploiting fungi in bioremediation of hazardous chemicals. Nat. Rev. Genet. 2011, 9, 177–192. [Google Scholar] [CrossRef]
- Sintakindi, A.; Ankamwar, B. Fungal biosorption as an alternative for the treatment of dyes in waste waters: A review. Environ. Technol. Rev. 2021, 10, 26–43. [Google Scholar] [CrossRef]
- Lu, Y.; Phillips, D.R.; Lu, L.; Hardin, I.R. Determination of the degradation products of selected sulfonated phenylazonaphthol dyes treated by white rot fungus Pleurotus ostreatus by capillary electrophoresis coupled with electrospray ionization ion trap mass spectrometry. J. Chromatogr. A 2008, 1208, 223–231. [Google Scholar] [CrossRef] [PubMed]
- Diwaniyan, S.; Kharb, D.; Raghukumar, C.; Kuhad, R.C. Decolorization of Synthetic Dyes and Textile Effluents by Basidiomycetous Fungi. Water Air Soil Pollut. 2009, 210, 409–419. [Google Scholar] [CrossRef]
- Singh, S.N.; Mishra, S.; Jauhari, N. Degradation of Anthroquinone Dyes Stimulated by Fungi. In Microbial Degradation of Synthetic Dyes in Wastewaters; Springer: Berlin/Heidelberg, Germany, 2014; pp. 333–356. [Google Scholar]
- Padmavathy, S.; Sandhya, S.; Swaminathan, K.; Subrahmanyam, Y.V.; Chakrabarti, T.; Kaul, S.N. Aerobic decolorization of reactive azo dyes in presence of various cosubstrates. Chem. Biochem. 2003, 17, 147–151. [Google Scholar]
- Kasinath, A.; Novotný, Č.; Svobodová, K.; Patel, K.C.; Šašek, V. Decolorization of synthetic dyes by Irpex lacteus in liquid cultures and packed-bed bioreactor. Enzym. Microb. Technol. 2003, 32, 167–173. [Google Scholar] [CrossRef]
- Vyas, R.M.; Molitoris, H.P. Involvement of an extracellular H2O2-dependent ligninolytic activity of the white rot fungus Pleurotus ostreatus in the decolorization of remazol brilliant blue R. Appl. Environ. Microbiol. 1995, 61, 3919–3927. [Google Scholar] [CrossRef] [Green Version]
- Wang, Y.; Yu, J. Adsorption and degradation of synthetic dyes on the mycelium of Trametes versicolor. Water Sci. Technol. 1998, 38. [Google Scholar] [CrossRef]
- Kim, S.J.; Shoda, M. Purification and Characterization of a Novel Peroxidase from Geotrichum candidum Dec 1 Involved in Decolorization of Dyes. Appl. Environ. Microbiol. 1999, 65, 1029–1035. [Google Scholar] [CrossRef] [Green Version]
- Balan, D.S.; Monteiro, R.T. Decolorization of textile indigo dye by ligninolytic fungi. J. Biotechnol. 2001, 89, 141–145. [Google Scholar] [CrossRef]
- Nyanhongo, G.S.; Gomes, J.; Gübitz, G.; Zvauya, R.; Read, J.; Steiner, W. Decolorization of textile dyes by laccases from a newly isolated strain of Trametes modesta. Water Res. 2002, 36, 1449–1456. [Google Scholar] [CrossRef]
- Riegas-Villalobos, A.; Martínez-Morales, F.; Tinoco-Valencia, R.; Serrano-Carreón, L.; Bertrand, B.; Trejo-Hernández, M.R. Efficient removal of azo-dye Orange II by fungal biomass absorption and laccase enzymatic treatment. 3 Biotech. 2020, 10, 1–10. [Google Scholar] [CrossRef]
- Fu, Y.; Viraraghavan, T. Fungal decolorization of dye wastewaters: A review. Bioresour. Technol. 2001, 79, 251–262. [Google Scholar] [CrossRef]
- Eichlerová, I.; Baldrian, P. Ligninolytic Enzyme Production and Decolorization Capacity of Synthetic Dyes by Saprotrophic White Rot, Brown Rot, and Litter Decomposing Basidiomycetes. J. Fungi 2020, 6, 301. [Google Scholar] [CrossRef] [PubMed]
- Merino-Restrepo, A.; Mejía, F.; Velásquez-Quintero, C.; Hormaza-Anaguano, A. Evaluation of several white-rot fungi for the decolorization of a binary mixture of anionic dyes and characterization of the residual biomass as potential organic soil amendment. J. Environ. Manag. 2019, 254, 109805. [Google Scholar] [CrossRef] [PubMed]
- Durán, N.; Esposito, E. Potential applications of oxidative enzymes and phenoloxidase-like compounds in wastewater and soil treatment: A review. Appl. Catal. B Environ. 2000, 28, 83–99. [Google Scholar] [CrossRef]
- Asgher, M.; Bhatti, H.N.; Ashraf, M.; Legge, R. Recent developments in biodegradation of industrial pollutants by white rot fungi and their enzyme system. Biodegradation 2008, 19, 771–783. [Google Scholar] [CrossRef]
- Majeau, J.-A.; Brar, S.K.; Tyagi, R.D. Laccases for removal of recalcitrant and emerging pollutants. Bioresour. Technol. 2010, 101, 2331–2350. [Google Scholar] [CrossRef]
- Dhouib, A.; Hamza, M.; Zouari, H.; Mechichi, T.; Labat, M.; Martinez, M.; Sayadi, S. Autochthonous fungal strains with high ligninolytic activities from Tunisian biotopes. Afr. J. Biotechnol. 2005, 4, 431–436. [Google Scholar] [CrossRef]
- González, I.; Nava, S.; Díaz, R.; Garrido, V.; Tecuitl, S.; Bibbins, M. Determination of oxidases activity produced by Oxyporus latemarginatus grown in submerged fermentation in the presence and the absence of the yellow azo dye. Rev. Latinoam. Ambiente Cienc. 2015, 6, 72–88. [Google Scholar]
- Azin, E.; Moghimi, H. Efficient mycosorption of anionic azo dyes by Mucor circinelloides: Surface functional groups and removal mechanism study. J. Environ. Chem. Eng. 2018, 6, 4114–4123. [Google Scholar] [CrossRef]
- Li, X.; Lan, X.; Feng, X.; Luan, X.; Cao, X.; Cui, Z. Biosorption capacity of Mucor circinelloides bioaugmented with Solanum nigrum L. for the cleanup of lead, cadmium and arsenic. Ecotoxicol. Environ. Saf. 2021, 212, 112014. [Google Scholar] [CrossRef]
- Karp, S.G.; Faraco, V.; Amore, A.; Letti, L.A.J.; Soccol, V.T.; Soccol, C.R. Statistical Optimization of Laccase Production and Delignification of Sugarcane Bagasse by Pleurotus ostreatus in Solid-State Fermentation. BioMed Res. Int. 2015, 2015, 1–8. [Google Scholar] [CrossRef] [Green Version]
- Thiribhuvanamala, G.; Kalaiselvi, G.; Parthasarathy, S.; Madhavan, S.; Prakasam, V. Extracellular secretion of lignocellulolytic enzymes by diversewhite rot asidiomycetes fungi. Ann. Phytomedicine Int. J. 2017, VI, 20–29. [Google Scholar] [CrossRef]
- Xu, X.; Xu, Z.; Shi, S.; Lin, M. Lignocellulose degradation patterns, structural changes, and enzyme secretion by Inonotus obliquus on straw biomass under submerged fermentation. Bioresour. Technol. 2017, 241, 415–423. [Google Scholar] [CrossRef]
- Garrido-Bazán, V.; Téllez-Téllez, M.; Herrera-Estrella, A.; Díaz-Godínez, G.; Nava-Galicia, S.; Villalobos-López, M.; Arroyo-Becerra, A.; Bibbins-Martínez, M. Effect of textile dyes on activity and differential regulation of laccase genes from Pleurotus ostreatus grown in submerged fermentation. AMB Express 2016, 6, 93. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cuamatzi-Flores, J.; Esquivel-Naranjo, E.U.; Nava-Galicia, S.; Munguia, A.L.; Arroyo-Becerra, A.; Villalobos-López, M.A.; Bibbins-Martínez, M. Differential regulation of Pleurotus ostreatus dye peroxidases gene expression in response to dyes and potential application of recombinant Pleos-DyP1 in decolorization. PLoS ONE 2019, 14, e0209711. [Google Scholar] [CrossRef] [PubMed] [Green Version]



| Colour Index Name | Chemical Structure | Chemical Class | λMax (nm) | Dye Content (%) | Solubility |
|---|---|---|---|---|---|
| Indigo |  | Indigoid | 298 | 96 | Insoluble |
| Remazol (reactive) Yellow145 | 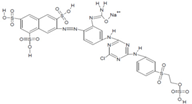 | Azo | 300 | Soluble in H2O 80 g/L | |
| Remazol (reactive) Red3B |  | Azo | 500 | 100 | No data available |
| Remazol (reactive) Brilliant Blue R |  | Anthraquinone | 600 | 50 | 1 mg/mL H2O |
| Disperse Black 1 |  | Azo | 300 | 79.3 mg/L H2O2 soluble in EtOH | |
| Disperse Yellow 3 | 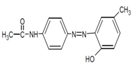 | Azo | 400 | 30 | 1 mg/mL formic acid:95% EtOH(1:1) |
| Disperse Red 19 | 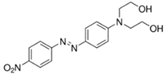 | Azo | 495 | 97 | 0.01 g/L in 50% EtOH |
| Acetyl yellow G (AYG) |  | Azo | 390 | 95 | No data available |
| Strain | Fermentation Period (Days) | |||
|---|---|---|---|---|
| 8 | 16 | 24 | * | |
| MAPO1 | Ng | 75.8 | 75.8 | b,c,d |
| MAPO2 | 16.6 | 25.0 | 25.0 | b,c,d,e |
| MAPO3 | 100.0 | 100.0 | 100.0 | a |
| MAPO4 | 59.1 | 66.6 | 66.6 | a,b,c |
| MAPO5 | 31.6 | 33.3 | 33.3 | c,d,e |
| MAPO6 | Ng | 8.3 | 8.3 | e |
| Strains | Remazol Brilliant Blue R | Remazol Red | Remazol Yellow | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Fermentation Period (Days) | ||||||||||||
| 8 | 16 | 24 | * | 8 | 16 | 24 | * | 8 | 16 | 24 | * | |
| MAPO1 | 35.9 | 38.5 | 45.1 | c | 25.0 | 25.0 | 24.5 | a,b | 0.0 | 10.6 | 15.5 | b |
| MAPO2 | Ng | 3.5 | 42.9 | d | 22.5 | 27.5 | 24.7 | a,b | 0.0 | 2.6 | 20.0 | b |
| MAPO3 | 73.2 | 82.0 | 100.0 | b | 25.0 | 25.0 | 22.5 | a,b | 6.6 | 6.6 | 15.5 | b |
| MAPO4 | 38.5 | 43.4 | 73.2 | b,c | 25.0 | 20.0 | 25.0 | a,b | 22.2 | 28.8 | 33.3 | a |
| MAPO5 | 74.1 | 95.6 | 100.0 | a | 2.50 | 22.5 | 25.0 | b | Ng | 20.0 | 10.6 | b |
| MAPO6 | Ng | Ng | 12.2 | d | 27.50 | 35.0 | 37.0 | a | 2.2 | 6.6 | 11.1 | b |
| Strains | Disperse Red | Disperse Yellow | Disperse Black | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Fermentation Period (Days) | ||||||||||||
| 8 | 16 | 24 | * | 8 | 16 | 24 | * | 8 | 16 | 24 | * | |
| MAPO1 | 58.3 | 44.1 | 47.2 | b | 2.5 | 12.5 | 60.0 | a,b | 27.8 | 32.3 | 50.0 | b,c |
| MAPO2 | 40.5 | 41.6 | 69.4 | a,b | 2.5 | 12.5 | 75.0 | a,b | 0.0 | 7.6 | 69.2 | b,c |
| MAPO3 | 64.7 | 65.2 | 75.0 | a,b | 2.5 | 12.5 | 68.7 | a,b | 25.7 | 45.3 | 65.3 | a,b,c |
| MAPO4 | 61.1 | 66.6 | 69.7 | a,b | 2.5 | 10.0 | 45.2 | b | 30.7 | 31.5 | 42.6 | b,c |
| MAPO5 | 58.8 | 58.3 | 61.1 | a,b | 2.5 | 27.5 | 80.0 | a | 28.0 | 42.3 | 88.4 | a,b |
| MAPO6 | 15.5 | 58.3 | 69.2 | b | 2.5 | 5.0 | 47.5 | b | Ng | Ng | Ng | c |
| Strains | Laccase | Phenoloxidase | Tyrosinase | Peroxidase | |
|---|---|---|---|---|---|
| Guaiacol | O-ANISIDINE | Tannic ACID | p-CRESOL | Pyrogallol and H2O2 | |
| MAPO1 | − | − | − | + | + |
| MAPO2 | − | − | − | + | + |
| MAPO3 | + | + | − | − | + |
| MAPO4 | + | + | − | − | + |
| MAPO5 | + | + | − | − | + |
| MAPO6 | − | − | − | + | + |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Juárez-Hernández, J.; Castillo-Hernández, D.; Pérez-Parada, C.; Nava-Galicia, S.; Cuervo-Parra, J.A.; Surian-Cruz, E.; Díaz-Godínez, G.; Sánchez, C.; Bibbins-Martínez, M. Isolation of Fungi from a Textile Industry Effluent and the Screening of Their Potential to Degrade Industrial Dyes. J. Fungi 2021, 7, 805. https://doi.org/10.3390/jof7100805
Juárez-Hernández J, Castillo-Hernández D, Pérez-Parada C, Nava-Galicia S, Cuervo-Parra JA, Surian-Cruz E, Díaz-Godínez G, Sánchez C, Bibbins-Martínez M. Isolation of Fungi from a Textile Industry Effluent and the Screening of Their Potential to Degrade Industrial Dyes. Journal of Fungi. 2021; 7(10):805. https://doi.org/10.3390/jof7100805
Chicago/Turabian StyleJuárez-Hernández, Juvenal, Dalia Castillo-Hernández, Cristhian Pérez-Parada, Soley Nava-Galicia, Jaime Alioscha Cuervo-Parra, Edy Surian-Cruz, Gerardo Díaz-Godínez, Carmen Sánchez, and Martha Bibbins-Martínez. 2021. "Isolation of Fungi from a Textile Industry Effluent and the Screening of Their Potential to Degrade Industrial Dyes" Journal of Fungi 7, no. 10: 805. https://doi.org/10.3390/jof7100805
APA StyleJuárez-Hernández, J., Castillo-Hernández, D., Pérez-Parada, C., Nava-Galicia, S., Cuervo-Parra, J. A., Surian-Cruz, E., Díaz-Godínez, G., Sánchez, C., & Bibbins-Martínez, M. (2021). Isolation of Fungi from a Textile Industry Effluent and the Screening of Their Potential to Degrade Industrial Dyes. Journal of Fungi, 7(10), 805. https://doi.org/10.3390/jof7100805







