Replicative Aging in Pathogenic Fungi
Abstract
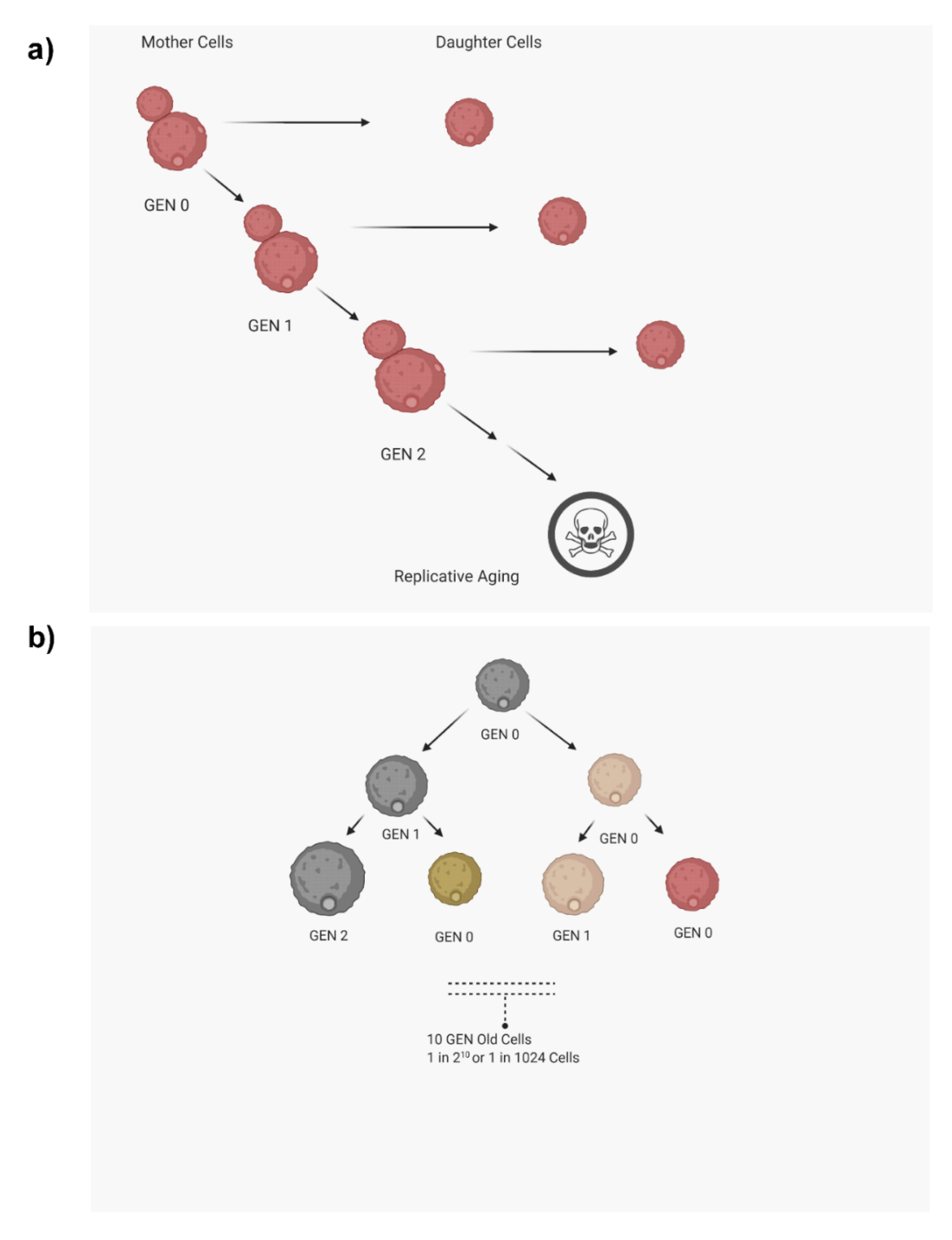
1. Introduction
2. Methods to Analyze Replicative Life Span (RLS)
2.1. Microdissection Method
- The method is labor intensive and time consuming. As mentioned above, RLS analysis can take as long as four weeks. Further, this method creates a discontinuity in the analysis, as at the end of the day the culture plates are refrigerated to slow down the replication time. This can cause unnecessary stress on the cells resulting in erroneous RLS determination.
- Since this method is time consuming and often lasts one to four weeks, the culture media can degrade or get contaminated, causing defects in cell growth affecting RLS.
- Only a small number of cells (around 30–40) can be used to analyze RLS. RLS varies from cell to cell within the same strain, which is referred to as stochasticity of life span. Hence, since a large number of cells are required to accurately analyze the stochasticity of RLS, this method is not ideal for that analysis.
- This method requires proper identification of mother and daughter cells after each budding event. For the first few generations, both mother and daughter cells are similar in size. This makes identifying daughter cells difficult, causing erroneous RLS analysis.
2.2. High-Throughput Microfluidics Method
- Extreme longevity RLS is difficult to assess with the help of this device. The device may get clogged due to a cell overgrowing during prolonged runs. Further, the method requires a continuous supply of fresh media through syringes. The volumes of syringes are limited, and if the cells have excessively long life spans, the device may run out of fresh media. As a result, buckets may get clogged.
- The microscope is sensitive to any liquid spills, so the system needs to be continuously monitored to prevent any leaks.
- Any extreme phenotypic change (hyphal formation or titan cell) will clog the system and abort the recording of RLS on that specific cell.
- There are still no reliable programs that permit automatic determination of bud count and RLS computation. Budding events are therefore still counted manually by reviewing the video images.
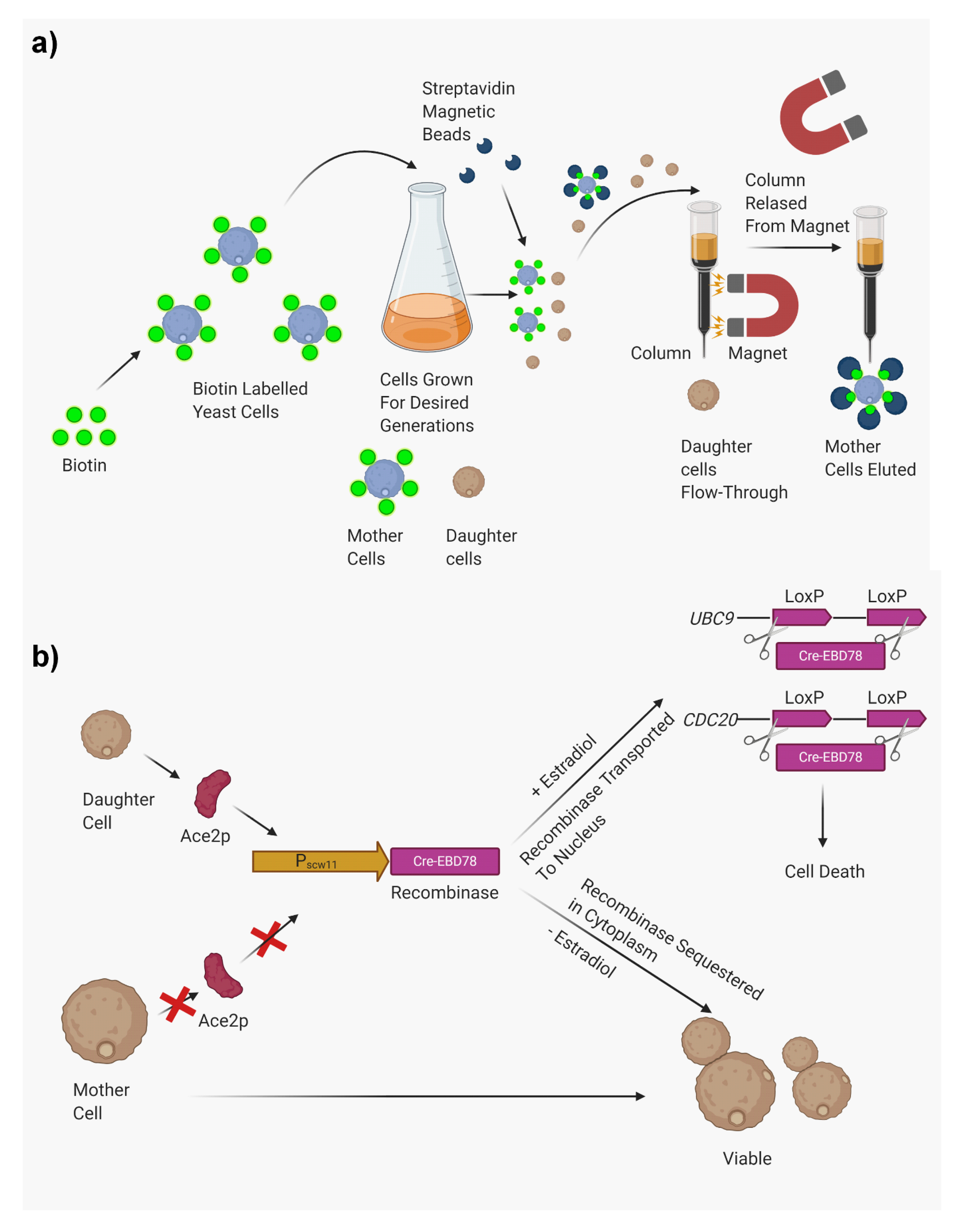
2.3. Biotin Label and Magnetic/FACS Sorting
- Encapsulated fungi such as C. neoformans may shield the biotin-labeled cell surface proteins, which ultimately impairs the binding of streptavidin-conjugated beads, leading to low yield of older mother cells.
- Expense is considerable, especially when yield remains low.
- In S. cerevisiae, the age of the isolated population can be easily verified. S. cerevisiae produces permanent scars in the cell wall each time they produce daughter cells. These scars can be visualized by staining with calcofluor white, and the generational age of the isolated population can be determined by counting the bud scars. However, cell wall scars heal in C. neoformans and any nonhealed scars are obscured by capsules. In C. glabrata, the bud scar count is more reliable.
2.4. Centrifugation
2.4.1. Sucrose Gradient Centrifugation
- The technique is labor intensive and requires multiple rounds of manipulation to isolate cells of the desired age.
2.4.2. Centrifugal Elutriation
- Elutriation does not often deliver a pure population if used repeatedly (cells older than 10 generations).
- Further, elutriation is not reliable for fungal populations that exhibit cell size heterogeneity at baseline.
2.5. Mother Enrichment Program (Genetic Mutation)
- The technique requires genetic manipulations that need to be introduced in the desired strains. This can be difficult in organisms such as C. neoformans, which are more difficult to transform than S. cerevisiae.
- Mutations in MEP strains can arise that can prevent the selection of the transformants.
- The technique is labor intensive.
- Purity has to be verified. In S. cerevisiae, it was found that the M-phase-arrested daughter cells do not senesce immediately, were metabolically active, and continued growth in the arrested state for at least 24 h. These cells produced 6.9 progenies before lysing in the presence of estradiol.
3. Phenotypic Variations Associated with Aging
3.1. Phenotypic Differences between Mother and Daughter Cells in Unicellular Yeasts
3.2. Antifungal Resistance in Older Fungal Cells
3.3. In Vitro and In Vivo Virulence
4. Future Directions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Steffen, K.K.; Kennedy, B.K.; Kaeberlein, M. Measuring replicative life span in the budding yeast. J. Vis. Exp. 2009, 28, e1209. [Google Scholar] [CrossRef] [PubMed]
- He, C.; Zhou, C.; Kennedy, B.K. The yeast replicative aging model. Biochim. Biophys. Acta. Mol. Basis Dis. 2018, 1864, 2690–2696. [Google Scholar] [CrossRef] [PubMed]
- Azbarova, A.V.; Galkina, K.V.; Sorokin, M.I.; Severin, F.F.; Knorre, D.A. The contribution of Saccharomyces cerevisiae replicative age to the variations in the levels of Trx2p, Pdr5p, Can1p and Idh isoforms. Sci. Rep. 2017, 7, 13220. [Google Scholar] [CrossRef] [PubMed]
- Okada, M.; Kusunoki, S.; Ishibashi, Y.; Kito, K. Proteomics analysis for asymmetric inheritance of preexisting proteins between mother and daughter cells in budding yeast. Genes Cells Devoted Mol. Cell. Mech. 2017, 22, 591–601. [Google Scholar] [CrossRef]
- Leupold, S.; Hubmann, G.; Litsios, A.; Meinema, A.C.; Takhaveev, V.; Papagiannakis, A.; Niebel, B.; Janssens, G.; Siegel, D.; Heinemann, M. Saccharomyces cerevisiae goes through distinct metabolic phases during its replicative lifespan. Elife 2019, 8, e41046. [Google Scholar] [CrossRef]
- Bouklas, T.; Pechuan, X.; Goldman, D.L.; Edelman, B.; Bergman, A.; Fries, B.C. Old Cryptococcus neoformans cells contribute to virulence in chronic cryptococcosis. mBio 2013, 4. [Google Scholar] [CrossRef]
- Coody, T.K.; Hughes, A.L. Advancing the aging biology toolkit. Elife 2018, 7, e42976. [Google Scholar] [CrossRef]
- Orner, E.P.; Zhang, P.; Jo, M.C.; Bhattacharya, S.; Qin, L.; Fries, B.C. High-Throughput Yeast Aging Analysis for Cryptococcus (HYAAC) microfluidic device streamlines aging studies in Cryptococcus neoformans. Commun. Biol. 2019, 2, 256. [Google Scholar] [CrossRef]
- O’Laughlin, R.; Jin, M.; Li, Y.; Pillus, L.; Tsimring, L.S.; Hasty, J.; Hao, N. Advances in quantitative biology methods for studying replicative aging in Saccharomyces cerevisiae. Transl. Med. Aging 2020, 4, 151–160. [Google Scholar] [CrossRef]
- Chen, K.L.; Crane, M.M.; Kaeberlein, M. Microfluidic technologies for yeast replicative lifespan studies. Mech. Ageing Dev. 2017, 161, 262–269. [Google Scholar] [CrossRef]
- Zhang, Y.; Luo, C.; Zou, K.; Xie, Z.; Brandman, O.; Ouyang, Q.; Li, H. Single cell analysis of yeast replicative aging using a new generation of microfluidic device. PLoS ONE 2012, 7, e48275. [Google Scholar] [CrossRef] [PubMed]
- Crane, M.M.; Clark, I.B.; Bakker, E.; Smith, S.; Swain, P.S. A microfluidic system for studying ageing and dynamic single-cell responses in budding yeast. PLoS ONE 2014, 9, e100042. [Google Scholar] [CrossRef] [PubMed]
- Ryley, J.; Pereira-Smith, O.M. Microfluidics device for single cell gene expression analysis in Saccharomyces cerevisiae. Yeast 2006, 23, 1065–1073. [Google Scholar] [CrossRef]
- Jo, M.C.; Liu, W.; Gu, L.; Dang, W.; Qin, L. High-throughput analysis of yeast replicative aging using a microfluidic system. Proc. Natl. Acad. Sci. USA 2015, 112, 9364–9369. [Google Scholar] [CrossRef] [PubMed]
- Liu, P.; Acar, M. The generational scalability of single-cell replicative aging. Sci. Adv. 2018, 4, eaao4666. [Google Scholar] [CrossRef]
- Bhattacharya, S.; Fries, B.C. Enhanced Efflux Pump Activity in Old Candida glabrata Cells. Antimicrob. Agents Chemother. 2018, 62. [Google Scholar] [CrossRef]
- Bhattacharya, S.; Holowka, T.; Orner, E.P.; Fries, B.C. Gene Duplication Associated with Increased Fluconazole Tolerance in Candida auris cells of Advanced Generational Age. Sci. Rep. 2019, 9, 5052. [Google Scholar] [CrossRef]
- Bouklas, T.; Alonso-Crisostomo, L.; Szekely, T., Jr.; Diago-Navarro, E.; Orner, E.P.; Smith, K.; Munshi, M.A.; Del Poeta, M.; Balazsi, G.; Fries, B.C. Generational distribution of a Candida glabrata population: Resilient old cells prevail, while younger cells dominate in the vulnerable host. PLoS Pathog. 2017, 13, e1006355. [Google Scholar] [CrossRef]
- Sinclair, D.; Mills, K.; Guarente, L. Aging in Saccharomyces cerevisiae. Annu. Rev. Microbiol. 1998, 52, 533–560. [Google Scholar] [CrossRef]
- Fu, X.H.; Meng, F.L.; Hu, Y.; Zhou, J.Q. Candida albicans, a distinctive fungal model for cellular aging study. Aging Cell 2008, 7, 746–757. [Google Scholar] [CrossRef]
- Jain, N.; Cook, E.; Xess, I.; Hasan, F.; Fries, D.; Fries, B.C. Isolation and characterization of senescent Cryptococcus neoformans and implications for phenotypic switching and pathogenesis in chronic cryptococcosis. Eukaryot. Cell 2009, 8, 858–866. [Google Scholar] [CrossRef] [PubMed]
- Woldringh, C.L.; Fluiter, K.; Huls, P.G. Production of senescent cells of Saccharomyces cerevisiae by centrifugal elutriation. Yeast 1995, 11, 361–369. [Google Scholar] [CrossRef] [PubMed]
- Lindstrom, D.L.; Gottschling, D.E. The mother enrichment program: A genetic system for facile replicative life span analysis in Saccharomyces cerevisiae. Genetics 2009, 183, 413–422. [Google Scholar] [CrossRef] [PubMed]
- Ghavidel, A.; Baxi, K.; Prusinkiewicz, M.; Swan, C.; Belak, Z.R.; Eskiw, C.H.; Carvalho, C.E.; Harkness, T.A. Rapid Nuclear Exclusion of Hcm1 in Aging Saccharomyces cerevisiae Leads to Vacuolar Alkalization and Replicative Senescence. Genes Genomes Genet. 2018, 8, 1579–1592. [Google Scholar] [CrossRef]
- Chen, K.; Shen, W.; Zhang, Z.; Xiong, F.; Ouyang, Q.; Luo, C. Age-dependent decline in stress response capacity revealed by proteins dynamics analysis. Sci. Rep. 2020, 10, 15211. [Google Scholar] [CrossRef]
- Sarnoski, E.A.; Liu, P.; Acar, M. A High-Throughput Screen for Yeast Replicative Lifespan Identifies Lifespan-Extending Compounds. Cell Rep. 2017, 21, 2639–2646. [Google Scholar] [CrossRef]
- Pal, S.; Postnikoff, S.D.; Chavez, M.; Tyler, J.K. Impaired cohesion and homologous recombination during replicative aging in budding yeast. Sci. Adv. 2018, 4, eaaq0236. [Google Scholar] [CrossRef]
- Henderson, K.A.; Hughes, A.L.; Gottschling, D.E. Mother-daughter asymmetry of pH underlies aging and rejuvenation in yeast. Elife 2014, 3, e03504. [Google Scholar] [CrossRef]
- Orner, E.P.; Bhattacharya, S.; Kalenja, K.; Hayden, D.; Del Poeta, M.; Fries, B.C. Cell Wall-Associated Virulence Factors Contribute to Increased Resilience of Old Cryptococcus neoformans Cells. Front. Microbiol. 2019, 10, 2513. [Google Scholar] [CrossRef]
- Lord, P.G.; Wheals, A.E. Variability in individual cell cycles of Saccharomyces cerevisiae. J. Cell Sci. 1981, 50, 361–376. [Google Scholar]
- Janssens, G.E.; Veenhoff, L.M. Evidence for the hallmarks of human aging in replicatively aging yeast. Microb. Cell 2016, 3, 263–274. [Google Scholar] [CrossRef] [PubMed]
- Sorokin, M.I.; Knorre, D.A.; Severin, F.F. Early manifestations of replicative aging in the yeast Saccharomyces cerevisiae. Microb. Cell 2014, 1, 37–42. [Google Scholar] [CrossRef] [PubMed]
- Zadrag-Tecza, R.; Kwolek-Mirek, M.; Bartosz, G.; Bilinski, T. Cell volume as a factor limiting the replicative lifespan of the yeast Saccharomyces cerevisiae. Biogerontology 2009, 10, 481–488. [Google Scholar] [CrossRef] [PubMed]
- Neumann, G.; Veeranagouda, Y.; Karegoudar, T.B.; Sahin, O.; Mausezahl, I.; Kabelitz, N.; Kappelmeyer, U.; Heipieper, H.J. Cells of Pseudomonas putida and Enterobacter sp. adapt to toxic organic compounds by increasing their size. Extremophiles 2005, 9, 163–168. [Google Scholar] [CrossRef] [PubMed]
- Aldridge, B.B.; Fernandez-Suarez, M.; Heller, D.; Ambravaneswaran, V.; Irimia, D.; Toner, M.; Fortune, S.M. Asymmetry and aging of mycobacterial cells lead to variable growth and antibiotic susceptibility. Science 2012, 335, 100–104. [Google Scholar] [CrossRef]
- Knorre, D.A.; Kulemzina, I.A.; Sorokin, M.I.; Kochmak, S.A.; Bocharova, N.A.; Sokolov, S.S.; Severin, F.F. Sir2-dependent daughter-to-mother transport of the damaged proteins in yeast is required to prevent high stress sensitivity of the daughters. Cell Cycle 2010, 9, 4501–4505. [Google Scholar] [CrossRef][Green Version]
- Delaney, J.R.; Sutphin, G.L.; Dulken, B.; Sim, S.; Kim, J.R.; Robison, B.; Schleit, J.; Murakami, C.J.; Carr, D.; An, E.H.; et al. Sir2 deletion prevents lifespan extension in 32 long-lived mutants. Aging Cell 2011, 10, 1089–1091. [Google Scholar] [CrossRef]
- Yang, J.; Dungrawala, H.; Hua, H.; Manukyan, A.; Abraham, L.; Lane, W.; Mead, H.; Wright, J.; Schneider, B.L. Cell size and growth rate are major determinants of replicative lifespan. Cell Cycle 2011, 10, 144–155. [Google Scholar] [CrossRef]
- Bouklas, T.; Masone, L.; Fries, B.C. Differences in Sirtuin Regulation in Response to Calorie Restriction in Cryptococcus neoformans. J. Fungi 2018, 4, 26. [Google Scholar] [CrossRef]
- Bouklas, T.; Jain, N.; Fries, B.C. Modulation of Replicative Lifespan in Cryptococcus neoformans: Implications for Virulence. Front. Microbiol. 2017, 8, 98. [Google Scholar] [CrossRef]
- Li, L.; Zaragoza, O.; Casadevall, A.; Fries, B.C. Characterization of a flocculation-like phenotype in Cryptococcus neoformans and its effects on pathogenesis. Cell Microbiol. 2006, 8, 1730–1739. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Feldmesser, M.; Kress, Y.; Casadevall, A. Dynamic changes in the morphology of Cryptococcus neoformans during murine pulmonary infection. Microbiology 2001, 147, 2355–2365. [Google Scholar] [CrossRef] [PubMed]
- Fries, B.C.; Bhattacharya, S.; Orner, E.; Bouklas, T. Replicative Aging in Candida auris Has an Effect on Antifungal Resistance. In Open Forum Infectious Diseases, US; Oxford University Press: Oxford, UK, 2017; Volume 4, p. S115. [Google Scholar] [CrossRef][Green Version]
- Sudbery, P.E. Growth of Candida albicans hyphae. Nat. Rev. Microbiol. 2011, 9, 737–748. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Rubio, R.; de Oliveira, H.C.; Rivera, J.; Trevijano-Contador, N. The Fungal Cell Wall: Candida, Cryptococcus, and Aspergillus Species. Front. Microbiol. 2019, 10, 2993. [Google Scholar] [CrossRef] [PubMed]
- Powell, C.D.; Quain, D.E.; Smart, K.A. Chitin scar breaks in aged Saccharomyces cerevisiae. Microbiology 2003, 149, 3129–3137. [Google Scholar] [CrossRef] [PubMed]
- Zaragoza, O.; Telzak, A.; Bryan, R.A.; Dadachova, E.; Casadevall, A. The polysaccharide capsule of the pathogenic fungus Cryptococcus neoformans enlarges by distal growth and is rearranged during budding. Mol. Microbiol. 2006, 59, 67–83. [Google Scholar] [CrossRef]
- McFadden, D.; Zaragoza, O.; Casadevall, A. The capsular dynamics of Cryptococcus neoformans. Trends Microbiol. 2006, 14, 497–505. [Google Scholar] [CrossRef]
- Frieman, M.B.; McCaffery, J.M.; Cormack, B.P. Modular domain structure in the Candida glabrata adhesin Epa1p, a beta1,6 glucan-cross-linked cell wall protein. Mol. Microbiol. 2002, 46, 479–492. [Google Scholar] [CrossRef]
- Cormack, B.P.; Ghori, N.; Falkow, S. An adhesin of the yeast pathogen Candida glabrata mediating adherence to human epithelial cells. Science 1999, 285, 578–582. [Google Scholar] [CrossRef]
- Delaney, J.R.; Murakami, C.; Chou, A.; Carr, D.; Schleit, J.; Sutphin, G.L.; An, E.H.; Castanza, A.S.; Fletcher, M.; Goswami, S.; et al. Dietary restriction and mitochondrial function link replicative and chronological aging in Saccharomyces cerevisiae. Exp. Gerontol. 2013, 48, 1006–1013. [Google Scholar] [CrossRef]
- McFaline-Figueroa, J.R.; Vevea, J.; Swayne, T.C.; Zhou, C.; Liu, C.; Leung, G.; Boldogh, I.R.; Pon, L.A. Mitochondrial quality control during inheritance is associated with lifespan and mother-daughter age asymmetry in budding yeast. Aging Cell 2011, 10, 885–895. [Google Scholar] [CrossRef] [PubMed]
- Hughes, A.L.; Gottschling, D.E. An early age increase in vacuolar pH limits mitochondrial function and lifespan in yeast. Nature 2012, 492, 261–265. [Google Scholar] [CrossRef] [PubMed]
- Aguilaniu, H.; Gustafsson, L.; Rigoulet, M.; Nystrom, T. Asymmetric inheritance of oxidatively damaged proteins during cytokinesis. Science 2003, 299, 1751–1753. [Google Scholar] [CrossRef] [PubMed]
- Erjavec, N.; Larsson, L.; Grantham, J.; Nystrom, T. Accelerated aging and failure to segregate damaged proteins in Sir2 mutants can be suppressed by overproducing the protein aggregation-remodeling factor Hsp104p. Genes Dev. 2007, 21, 2410–2421. [Google Scholar] [CrossRef]
- Lin, S.S.; Manchester, J.K.; Gordon, J.I. Enhanced gluconeogenesis and increased energy storage as hallmarks of aging in Saccharomyces cerevisiae. J. Biol. Chem. 2001, 276, 36000–36007. [Google Scholar] [CrossRef]
- Lesur, I.; Campbell, J.L. The transcriptome of prematurely aging yeast cells is similar to that of telomerase-deficient cells. Mol. Biol. Cell 2004, 15, 1297–1312. [Google Scholar] [CrossRef]
- Grzelak, A.; Macierzynska, E.; Bartosz, G. Accumulation of oxidative damage during replicative aging of the yeast Saccharomyces cerevisiae. Exp. Gerontol. 2006, 41, 813–818. [Google Scholar] [CrossRef]
- Nosanchuk, J.D.; Gomez, B.L.; Youngchim, S.; Diez, S.; Aisen, P.; Zancope-Oliveira, R.M.; Restrepo, A.; Casadevall, A.; Hamilton, A.J. Histoplasma capsulatum synthesizes melanin-like pigments in vitro and during mammalian infection. Infect. Immun. 2002, 70, 5124–5131. [Google Scholar] [CrossRef]
- Nosanchuk, J.D.; Rosas, A.L.; Lee, S.C.; Casadevall, A. Melanisation of Cryptococcus neoformans in human brain tissue. Lancet 2000, 355, 2049–2050. [Google Scholar] [CrossRef]
- Youngchim, S.; Hay, R.J.; Hamilton, A.J. Melanization of Penicillium marneffei in vitro and in vivo. Microbiology 2005, 151, 291–299. [Google Scholar] [CrossRef]
- Youngchim, S.; Morris-Jones, R.; Hay, R.J.; Hamilton, A.J. Production of melanin by Aspergillus fumigatus. J. Med. Microbiol. 2004, 53, 175–181. [Google Scholar] [CrossRef] [PubMed]
- Nosanchuk, J.D.; Casadevall, A. The contribution of melanin to microbial pathogenesis. Cell Microbiol. 2003, 5, 203–223. [Google Scholar] [CrossRef] [PubMed]
- Zhu, X.; Williamson, P.R. Role of laccase in the biology and virulence of Cryptococcus neoformans. FEMS Yeast Res. 2004, 5, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Aisen, P.; Casadevall, A. Cryptococcus neoformans melanin and virulence: Mechanism of action. Infect. Immun. 1995, 63, 3131–3136. [Google Scholar] [CrossRef] [PubMed]
- van Duin, D.; Casadevall, A.; Nosanchuk, J.D. Melanization of Cryptococcus neoformans and Histoplasma capsulatum reduces their susceptibilities to amphotericin B and caspofungin. Antimicrob. Agents Chemother. 2002, 46, 3394–3400. [Google Scholar] [CrossRef] [PubMed]
- Nosanchuk, J.D.; Casadevall, A. Budding of melanized Cryptococcus neoformans in the presence or absence of L-dopa. Microbiology 2003, 149, 1945–1951. [Google Scholar] [CrossRef] [PubMed]
- Chrissian, C.; Camacho, E.; Kelly, J.E.; Wang, H.; Casadevall, A.; Stark, R.E. Solid-state NMR spectroscopy identifies three classes of lipids in Cryptococcus neoformans melanized cell walls and whole fungal cells. J. Biol. Chem. 2020, 295, 15083–15096. [Google Scholar] [CrossRef] [PubMed]
- Bhattacharya, S.; Sae-Tia, S.; Fries, B.C. Candidiasis and Mechanisms of Antifungal Resistance. Antibiotics 2020, 9, 312. [Google Scholar] [CrossRef]

| Organism | Centrifugation | Microdissection | Biotin Label/Magnetic/FACS | High Throughput | Mother Enrichment |
|---|---|---|---|---|---|
| Saccharomyces cerevisiae | YES [19] | YES [19] | YES [19] | YES [12] | YES [23,28] |
| Cryptococcus neoformans | YES [21] | YES [6] | YES [21] | YES [29] | NO |
| Candida albicans | YES [20] | NO | NO | NO | NO |
| Candida glabrata | YES [18] | YES [18] | YES [18] | NO | NO |
| Candida auris | NO | YES [17] | YES [17] | NO | NO |
| S. cerevisiae | C. neoformans | C. albicans | C. glabrata | C. auris | |
|---|---|---|---|---|---|
| Cell size | Increases | Increases | Increases | Increases | Increases |
| Cell wall | Weakens | Thickens | Thickens | Thickens | Thickens |
| Capsule | N/A | Increases | N/A | N/A | N/A |
| Cell shape | Oval | Round | Filamentous | Pseudofilamentous | Round |
| Cytosolic components | Increased glycogen, oxidized proteins | Not tested | Increased glycogen, oxidized proteins | Not tested | Not tested |
| Melanin | N/A | Increases | Not tested | Not tested | Not tested |
| Arrest Cell phenotype | Unbudded | Unbudded, Budded | Hyphal | Unbudded, budded, Pseudohyphal | Unbudded |
| Antiphagocytic ability | Not tested | Increases | Increases | Increases | Increases |
| Hydrogen peroxide tolerance | Not tested | Increases | Increases | Increases | Increases |
| Antifungal resistance | Not tested | Increases | Increases | Increases | Increases |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bhattacharya, S.; Bouklas, T.; Fries, B.C. Replicative Aging in Pathogenic Fungi. J. Fungi 2021, 7, 6. https://doi.org/10.3390/jof7010006
Bhattacharya S, Bouklas T, Fries BC. Replicative Aging in Pathogenic Fungi. Journal of Fungi. 2021; 7(1):6. https://doi.org/10.3390/jof7010006
Chicago/Turabian StyleBhattacharya, Somanon, Tejas Bouklas, and Bettina C. Fries. 2021. "Replicative Aging in Pathogenic Fungi" Journal of Fungi 7, no. 1: 6. https://doi.org/10.3390/jof7010006
APA StyleBhattacharya, S., Bouklas, T., & Fries, B. C. (2021). Replicative Aging in Pathogenic Fungi. Journal of Fungi, 7(1), 6. https://doi.org/10.3390/jof7010006






