Revealing of Non-Cultivable Bacteria Associated with the Mycelium of Fungi in the Kerosene-Degrading Community Isolated from the Contaminated Jet Fuel
Abstract
1. Introduction
2. Materials and Methods
2.1. Sampling and Cultures Isolation
2.2. Morphological Features of Micromycetes
2.3. Growth Ability on the TS-1 Jet Fuel
2.4. Express Assay for the Detection of 16S/18S rRNA Genes
2.5. Next-Generation 16S rRNA Amplicon Sequencing (16S rRNA Metabarcoding) and Bioinformatic Analysis
3. Results
3.1. Micromycetes Isolated from the TS-1 Jet Fuel
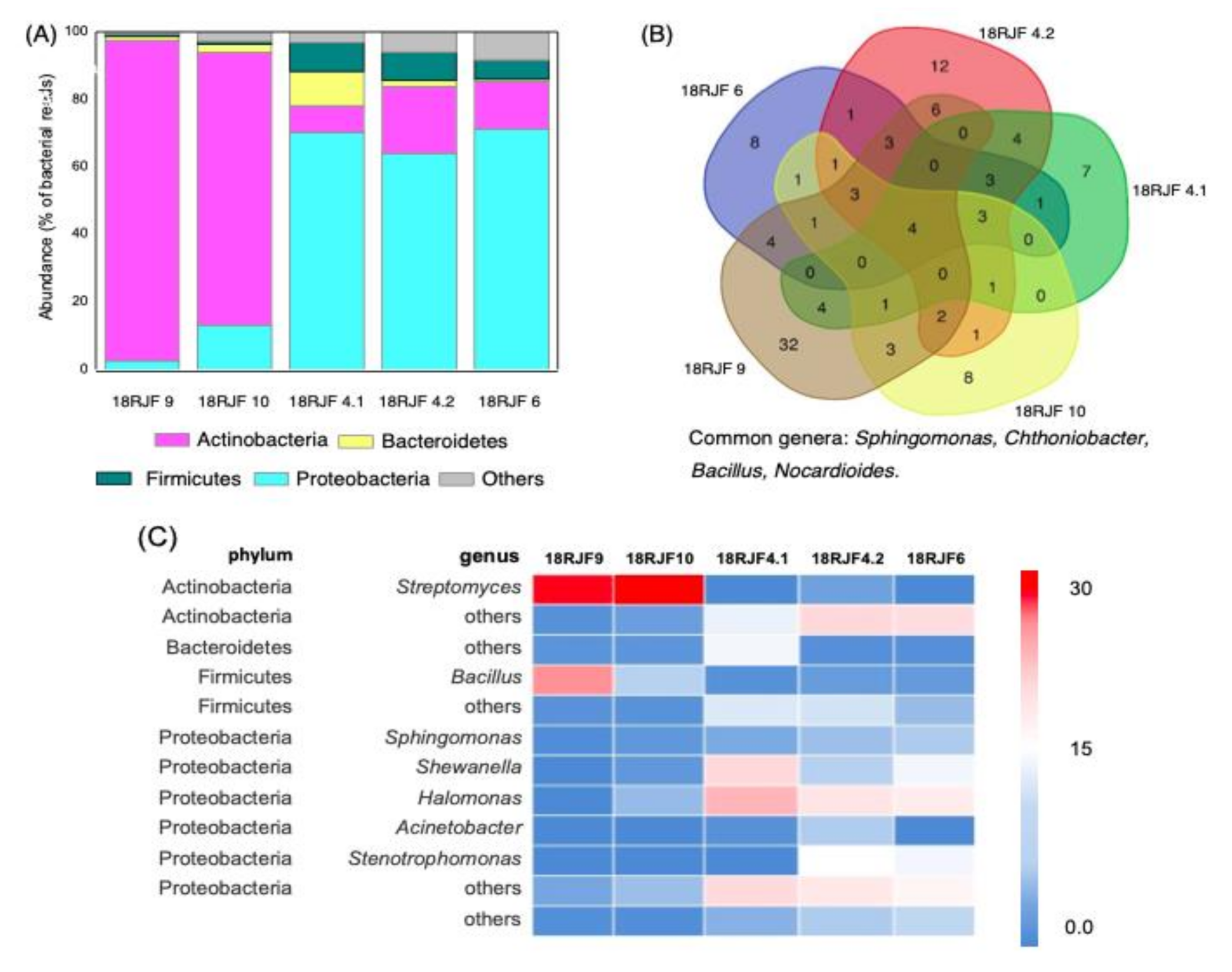
3.2. Bacterial Component of the Micromycetes’ Biomass Isolated from Jet Fuel Communities Revealed by 16S rRNA Data
3.3. The Growth Capacity of Isolated Communities on the TS-1 Jet Fuel
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Iverson, W.P. Microbial corrosion of metals. In Advances in Applied Microbiology; Academic Press: Cambridge, MA, USA, 1987; Volume 32, pp. 1–36. [Google Scholar]
- Gerasimenko, A.A.; Yampol’skaya, T.E. Exfoliation corrosion of aluminum alloys II. Protection methods, their effectiveness and improvement. Prot. Met. 2000, 36, 397–407. [Google Scholar] [CrossRef]
- Itah, A.Y.; Brooks, A.A.; Ogar, B.O.; Okure, A.B. Biodegradation of international jet A-1 aviation fuel by microorganisms isolated from aircraft tank and joint hydrant storage systems. Bull. Environ. Contam. Toxicol. 2009, 83, 318–327. [Google Scholar] [CrossRef]
- Buddie, A.G.; Bridge, P.D.; Kelley, J.; Ryan, M.J. Candida keroseneae sp. nov., a novel contaminant of aviation kerosene. Lett. Appl. Microbiol. 2011, 52, 70–75. [Google Scholar] [CrossRef] [PubMed]
- Soriano, A.U.; Martins, L.F.; Ventura, E.S.D.; de Landa, F.H.G.T.; da Auraujo Valoni, E.; Faria, F.R.D.; Ferreira, R.F.; Clara, M.; Fallera, K.; Valério, R.R.; et al. Microbiological aspects of biodiesel and biodiesel/diesel blends biodeterioration. Int. Biodeterior. Biodegrad. 2015, 99, 102–114. [Google Scholar] [CrossRef]
- Passman, F.J. Microbial contamination and its control in fuels and fuel systems since 1980—A review. Int. Biodeterior. Biodegrad. 2013, 81, 88–104. [Google Scholar] [CrossRef]
- Rodríguez-Rodríguez, C.E.; Rodríguez, E.; Blanco, R.; Cordero, I.; Segura, D. Fungal contamination of stored automobile-fuels in a tropical environment. J. Environ. Sci. 2010, 22, 1595–1601. [Google Scholar] [CrossRef]
- Varjani, S.J. Microbial degradation of petroleum hydrocarbons. Bioresour. Technol. 2017, 223, 277–286. [Google Scholar] [CrossRef]
- Koshlaf, E.; Ball, A. Soil bioremediation approaches for petroleum hydrocarbon polluted environments. Aims Microbiol. 2017, 3, 25–49. [Google Scholar] [CrossRef]
- Leahy, J.G.; Colwell, R.R. Microbial degradation of hydrocarbons in the environment. Microbiol. Mol. Biol. Rev. 1990, 54, 305–315. [Google Scholar] [CrossRef]
- Bücker, F.; de Moura, T.M.; da Cunha, M.E.; de Quadros, P.D.; Beker, S.A.; Cazarolli, J.C.; Caramão, E.B.; Frazzon, A.P.G.; Bento, F.M. Evaluation of the deteriogenic microbial community using qPCR, n-alkanes and FAMEs biodegradation in diesel, biodiesel and blends (B5, B10, and B50) during storage. Fuel 2018, 233, 911–917. [Google Scholar] [CrossRef]
- Das, N.; Chandran, P. Microbial degradation of petroleum hydrocarbon contaminants: An overview. Biotechnol. Res. Int. 2011, 2011, 941810. [Google Scholar] [CrossRef] [PubMed]
- Shabir, G.; Afzal, M.; Anwar, F.; Tahseen, R.; Khalid, Z.M. Biodegradation of kerosene in soil by a mixed bacterial culture under different nutrient conditions. Int. Biodeterior. Biodegrad. 2008, 61, 161–166. [Google Scholar] [CrossRef]
- Boychenko, S.; Shkilnuk, I.; Turchak, V. The problems of biopollution with jet fuels and the way of achieving solution. Transport. 2008, 23, 253–257. [Google Scholar] [CrossRef][Green Version]
- Sørensen, G.; Pedersen, D.V.; Nørgaard, A.K.; Sørensen, K.B.; Nygaard, S.D. Microbial growth studies in biodiesel blends. Bioresour. Technol. 2011, 102, 5259–5264. [Google Scholar] [CrossRef] [PubMed]
- Shkilniuk, I.; Boichenko, S. Methodically organizational principles of biological stability providing of aviation fuels. Pr. Inst. Lotnictwa 2014, 237, 76–83. [Google Scholar] [CrossRef]
- Bücker, F.; Barbosa, C.S.; Quadros, P.D.; Bueno, M.K.; Fiori, P.; te Huang, C.; Frazzon, A.P.G.; Ferrão, M.F.; de Oliveira Camargo, F.A.; Bento, F.M. Fuel biodegradation and molecular characterization of microbial biofilms in stored diesel/biodiesel blend B10 and the effect of biocide. Int. Biodeterior. Biodegrad. 2014, 95, 346–355. [Google Scholar] [CrossRef]
- Chiciudean, I.; Mereuţă, I.; Ionescu, R.; Vassu, T.; Tănase, A.M.; Stoica, I. Jet A-1 Bacterial contamination: A case study of cultivable bacteria diversity, alkane degradation and biofilm formation. Pol. J. Environ. Stud. 2019, 28, 4139–4146. [Google Scholar]
- Atlas, R.M. Microbial degradation of petroleum hydrocarbons: An environmental perspective. Microbiol. Rev. 1981, 45, 180–209. [Google Scholar] [CrossRef]
- Chaillan, F.; Le Flèche, A.; Bury, E.; Phantavong, Y.H.; Grimont, P.; Saliot, A.; Oudot, J. Identification and biodegradation potential of tropical aerobic hydrocarbon-degrading microorganisms. Res. Microbiol. 2004, 155, 587–595. [Google Scholar] [CrossRef]
- De Azambuja, A.O.; Bücker, F.; de Quadros, P.D.; Zhalnina, K.; Dias, R.; Vacaro, B.B.; Bento, F.M. Microbial community composition in Brazilian stored diesel fuel of varying sulfur content, using high-throughput sequencing. Fuel 2017, 189, 340–349. [Google Scholar] [CrossRef]
- Pereira, E.; Napp, A.P.; Allebrandt, S.; Barbosa, R.; Reuwsaat, J.; Lopes, W.; Maria do Carmo, R.P. Biodegradation of aliphatic and polycyclic aromatic hydrocarbons in seawater by autochthonous microorganisms. Int. Biodeterior. Biodegrad. 2019, 145, 104789. [Google Scholar] [CrossRef]
- Hasan, I. Biodegradation of kerosene by Aspergillus niger and Rhizopus stolonifer. Appl. Environ. Microbiol. 2014, 2, 31–36. [Google Scholar]
- Lotfinasabasl, S.; Gunale, V.R.; Rajurkar, N.S. Assessment of petroleum hydrocarbon degradation from soil and tarball by fungi. Biosci. Discov. 2012, 3, 186–192. [Google Scholar]
- Dhar, K.; Dutta, S.; Anwar, M.N. Biodegradation of petroleum hydrocarbon by indigenous fungi isolated from ship breaking yards of Bangladesh. Int. Res. J. Biol. Sci. 2014, 3, 22–30. [Google Scholar]
- Mnif, S.; Chamkha, M.; Sayadi, S. Isolation and characterization of Halomonas sp. strain C2SS100, a hydrocarbon-degrading bacterium under hypersaline conditions. J. Appl. Microbiol. 2009, 107, 785–794. [Google Scholar] [CrossRef]
- Ameen, F.; Moslem, M.; Hadi, S.; Al-Sabri, A.E. Biodegradation of diesel fuelhydrocarbons by mangrove fungi from Red Sea Coast of Saudi Arabia. Saudi J. Biol. Sci. 2016, 23, 211–218. [Google Scholar]
- Boonchan, S.; Britz, M.L.; Stanley, G.A. Degradation and mineralization of high-molecular-weight polycyclic aromatic hydrocarbons by defined fungal-bacterial cocultures. Appl. Environ. Microbiol. 2000, 66, 1007–1019. [Google Scholar] [CrossRef]
- White, J.; Gilbert, J.; Hill, G.; Hill, E.; Huse, S.M.; Weightman, A.J.; Mahenthiralingam, E. Culture-independent analysis of bacterial fuel contamination provides insight into the level of concordance with the standard industry practice of aerobic cultivation. Appl. Environ. Microbiol. 2011, 77, 4527–4538. [Google Scholar] [CrossRef]
- Boer, W.D.; Folman, L.B.; Summerbell, R.C.; Boddy, L. Living in a fungal world: Impact of fungi on soil bacterial niche development. FEMS Microbiol. Rev. 2005, 29, 795–811. [Google Scholar]
- Furuno, S.; Päzolt, K.; Rabe, C.; Neu, T.R.; Harms, H.; Wick, L.Y. Fungal mycelia allow chemotactic dispersal of polycyclic aromatic hydrocarbon-degrading bacteria in water-unsaturated systems. Environ. Microbiol. 2010, 12, 1391–1398. [Google Scholar]
- Wick, L.Y.; Furuno, S.; Harms, H. Fungi as transport vectors for contaminants and contaminant-degrading bacteria. In Handbook of Hydrocarbon and Lipid Microbiology; Springer: Berlin/Heidelberg, Germany, 2010; pp. 1555–1561. [Google Scholar]
- Partida-Martinez, L.P.; Hertweck, C. Pathogenic fungus harbours endosymbiotic bacteria for toxin production. Nature 2005, 437, 884–888. [Google Scholar] [CrossRef] [PubMed]
- Wick, L.Y.; Remer, R.; Würz, B.; Reichenbach, J.; Braun, S.; Schäfer, F.; Harms, H. Effect of fungal hyphae on the access of bacteria to phenanthrene in soil. Environ. Sci. Technol. 2007, 41, 500–505. [Google Scholar] [CrossRef] [PubMed]
- Furuno, S.; Remer, R.; Chatzinotas, A.; Harms, H.; Wick, L.Y. Use of mycelia as paths for the isolation of contaminant-degrading bacteria from soil. Microb. Biotechnol. 2012, 5, 142–148. [Google Scholar] [CrossRef] [PubMed]
- Banitz, T.; Johst, K.; Wick, L.Y.; Schamfuß, S.; Harms, H.; Frank, K. Highways versus pipelines: Contributions of two fungal transport mechanisms to efficient bioremediation. Environ. Microbiol. Rep. 2013, 5, 211–218. [Google Scholar] [CrossRef]
- Schamfuß, S.; Neu, T.R.; Harms, H.; van der Meer, J.R.; Tecon, R.; Wick, L.Y. Mycelial networks enhance the bioavailability of PAH in water unsaturated environments. Environ. Sci. Technol. 2013, 47, 6908–6915. [Google Scholar] [CrossRef]
- Wick, L.; De Munain, A.; Springael, D.; Harms, H. Responses of mycobacterium sp. LB501T to the low bioavailability of solid anthracene. Appl. Microbiol. Biotechnol. 2002, 58, 378–385. [Google Scholar]
- Ferradji, F.Z.; Mnif, S.; Badis, A.; Rebbani, S.; Fodil, D.; Eddouaouda, K.; Sayadi, S. Naphthalene and crude oil degradation by biosurfactant producing Streptomyces spp. isolated from Mitidja plain soil (North of Algeria). Int. Biodeterior. Biodegrad. 2014, 86, 300–308. [Google Scholar] [CrossRef]
- Rapp, P.; Backhaus, S. Formation of extracellular lipases by filamentous fungi, yeasts, and bacteria. Enzym. Microb. Technol. 1992, 14, 938–943. [Google Scholar] [CrossRef]
- Thom, C.; Church, M.B. The Aspergilli; Williams and Wilkins Co.: Baltimore, MD, USA, 1926; p. 272. [Google Scholar]
- Matlakowska, R.; Sklodowska, A. The culturable bacteria isolated from organic-rich black shale potentially useful in biometallurgical procedures. J. Aapplied Microbiol. 2009, 107, 858–866. [Google Scholar] [CrossRef]
- Evans, C.G.T.; Herbert, D.; Tempest, D.W. Chapter XIII the continuous cultivation of microorganisms: 2, construction of a chemostat. In Methods in Microbiology; Academic Press: Cambridge, MA, USA, 1970; Volume 2, pp. 277–327. [Google Scholar]
- Lobakova, E.; Vasilieva, S.; Kashcheeva, P.; Ivanova, E.; Dolnikova, G.; Chekanov, K.; Dedov, A. New bio-hybrid materials for bioremoval of crude oil spills from marine waters. Int. Biodeterior. Biodegrad. 2016, 108, 99–107. [Google Scholar] [CrossRef]
- Shapiro, T.; Dolnikova, G.; Nemtseva, N.; Sanjieva, D.; Lobakova, E. Identification and physiological characterisationtics of athe consortium of hydrocarbon-oxidizing bacteria of oil and oilpetroleum products. J. Microbiol. Epidemiol. Immunobiol. 2018, 107–113. [Google Scholar] [CrossRef]
- Ivanova, E.A.; Lobakova, E.S.; Idiatulov, R.K.; Shapiro, T.N.; Sandzhieva, D.A.; Kuznetsova, O.V.; Zaitseva, Y.u.N.; Dzhabrailova, K.h.S.; Dedov, A.G. Biocomposite materials for purification of aqueous media contaminated with hydrocarbons. Pet. Chem. 2019, 59, 420–426. [Google Scholar] [CrossRef]
- Gorelova, O.A.; Kosevich, I.A.; Baulina, O.I.; Fedorenko, T.A.; Torshkhoeva, A.Z.; Lobakova, E.S. Associations between the White Sea invertebrates and oxygen-evolving phototrophic microorganisms. Mosc. Univ. Biol. Sci. Bull. 2009, 64, 16–22. [Google Scholar] [CrossRef]
- Raper, K.B.; Fennel, D.I. The Genus Aspergillus; Williams & Wilkins: Baltimore, MD, USA, 1965; p. 686. [Google Scholar]
- Raper, K.B.; Thom, C.; Fennell, D.I. A Manual of the Penicillia; Hefner Publishing Co.: New York, NY, USA, 1949; p. 875. [Google Scholar]
- Pitt, G.I. A Laboratory Guide to Common Penicillium Species, 2nd ed.; CSIRO Food Research Laboratory: North Ryde, NSW, Australia, 1968; p. 188. [Google Scholar]
- Klich, M. Identification of Common Aspergillus Species; CBS: New York, NY, USA, 2002; p. 116. [Google Scholar]
- Samson, R.A.; Frisvard, J.C. Penicillium subgenus Penicillium: New taxonomic schemes, mycotoxins and other extrolites. Stud. Mycol. 2004, 49, 251. [Google Scholar]
- Yilmaz, N.; Houbraken, J.; Hoekstra, E.S.; Frisvad, J.C.; Visagie, C.M.; Samson, R.A. Delimitation and characterisation of Talaromyces purpurogenus and related species. Pers. Mol. Phylogeny Evol. Fungi 2012, 29, 39–54. [Google Scholar] [CrossRef] [PubMed]
- Yilmaz, N.; Visagie, C.M.; Houbraken, J.; Frisvad, J.C.; Samson, R.A. Polyphasic taxonomy of the genus Talaromyces. Stud. Mycol. 2014, 78, 175–341. [Google Scholar] [CrossRef] [PubMed]
- Chen, A.J.; Frisvad, J.C.; Sun, B.D.; Varga, J.; Kocsubé, S.; Dijksterhuis, J.; Kim, D.H.; Hong, S.B.; Houbraken, J.; Samson, R. Aspergillus section Nidulantes (formerly Emericella): Polyphasic taxonomy, chemistry and biology. Stud. Mycol. 2016, 84, 1–118. [Google Scholar] [CrossRef]
- Vasilieva, A.A.; Chekunova, L.N.; Polyakova, A.V. Effect of temperature on growth and viability of Hormoconis resinae and Phialophora sp. developing in the aviation fuel. Mycol. Phytopatologiya 2009, 43, 312–316. [Google Scholar]
- Wang, Y.; Tian, R.M.; Gao, Z.M.; Bougouffa, S.; Qian, P.-Y. Optimal eukaryotic 18S and universal 16S/18S ribosomal RNA primers and their application in a study of symbiosis. PLoS ONE 2014, 9, e90053. [Google Scholar] [CrossRef]
- Chekanov, K.; Kublanovskaya, A.; Lobakova, E. Eukaryotic sequences in the 16Sr RNA metagenomic dataset of algal–bacterial consortia of the White Sea coastal zone. J. Eukaryot. Microbiol. 2019, 66, 853–856. [Google Scholar] [CrossRef]
- Bates, S.T.; Berg-lyons, D.; Caporaso, J.G.; Walters, W.A.; Knight, R.; Fierer, N. Examining the global distribution of dominant archaeal populations in soil. ISME J. 2010, 5, 908–917. [Google Scholar] [CrossRef] [PubMed]
- Kublanovskaya, A.; Chekanov, K.; Solovchenko, A.; Lobakova, E. Cyanobacterial diversity in the algal–bacterial consortia from Subarctic regions: New insights from the rock baths at White Sea Coast. Hydrobiologia 2019, 830, 17–31. [Google Scholar] [CrossRef]
- Caporaso, J.G.; Kuczynski, J.; Stombaugh, J.; Bittinger, K.; Bushman, F.D.; Costello, E.K.; Fierer, N.; Peña, A.G.; Goodrich, J.K.; Gordon, J.I.; et al. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 2010, 7, 335–336. [Google Scholar] [CrossRef] [PubMed]
- Altschul, S.F.; Madden, T.L.; Schäffer, A.A.; Zhang, J.; Zhang, Z.; Miller, W.; Lipman, D.J. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 1997, 25, 3389–3402. [Google Scholar] [CrossRef] [PubMed]
- Huse, S.M.; Welch, D.B.M.; Voorhis, A.; Shipunova, A.; Morrison, H.G.; Eren, A.M.; Sogin, M.L. VAMPS: A website for visualization and analysis of microbial population structures. BMC Bioinform. 2014, 15, 41. [Google Scholar] [CrossRef] [PubMed]
- Beccati, A.; Gerken, J.; Quast, C.; Yilmaz, P.; Glöckner, F.O. SILVA tree viewer: Interactive web browsing of the SILVA phylogenetic guide trees. BMC Bioinform. 2017, 18, 433. [Google Scholar] [CrossRef]
- Gaylarde, C.C.; Bento, F.M.; Kelley, J. Microbial contamination of stored hydrocarbon fuels and its control. Rev. Microbiol. 1999, 30, 1–10. [Google Scholar] [CrossRef]
- Rauch, M.E.; Graef, H.W.; Rozenzhak, S.M.; Jones, S.E.; Bleckmann, C.A.; Kruger, R.L.; Naik, R.R.; Stone, M.O. Characterization of microbial contamination in United States Air Force aviation fuel tanks. J. Ind. Microbiol. Biotechnol. 2006, 33, 29–36. [Google Scholar] [CrossRef]
- Yemashova, N.A.; Murygina, V.P.; Zhukov, D.V.; Zakharyantz, A.A.; Gladchenko, M.A.; Appanna, V.; Kalyuzhnyi, S.V. Biodeterioration of crude oil and oil derived products: A review. Rev. Environ. Sci. Bio Technol. 2007, 6, 315–337. [Google Scholar] [CrossRef]
- Martin-Sanchez, P.M.; Becker, R.; Gorbushina, A.A.; Toepel, J. An improved test for the evaluation of hydrocarbon degradation capacities of diesel-contaminating microorganisms. Int. Biodeterior. Biodegrad. 2018, 129, 89–94. [Google Scholar]
- Korshunova, T.; Chetverikov, S.; Bakaeva, M.; Kuzina, E.; Rafikova, G.; Chetverikova, D.; Loginov, O. Microorganisms in the elimination of oil pollution consequences. Appl. Biochem. Microbiol. 2019, 55, 344–354. [Google Scholar] [CrossRef]
- Mitchell, R.; Alexander, M. Lysis of soil fungi by bacteria. Can. J. Microbiol. 1963, 9, 169–177. [Google Scholar] [CrossRef]
- Balachandran, C.; Duraipandiyan, V.; Balakrishna, K.; Ignacimuthu, S. Petroleum and polycyclic aromatic hydrocarbons (PAHs) degradation and naphthalene metabolism in Streptomyces sp. (ERI-CPDA-1) isolated from oil contaminated soil. Bioresour. Technol. 2012, 112, 83–90. [Google Scholar] [CrossRef] [PubMed]
- Liu, R.; Zhang, Y.; Ding, R.; Li, D.; Gao, Y.; Yang, M. Comparison of archaeal and bacterial community structures in heavily oil-contaminated and pristine soils. J. Biosci. Bioeng. 2009, 108, 400–407. [Google Scholar] [CrossRef]
- Tapilatu, Y.H.; Grossi, V.; Acquaviva, M.; Militon, C.; Bertrand, J.C.; Cuny, P. Isolation of hydrocarbon-degrading extremely halophilic archaea from an uncontaminated hypersaline pond (Camargue, France). Extremophiles 2010, 14, 225–231. [Google Scholar] [CrossRef]
- Le Borgne, S.; Paniagua, D.; Vazquez-Duhalt, R. Biodegradation of organic pollutants by halophilic bacteria and archaea. J. Mol. Microbiol. Biotechnol. 2008, 15, 74–92. [Google Scholar] [CrossRef]
- Baoune, H.; El Hadj-Khelil, A.O.; Pucci, G.; Sineli, P.; Loucif, L.; Polti, M.A. Petroleum degradation by endophytic Streptomyces spp. isolated from plants grown in contaminated soil of southern Algeria. Ecotoxicol. Environ. Saf. 2018, 147, 602–609. [Google Scholar] [CrossRef]
- Chen, J.; Huang, P.T.; Zhang, K.Y.; Ding, F.R. Isolation of biosurfactant producers, optimization and properties of biosurfactant produced by Acinetobacter sp. from petroleum-contaminated soil. J. Appl. Microbiol. 2012, 112, 660–671. [Google Scholar] [CrossRef]
- Yanan, W.; Yingying, L.; Jianliang, X.; Ke, S.; Yu, G.; Xiaolong, X. Exploring the degradation potential of Halomonas bacteria from oil-contaminated marine environment. China Pet. Process. Petrochem. Technol. 2018, 20, 91–98. [Google Scholar]
- Bianciotto, V.; Bandi, C.; Minerdi, D.; Sironi, M.; Tichy, H.V.; Bonfante, P. An obligately endosymbiotic mycorrhizal fungus itself harbors obligately intracellular bacteria. Appl. Environ. Microbiol. 1996, 62, 3005–3010. [Google Scholar]
- Pion, M.; Spangenberg, J.E.; Simon, A.; Bindschedler, S.; Flury, C.; Chatelain, A.; Bshary, R.; Job, D.; Junier, P. Bacterial farming by the fungus Morchella crassipes. Proc. R. Soc. B Biol. Sci. 2013, 280, 20132242. [Google Scholar] [CrossRef] [PubMed]
- Haq, I.U.; Graupner, K.; Nazir, R.; van Elsas, J.D. The genome of the fungal-interactive soil bacterium Burkholderia terrae BS001—A plethora of outstanding interactive capabilities unveiled. Genome Biol. Evol. 2014, 6, 1652–1668. [Google Scholar] [CrossRef] [PubMed]
- Abbasian, F.; Lockington, R.; Mallavarapu, M.; Naidu, R. A comprehensive review of aliphatic hydrocarbon biodegradation by bacteria. Appl. Biochem. Biotechnol. 2015, 176, 670–699. [Google Scholar] [CrossRef] [PubMed]
- Young, D.; Rice, J.; Martin, R.; Lindquist, E.; Lipzen, A.; Grigoriev, I.; Hibbett, D. Degradation of bunker C fuel oil by white-rot fungi in sawdust cultures suggests potential applications in bioremediation. PLoS ONE 2015, 10, e0130381. [Google Scholar] [CrossRef]
- Johnsen, A.R.; Wick, L.Y.; Harms, H. Principles of microbial PAH-degradation in soil. Environ. Pollut. 2005, 133, 71–84. [Google Scholar] [CrossRef]
- Seneviratne, G.; Zavahir, J.S.; Bandara, W.M.M.S.; Weerasekara, M.L.M.A.W. Fungal-bacterial biofilms: Their development for novel biotechnological applications. World J. Microbiol. Biotechnol. 2008, 24, 739–743. [Google Scholar] [CrossRef]
- Costerton, J.W.; Lewandowski, Z.; DeBeer, D.; Caldwell, D.; Korber, D.; James, G. Biofilms, the customized microniche. J. Bacteriol. 1994, 176, 2137. [Google Scholar] [CrossRef]
- Herath, H.M.L.I.; Rajapaksha, A.U.; Vithanage, M.; Seneviratne, G. Developed fungal–bacterial biofilms as a novel tool for bioremoval of hexavelant chromium from wastewater. Chem. Ecol. 2014, 30, 418–427. [Google Scholar] [CrossRef]
- Pandit, A.; Adholeya, A.; Cahill, D.; Brau, L.; Kochar, M. Microbial biofilms in nature: Unlocking their potential for agricultural applications. J. Appl. Microbiol. 2020, 129, 199–211. [Google Scholar] [CrossRef]
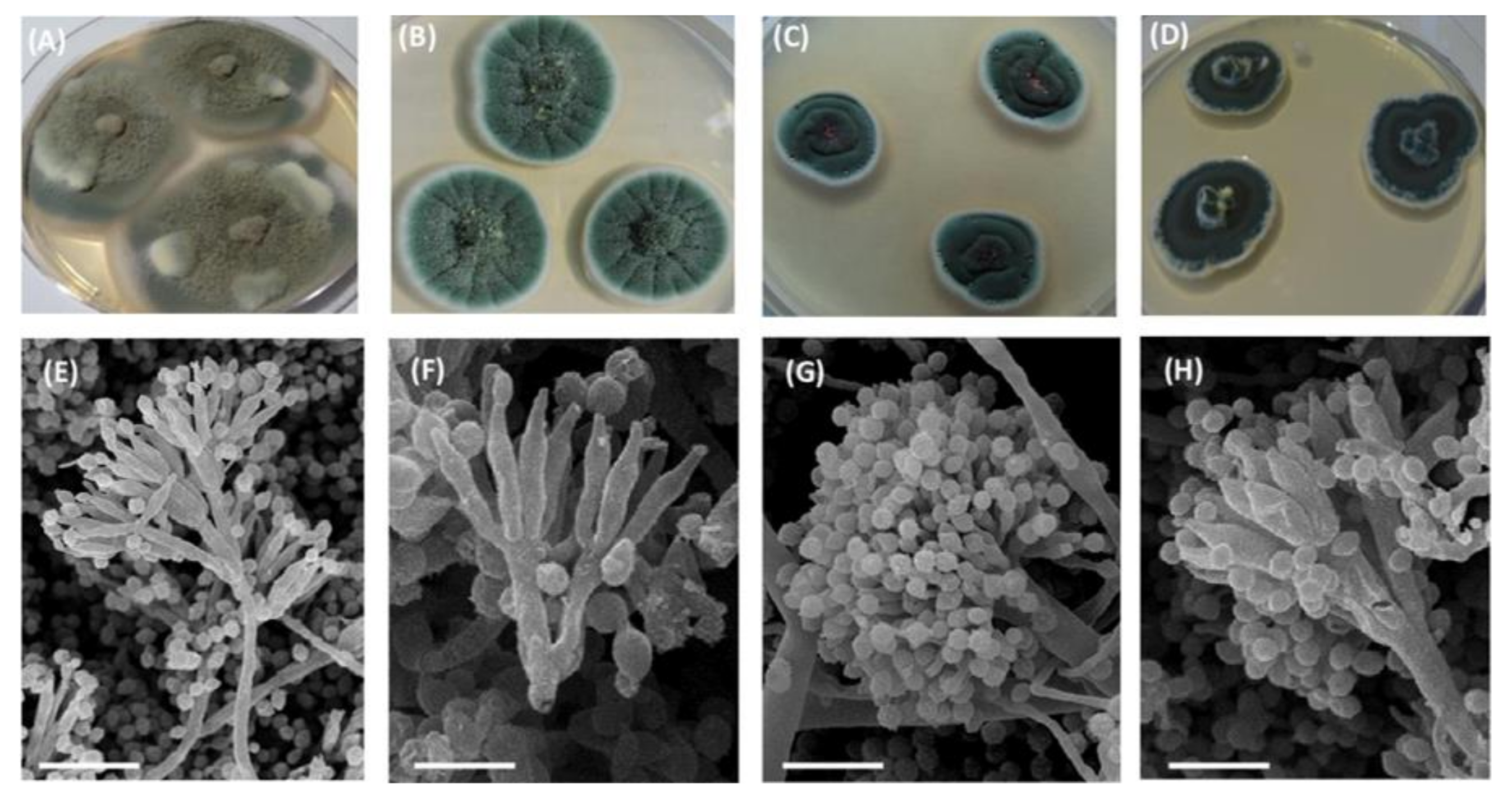

| Isolate | Cultural Characteristics | Morphological Characteristics | |
|---|---|---|---|
| Mycelial Morphology | Sporulation Characteristics | ||
| 18RJF6 | Widely grown, velvety surface with individual | 1–2 mm height | Sporulation: grayish-green, in some cases, the edge of the colonies lighter |
| Conidiophores: 100–150 μm × 2.5–3 μm Typically, biverticillate and symmetrical; each conidiophore contained 3–5 metules and lanceolate phialides | |||
| 18RJF9 | felted areas; smooth reverse side; with exudate; reddish brown | ||
| 18RJF10 | Elliptic conidia: 2–3 µm × 1.5–2.5 μm; smooth or slightly rough | ||
| 18RJF2 | Colonies of 10–20 mm in diameter with low growth; velvety surface, rugose | 1–2 mm height | Sporulation: bright green |
| Conidiophores: 40–110 μm × 2–3 μm | |||
| Typically, biverticillate and symmetrical, as a rule, with additional branches (10–25 μm length); each conidiophore contained 5 or 6 metals and were from lanceolate to flask-shaped phialides | |||
| Elliptic to fusiform conidia: 2.5–6 μm × 2.5–4 μm; smooth or slightly rough | |||
| 18RJF4.1 | Colonies of 2.5–3.5 mm in diameter; moderate growth rate; velvety surface; radially folded; colorless or yellow drops of exudate | 1–3 mm height | Sporulation: yellow-green |
| Conidiophores: 250–500 μm × 2.5–3.5 μm | |||
| Typically, terverticillate and asymmetric, with a pressed lateral twig; each conidiophore contained 5 or 6 metules and from 3 to 6 bottle-shaped phialides | |||
| Subspherical to ellipsoidal conidia: 3.0–4.0 μm × 2.8–3.8 μm; smooth | |||
| 18RJF4.2 | Colonies of 10–20 mm in diameter with slow growth rate; strongly folded with a well-defined edge; | 2–4 mm height | Sporulation: blue-green |
| Conidiophores formed on the substrate mycelium only: 500 μm × 5–8 μm | |||
| the reverse side radially folded, initially without a specific coloration and then wine-purple | The spore heads biseriate radial, up to 20 µm in diameter; metules (6–7 μm length), phialides 7–10 µm × 2.0–2.5 µm | ||
| Globular conidia: 2.5 µm × 3.5 µm; prickly, green in mass | |||
| Isolate | Isolation Medium | Micromycete | TS-1 Degradation Score | Mycelium Lysis |
|---|---|---|---|---|
| 18RJF2 | EM | Talaromyces rugulosus | 0 | − |
| 18RJF4.1 | CP | Penicillium chrysogenum | 3 | + |
| 18RJF4.2 | CP | Aspergillus sydowii | 5 | + |
| 18RJF6 | WA | Talaromyces amestolkiae | 5 | + |
| 18RJF9 | WA | Talaromyces amestolkiae | 3 | + |
| 18RJF10 | WA | Talaromyces amestolkiae | 2 | + |
| Isolate | Mitochondrial rRNA | GenBank ID | Bacteria with Putative Petroleum Destruction Activity |
|---|---|---|---|
| 18RJF2 | Talaromyces | MW393516 | None |
| 18RJF4.1 | Penicillium | MW393517 | Sphingomonas, Bacillus, Rhodococcus, Halomonas, Nocardioides |
| 18RJF4.2 | Aspergillus | MW393518 | Sphingomonas, Bacillus, Pseudomonas, Stenotrophomonas, Arthrobacter, Halomonas, Nocardioides |
| 18RJF6 | Talaromyces | MW393519 | Sphingomonas, Bacillus, Pseudomonas, Stenotrophomonas, Arthrobacter, Halomonas, Nocardioides |
| 18RJF9 | Talaromyces | MW393520 | Sphingomonas, Bacillus, Pseudomonas, Stenotrophomonas, Arthrobacter, Streptomyces, Nocardioides |
| 18RJF10 | Talaromyces | MW393521 | Sphingomonas, Bacillus, Arthrobacter, Halomonas, Streptomyces, Nocardioides |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shapiro, T.; Chekanov, K.; Alexandrova, A.; Dolnikova, G.; Ivanova, E.; Lobakova, E. Revealing of Non-Cultivable Bacteria Associated with the Mycelium of Fungi in the Kerosene-Degrading Community Isolated from the Contaminated Jet Fuel. J. Fungi 2021, 7, 43. https://doi.org/10.3390/jof7010043
Shapiro T, Chekanov K, Alexandrova A, Dolnikova G, Ivanova E, Lobakova E. Revealing of Non-Cultivable Bacteria Associated with the Mycelium of Fungi in the Kerosene-Degrading Community Isolated from the Contaminated Jet Fuel. Journal of Fungi. 2021; 7(1):43. https://doi.org/10.3390/jof7010043
Chicago/Turabian StyleShapiro, Tatiana, Konstantin Chekanov, Alina Alexandrova, Galina Dolnikova, Ekaterina Ivanova, and Elena Lobakova. 2021. "Revealing of Non-Cultivable Bacteria Associated with the Mycelium of Fungi in the Kerosene-Degrading Community Isolated from the Contaminated Jet Fuel" Journal of Fungi 7, no. 1: 43. https://doi.org/10.3390/jof7010043
APA StyleShapiro, T., Chekanov, K., Alexandrova, A., Dolnikova, G., Ivanova, E., & Lobakova, E. (2021). Revealing of Non-Cultivable Bacteria Associated with the Mycelium of Fungi in the Kerosene-Degrading Community Isolated from the Contaminated Jet Fuel. Journal of Fungi, 7(1), 43. https://doi.org/10.3390/jof7010043






