Expanding the Knowledge on the Skillful Yeast Cyberlindnera jadinii
Abstract
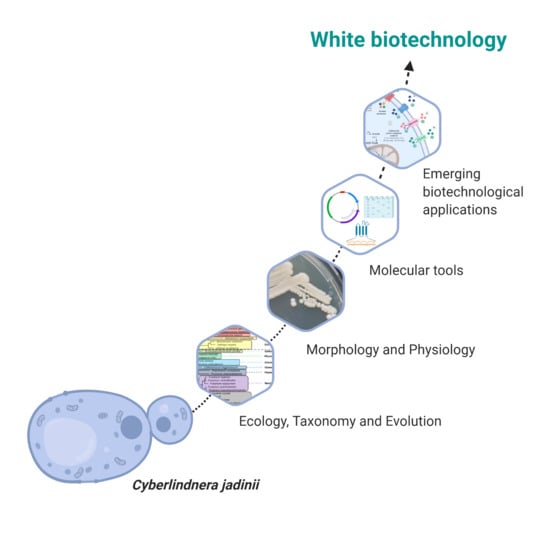
1. Introduction
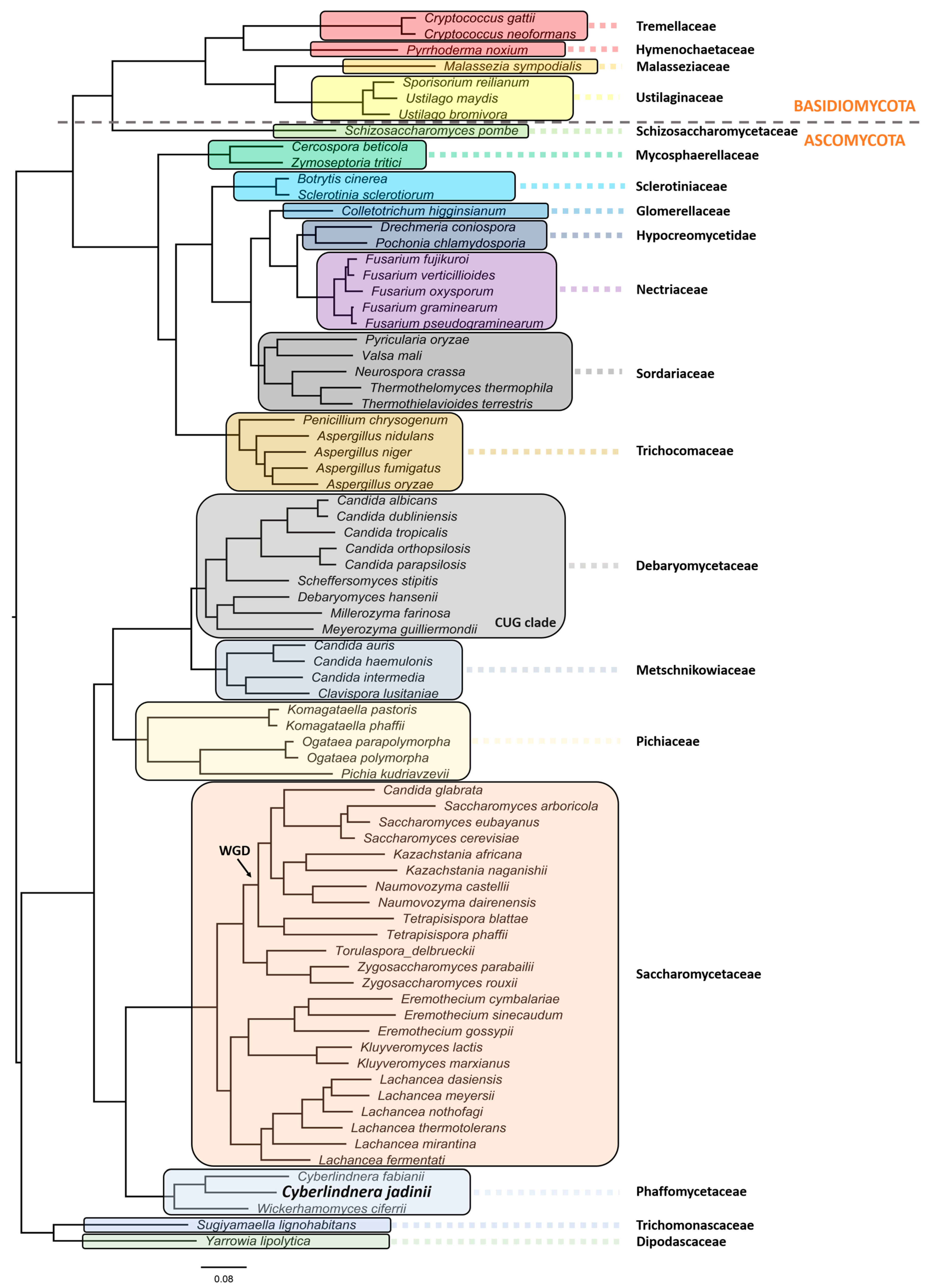
2. Ecology, Taxonomy, and Evolution
3. Life Cycle and Genome Organization
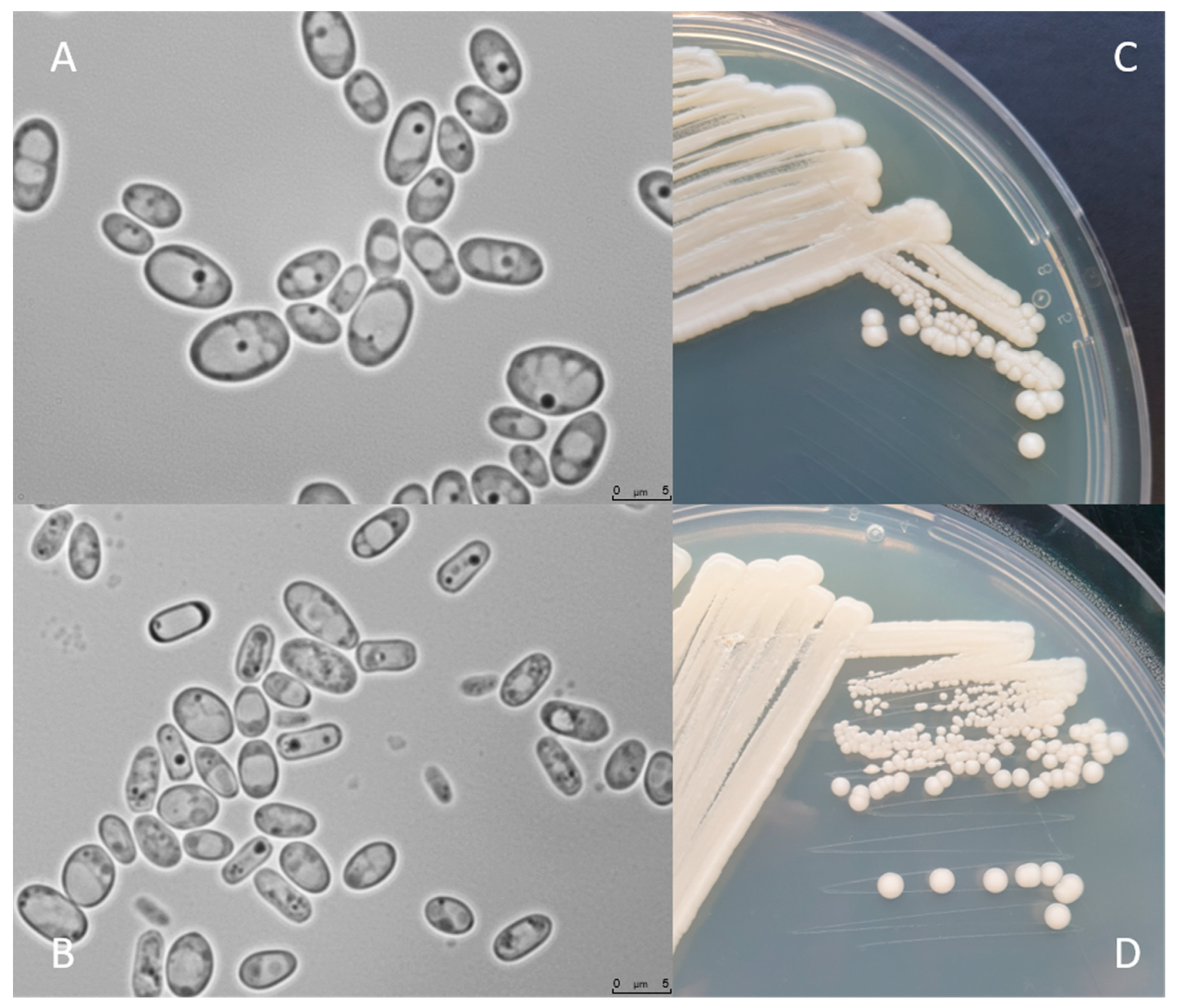
4. Morphology and Physiology
5. Molecular Tools for C. jadinii Manipulation
6. Emerging Biotechnological Applications
6.1. Therapeutic Applications
6.2. Bioproduction of Valuable Compounds Using Cost-Effective Carbon Sources
6.3. Industrial Applications—A Patent-View
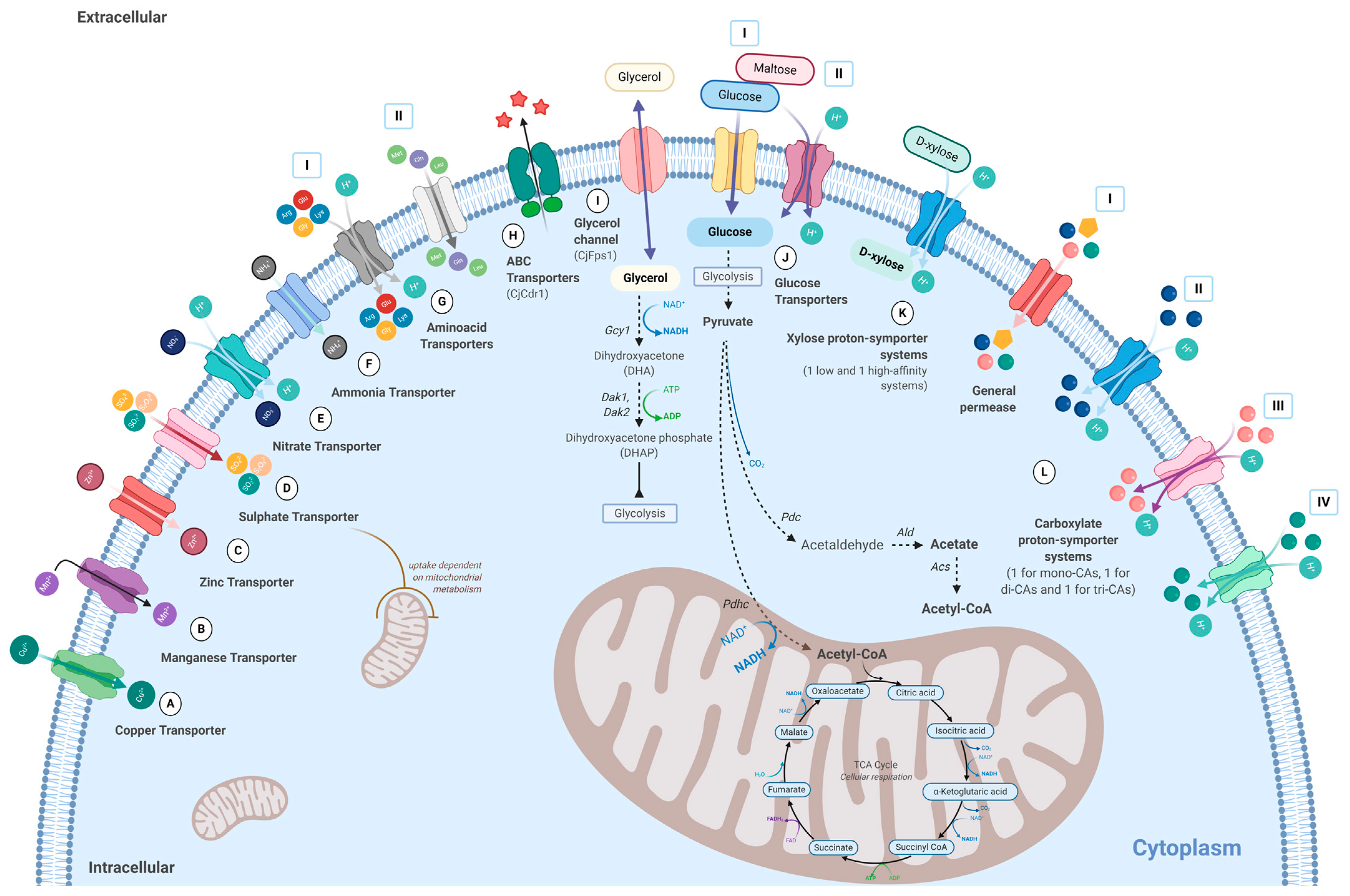
7. Membrane Transporters Characterized in C. jadinii
8. Conclusions and Future Perspectives
Funding
Conflicts of Interest
References
- Heux, S.; Meynial-Salles, I.; O’Donohue, M.; Dumon, C. White biotechnology: State of the art strategies for the development of biocatalysts for biorefining. Biotechnol. Adv. 2015, 33, 1653–1670. [Google Scholar] [CrossRef] [PubMed]
- Gong, Z.; Nielsen, J.; Zhou, Y.J. Engineering Robustness of Microbial Cell Factories. Biotechnol. J. 2017, 12. [Google Scholar] [CrossRef] [PubMed]
- Sibirny, A. Non-Conventional Yeasts: From Basic Research to Application; Springer: Berlin/Heidelberg, Germany, 2019. [Google Scholar]
- Martínez, E.; Santos, J.; Araujo, G.; Souza, S.; Rodrigues, R.; Canettieri, E. Production of Single Cell Protein (SCP) from Vinasse. In Fungal Biorefineries; Springer: Berlin/Heidelberg, Germany, 2018; pp. 215–238. [Google Scholar]
- Buerth, C.; Heilmann, C.; Klis, F.; de Koster, C.; Ernst, J.; Tielker, D. Growth-dependent secretome of Candida utilis. Microbiology 2011, 157, 2493–2503. [Google Scholar] [CrossRef] [PubMed]
- Barford, J. The technology of aerobic yeast growth. Yeast Biotechnol. 1987, 200–230. [Google Scholar] [CrossRef]
- Miura, Y.; Kettoku, M.; Kato, M.; Kobayashi, K.; Kondo, K. High level production of thermostable alpha-amylase from Sulfolobus solfataricus in high-cell density culture of the food yeast Candida utilis. J. Mol. Microbiol. Biotechnol. 1999, 1, 129–134. [Google Scholar] [PubMed]
- Bekatorou, A.; Psarianos, C.; Koutinas, A. Production of food grade yeasts. Food Technol. Biotechnol. 2006, 44, 407–415. [Google Scholar]
- Boze, H.; Moulin, G.; Galzy, P. Production of food and fodder yeasts. Crit. Rev. Biotechnol. 1992, 12, 65–86. [Google Scholar] [CrossRef]
- Buerth, C.; Tielker, D.; Ernst, J. Candida utilis and Cyberlindnera (Pichia) jadinii: Yeast relatives with expanding applications. Appl. Microbiol. Biotechnol. 2016, 100, 6981–6990. [Google Scholar] [CrossRef]
- Klein, R.; Faureau, M. Chapter 8: The Candida species: Biochemistry, molecular biology, and industrial applications. In Food Biotechnology: Microorganisms; Hui, Y.H., Khachatourians, G.G., Eds.; Wiley VCH: New York, NY, USA, 1995; p. 297. [Google Scholar]
- Kurtzman, C.; Robnett, C.; Basehoar-Powers, E. Phylogenetic relationships among species of Pichia, Issatchenkia and Williopsis determined from multigene sequence analysis, and the proposal of Barnettozyma gen. nov., Lindnera gen. nov. and Wickerhamomyces gen. nov. Fems Yeast Res. 2008, 8, 939–954. [Google Scholar] [CrossRef]
- Suh, S.; Blackwell, M.; Kurtzman, C.; Lachance, M. Phylogenetics of Saccharomycetales, the ascomycete yeasts. Mycologia 2006, 98, 1006–1017. [Google Scholar] [CrossRef]
- Barnett, J. A history of research on yeasts 8: Taxonomy. Yeast 2004, 21, 1141–1193. [Google Scholar] [CrossRef] [PubMed]
- Minter, D. Cyberlindnera, a replacement name for Lindnera Kurtzman et al., nom. illegit. Mycotaxon 2009, 110, 473–476. [Google Scholar] [CrossRef]
- Rupp, O.; Brinkrolf, K.; Buerth, C.; Kunigo, M.; Schneider, J.; Jaenicke, S.; Goesmann, A.; Pühler, A.; Jaeger, K.; Ernst, J. The structure of the Cyberlindnera jadinii genome and its relation to Candida utilis analyzed by the occurrence of single nucleotide polymorphisms. J. Biotechnol. 2015, 211, 20–30. [Google Scholar] [CrossRef] [PubMed]
- Murray, P.; Baron, E.; Pfaller, M.; Tenover, F.; Yolken, R. Manual of Clinical Microbiology, 6th ed.; American Society for Microbiology: Washington, DC, USA, 1995. [Google Scholar]
- Lodder, J.; Kreger-Van Rij, J.W. The Yeasts, A Taxonomic Study; North Holland Publishing Company: Amesterdam, The Netherlands, 1952. [Google Scholar]
- Daniel, H.; Lachance, M.; Kurtzman, C. On the reclassification of species assigned to Candida and other anamorphic ascomycetous yeast genera based on phylogenetic circumscription. Antonie Van Leeuwenhoek 2014, 106, 67–84. [Google Scholar] [CrossRef] [PubMed]
- Kurtzman, C.; Johnson, C.; Smiley, M. Determination of conspecificity of Candida utilis and Hansenula jadinii through DNA reassociation. Mycologia 1979, 71, 844–847. [Google Scholar] [CrossRef]
- Deak, T. Characteristics and properties of Foodborne yeasts. In Handbook of Food Spoilage Yeasts; Deak, T., Ed.; CRC Press: Boca Raton, FL, USA, 2007. [Google Scholar]
- Kurtzman, C.; Fell, J.; Boekhout, T. Definition, classification and nomenclature of the yeasts. In The Yeasts: A Taxonomic Study; Elsevier: Amsterdam, The Netherlands, 2011; pp. 3–5. [Google Scholar]
- Hawksworth, D. Managing and coping with names of pleomorphic fungi in a period of transition. Ima Fungus 2012, 3, 15–24. [Google Scholar] [CrossRef]
- Tomita, Y.; Ikeo, K.; Tamakawa, H.; Gojobori, T.; Ikushima, S. Genome and transcriptome analysis of the food-yeast Candida utilis. PLoS ONE 2012, 7, e37226. [Google Scholar] [CrossRef] [PubMed]
- Kondo, K.; Saito, T.; Kajiwara, S.; Takagi, M.; Misawa, N. A transformation system for the yeast Candida utilis: Use of a modified endogenous ribosomal protein gene as a drug-resistant marker and ribosomal DNA as an integration target for vector DNA. J. Bacteriol. 1995, 177, 7171–7177. [Google Scholar] [CrossRef]
- Ikushima, S.; Minato, T.; Kondo, K. Identification and application of novel autonomously replicating sequences (ARSs) for promoter-cloning and co-transformation in Candida utilis. Biosci. Biotechnol. Biochem. 2009, 73, 152–159. [Google Scholar] [CrossRef][Green Version]
- Belcarz, A.; Ginalska, G.; Lobarzewski, J.; Penel, C. The novel non-glycosylated invertase from Candida utilis (the properties and the conditions of production and purification). Biochim. Et Biophys. Acta Protein Struct. Mol. Enzymol. 2002, 1594, 40–53. [Google Scholar] [CrossRef]
- Fujino, S.; Akiyama, D.; Akaboshi, S.; Fujita, T.; Watanabe, Y.; Tamai, Y. Purification and characterization of phospholipase B from Candida utilis. Biosci. Biotechnol. Biochem. 2006, 70, 377–386. [Google Scholar] [CrossRef] [PubMed]
- Leão, C.; Van Uden, N. Transport of lactate and other short-chain monocarboxylates in the yeast Candida utilis. Appl. Microbiol. Biotechnol. 1986, 23, 389–393. [Google Scholar] [CrossRef]
- Cássio, F.; Leão, C. Low-and high-affinity transport systems for citric acid in the yeast Candida utilis. Appl. Environ. Microbiol. 1991, 57, 3623–3628. [Google Scholar] [CrossRef] [PubMed]
- Cássio, F.; Leão, C. A comparative study on the transport of L (-) malic acid and other short-chain carboxylic acids in the yeast Candida utilis: Evidence for a general organic acid permease. Yeast 1993, 9, 743–752. [Google Scholar] [CrossRef] [PubMed]
- Krahulec, J.; Lišková, V.; Boňková, H.; Lichvariková, A.; Šafranek, M.; Turňa, J. The ploidy determination of the biotechnologically important yeast Candida utilis. J. Appl. Genet. 2020, 61, 275–286. [Google Scholar] [CrossRef] [PubMed]
- Ross, I.; Parkin, M. Uptake of copper by Candida utilis. Mycol. Res. 1989, 93, 33–37. [Google Scholar] [CrossRef]
- Kurtzman, C. Chapter 42—Lindnera Kurtzman, Robnett & Basehoar-Powers (2008). In The Yeasts: A Taxonomic Study; Kurtzman, C., Fell, J., Boekhout, T., Eds.; Elsevier: Amsterdam, The Netherlands, 2011; pp. 521–543. [Google Scholar]
- Pinheiro, R.; Lopes, M.; Belo, I.; Mota, M. Candida utilis metabolism and morphology under increased air pressure up to 12 bar. Process Biochem. 2014, 49, 374–379. [Google Scholar] [CrossRef][Green Version]
- Hittinger, C.; Rokas, A.; Bai, F.; Boekhout, T.; Gonçalves, P.; Jeffries, T.; Kominek, J.; Lachance, M.; Libkind, D.; Rosa, C. Genomics and the making of yeast biodiversity. Curr. Opin. Genet. Dev. 2015, 35, 100. [Google Scholar] [CrossRef]
- Riley, R.; Haridas, S.; Wolfe, K.; Lopes, M.; Hittinger, C.; Göker, M.; Salamov, A.; Wisecaver, J.; Long, T.; Calvey, C. Comparative genomics of biotechnologically important yeasts. Proc. Natl. Acad. Sci. USA 2016, 113, 9882–9887. [Google Scholar] [CrossRef]
- Shen, X.; Opulente, D.; Kominek, J.; Zhou, X.; Steenwyk, J.; Buh, K.; Haase, M.; Wisecaver, J.; Wang, M.; Doering, D. Tempo and mode of genome evolution in the budding yeast subphylum. Cell 2018, 175, 1533–1545.e20. [Google Scholar] [CrossRef]
- Marcet-Houben, M.; Gabaldón, T. Evolutionary and functional patterns of shared gene neighbourhood in fungi. Nat. Microbiol. 2019, 4, 2383–2392. [Google Scholar] [CrossRef] [PubMed]
- Shen, X.; Zhou, X.; Kominek, J.; Kurtzman, C.; Hittinger, C.; Rokas, A. Reconstructing the backbone of the Saccharomycotina yeast phylogeny using genome-scale data. G3 GenesGenomesGenet. 2016, 6, 3927–3939. [Google Scholar] [CrossRef]
- Alves, R.; Sousa-Silva, M.; Vieira, D.; Soares, P.; Chebaro, Y.; Lorenz, M.; Casal, M.; Soares-Silva, I.; Paiva, S. Carboxylic Acid Transporters in Candida Pathogenesis. MBio 2020, 11, e00156-00120. [Google Scholar] [CrossRef] [PubMed]
- Thanh, V.; Thuy, N.; Huong, H.; Hien, D.; Hang, D.; Anh, D.; Hüttner, S.; Larsbrink, J.; Olsson, L. Surveying of acid-tolerant thermophilic lignocellulolytic fungi in Vietnam reveals surprisingly high genetic diversity. Sci. Rep. 2019, 9, 3674. [Google Scholar] [CrossRef] [PubMed]
- Ikushima, S.; Fujii, T.; Kobayashi, O. Efficient gene disruption in the high-ploidy yeast Candida utilis using the Cre-loxP system. Biosci. Biotechnol. Biochem. 2009, 73, 879–884. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Ikushima, S.; Fujii, T.; Kobayashi, O.; Yoshida, S.; Yoshida, A. Genetic engineering of Candida utilis yeast for efficient production of L-lactic acid. Biosci. Biotechnol. Biochem. 2009, 73, 1818–1824. [Google Scholar] [CrossRef] [PubMed]
- Freel, K.; Sarilar, V.; Neuvéglise, C.; Devillers, H.; Friedrich, A.; Schacherer, J. Genome Sequence of the Yeast Cyberlindnera fabianii (Hansenula fabianii). Genome. Announc. 2014, 2, e00638-00614. [Google Scholar] [CrossRef]
- Schneider, J.; Andrea, H.; Blom, J.; Jaenicke, S.; Rückert, C.; Schorsch, C.; Szczepanowski, R.; Farwick, M.; Goesmann, A.; Pühler, A. Draft genome sequence of Wickerhamomyces ciferrii NRRL Y-1031 F-60-10. Eukaryot Cell 2012, 11, 1582–1583. [Google Scholar] [CrossRef]
- Foury, F.; Roganti, T.; Lecrenier, N.; Purnelle, B. The complete sequence of the mitochondrial genome of Saccharomyces cerevisiae. Febs Lett. 1998, 440, 325–331. [Google Scholar] [CrossRef]
- Goffeau, A.; Barrell, B.; Bussey, H.; Davis, R.; Dujon, B.; Feldmann, H.; Galibert, F.; Hoheisel, J.; Jacq, C.; Johnston, M. Life with 6000 genes. Science 1996, 274, 546–567. [Google Scholar] [CrossRef]
- Coelho, M.; Amaral, P.; Belo, I. Yarrowia lipolytica: An industrial workhorse. In Current Research, Technology and Education Topics in Applied Microbiology and Microbial Biotechnology; Méndez-Vilas, A., Ed.; Formatex: Badajoz, Spain, 2010; Volume 2, pp. 930–940. [Google Scholar]
- Andreeva, E.; Pozmogova, I.; Rabotnova, I. Principle growth indices of a chemostat Candida utilis culture resistant to acid pH values. Mikrobiologiia 1979, 48, 481. [Google Scholar] [PubMed]
- Verduyn, C. Physiology of yeasts in relation to biomass yields. In Quantitative Aspects of Growth and Metabolism of Microorganisms; Springer: Berlin/Heidelberg, Germany, 1992; pp. 325–353. [Google Scholar]
- Ordaz, L.; Lopez, R.; Melchy, O.; Torre, M. Effect of high-cell-density fermentation of Candida utilis on kinetic parameters and the shift to respiro-fermentative metabolism. Appl. Microbiol. Biotechnol. 2001, 57, 374–378. [Google Scholar] [PubMed]
- Dashko, S.; Zhou, N.; Compagno, C.; Piškur, J. Why, when, and how did yeast evolve alcoholic fermentation? Fems Yeast Res. 2014, 14, 826–832. [Google Scholar] [CrossRef]
- Gomes, A.; Carmo, T.; Carvalho, L.; Bahia, F.; Parachin, N. Comparison of yeasts as hosts for recombinant protein production. Microorganisms 2018, 6, 38. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.; Zhang, J.; Dong, Y.; Wei, G.; Qi, B. Glutathione is involved in physiological response of Candida utilis to acid stress. Appl. Microbiol. Biotechnol. 2015, 99, 10669–10679. [Google Scholar] [CrossRef]
- Kunigo, M.; Buerth, C.; Tielker, D.; Ernst, J. Heterologous protein secretion by Candida utilis. Appl. Microbiol. Biotechnol. 2013, 97, 7357–7368. [Google Scholar] [CrossRef]
- Moftah, O.; Grbavčić, S.; Žuža, M.; Luković, N.; Bezbradica, D.; Knežević-Jugović, Z. Adding value to the oil cake as a waste from oil processing industry: Production of lipase and protease by Candida utilis in solid state fermentation. Appl. Biochem. Biotechnol. 2012, 166, 348–364. [Google Scholar] [CrossRef]
- Grbavčić, S.; Dimitrijević-Branković, S.; Bezbradica, D.; Šiler-Marinković, S.; Knežević, Z. Effect of fermentation conditions on lipase production by Candida utilis. J. Serb. Chem. Soc. 2007, 72, 757–765. [Google Scholar] [CrossRef]
- Tamakawa, H.; Ikushima, S.; Yoshida, S. Efficient production of L-lactic acid from xylose by a recombinant Candida utilis strain. J. Biosci. Bioeng. 2012, 113, 73–75. [Google Scholar] [CrossRef]
- Shimada, H.; Kondo, K.; Fraser, P.; Miura, Y.; Saito, T.; Misawa, N. Increased Carotenoid Production by the Food Yeast Candida utilis through Metabolic Engineering of the Isoprenoid Pathway. Appl. Environ. Microbiol. 1998, 64, 2676–2680. [Google Scholar] [CrossRef]
- Boňková, H.; Osadská, M.; Krahulec, J.; Lišková, V.; Stuchlík, S.; Turňa, J. Upstream regulatory regions controlling the expression of the Candida utilis maltase gene. J. Biotechnol. 2014, 189, 136–142. [Google Scholar] [CrossRef] [PubMed]
- Iwakiri, R.; Noda, Y.; Adachi, H.; Yoda, K. Isolation of the YAP1 homologue of Candida utilis and its use as an efficient selection marker. Yeast 2005, 22, 1079–1087. [Google Scholar] [CrossRef] [PubMed]
- Tamakawa, H.; Mita, T.; Yokoyama, A.; Ikushima, S.; Yoshida, S. Metabolic engineering of Candida utilis for isopropanol production. Appl. Microbiol. Biotechnol. 2013, 97, 6231–6239. [Google Scholar] [CrossRef] [PubMed]
- Kondo, K.; Miura, Y.; Sone, H.; Kobayashi, K. High-level expression of a sweet protein, monellin, in the food yeast Candida utilis. Nat. Biotechnol. 1997, 15, 453. [Google Scholar] [CrossRef]
- Kunigo, M.; Buerth, C.; Ernst, J. Secreted xylanase XynA mediates utilization of xylan as sole carbon source in Candida utilis. Appl. Microbiol. Biotechnol. 2015, 99, 8055–8064. [Google Scholar] [CrossRef]
- Miura, Y.; Kondo, K.; Shimada, H.; Saito, T.; Nakamura, K.; Misawa, N. Production of lycopene by the food yeast, Candida utilis that does not naturally synthesize carotenoid. Biotechnol. Bioeng. 1998, 58, 306–308. [Google Scholar] [CrossRef]
- Iwakiri, R.; Noda, Y.; Adachi, H.; Yoda, K. Isolation and characterization of promoters suitable for a multidrug-resistant marker CuYAP1 in the yeast Candida utilis. Yeast 2006, 23, 23–34. [Google Scholar] [CrossRef]
- Miura, Y.; Kondo, K.; Saito, T.; Shimada, H.; Fraser, P.; Misawa, N. Production of the carotenoids lycopene, β-carotene, and astaxanthin in the food yeast Candida utilis. Appl. Environ. Microbiol. 1998, 64, 1226–1229. [Google Scholar] [CrossRef]
- Sander, J.; Joung, J. CRISPR-Cas systems for editing, regulating and targeting genomes. Nat. Biotechnol. 2014, 32, 347. [Google Scholar] [CrossRef]
- Cai, P.; Gao, J.; Zhou, Y. CRISPR-mediated genome editing in non-conventional yeasts for biotechnological applications. Microb. Cell Factories 2019, 18, 63. [Google Scholar] [CrossRef]
- Stovicek, V.; Holkenbrink, C.; Borodina, I. CRISPR/Cas system for yeast genome engineering: Advances and applications. Fems Yeast Res. 2017, 17, fox030. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Lin, J.; Dai, Y. Method for Verifying Feasibility of CRISPR-Cas9 System for Knocking Out Candida utilis Target Gene. CN Patent Application Number 201910633290.6, 12 July 2019. [Google Scholar]
- Mukherjee, P.; Chandra, J.; Retuerto, M.; Sikaroodi, M.; Brown, R.; Jurevic, R.; Salata, R.; Lederman, M.; Gillevet, P.; Ghannoum, M. Oral mycobiome analysis of HIV-infected patients: Identification of Pichia as an antagonist of opportunistic fungi. PLoS Pathog. 2014, 10, e1003996. [Google Scholar] [CrossRef] [PubMed]
- De Jesus, M.; Rodriguez, A.; Yagita, H.; Ostroff, G.; Mantis, N. Sampling of Candida albicans and Candida tropicalis by langerin-positive dendritic cells in mouse Peyer’s patches. Immunol. Lett. 2015, 168, 64–72. [Google Scholar] [CrossRef] [PubMed]
- Thévenot, J.; Cordonnier, C.; Rougeron, A.; Le Goff, O.; Nguyen, H.; Denis, S.; Alric, M.; Livrelli, V.; Blanquet-Diot, S. Enterohemorrhagic Escherichia coli infection has donor-dependent effect on human gut microbiota and may be antagonized by probiotic yeast during interaction with Peyer’s patches. Appl. Microbiol. Biotechnol. 2015, 99, 9097–9110. [Google Scholar] [CrossRef]
- Buerth, C.; Mausberg, A.; Heininger, M.; Hartung, H.; Kieseier, B.; Ernst, J. Oral tolerance induction in experimental autoimmune encephalomyelitis with Candida utilis expressing the immunogenic MOG35-55 peptide. PLoS ONE 2016, 11, e0155082. [Google Scholar] [CrossRef] [PubMed]
- Feng, T.; Elson, C. Adaptive immunity in the host–microbiota dialog. Mucosal Immunol. 2011, 4, 15. [Google Scholar] [CrossRef] [PubMed]
- Singh, R.; Singh, T.; Singh, A. Chapter 9—Enzymes as Diagnostic Tools. In Advances in Enzyme Technology; Singh, R.S., Singhania, R.R., Pandey, A., Larroche, C., Eds.; Elsevier: Amsterdam, The Netherlands, 2019; pp. 225–271. [Google Scholar]
- Żymańczyk-Duda, E.; Brzezińska-Rodak, M.; Klimek-Ochab, M.; Duda, M.; Zerka, A. Yeast as a Versatile Tool in Biotechnology. Yeast Ind. Appl. 2017, 1, 3–40. [Google Scholar]
- Khan, T.; Daugulis, A. Application of solid–liquid TPPBs to the production of L-phenylacetylcarbinol from benzaldehyde using Candida utilis. Biotechnol. Bioeng. 2010, 107, 633–641. [Google Scholar] [CrossRef]
- Tripathi, C.; Agarwal, S.; Basu, S. Production of L-phenylacetylcarbinol by fermentation. J. Ferment. Bioeng. 1997, 84, 487–492. [Google Scholar] [CrossRef]
- Shin, H.; Rogers, P. Biotransformation of benzeldehyde to L-phenylacetylcarbinol, an intermediate in L-ephedrine production, by immobilized Candida utilis. Appl. Microbiol. Biotechnol. 1995, 44, 7–14. [Google Scholar] [CrossRef]
- Rogers, P.; Shin, H.; Wang, B. Biotransformation for L-ephedrine production. In Biotreatment, Downstream Processing and Modelling; Springer: Berlin/Heidelberg, Germany, 1997; pp. 33–59. [Google Scholar]
- Lamping, E.; Monk, B.; Niimi, K.; Holmes, A.; Tsao, S.; Tanabe, K.; Niimi, M.; Uehara, Y.; Cannon, R. Characterization of Three Classes of Membrane Proteins Involved in Fungal Azole Resistance by Functional Hyperexpression in Saccharomyces cerevisiae. Eukaryot. Cell 2007, 6, 1150–1165. [Google Scholar] [CrossRef] [PubMed]
- Watanasrisin, W.; Iwatani, S.; Oura, T.; Tomita, Y.; Ikushima, S.; Chindamporn, A.; Niimi, M.; Niimi, K.; Lamping, E.; Cannon, R. Identification and characterization of Candida utilis multidrug efflux transporter CuCdr1p. Fems Yeast Res. 2016, 16, fow042. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Hong, Y.; Chen, Y.; Farh, L.; Yang, W.; Liao, C.; Shiuan, D. Recombinant Candida utilis for the production of biotin. Appl. Microbiol. Biotechnol. 2006, 71, 211. [Google Scholar] [CrossRef]
- Li, Y.; Wei, G.; Chen, J. Glutathione: A review on biotechnological production. Appl. Microbiol. Biotechnol. 2004, 66, 233–242. [Google Scholar] [CrossRef] [PubMed]
- Kogan, G.; Šandula, J.; Šimkovicová, V. Glucomannan from Candida utilis. Folia Microbiol. 1993, 38, 219–224. [Google Scholar] [CrossRef] [PubMed]
- Ruszova, E.; Pavek, S.; Hajkova, V.; Jandova, S.; Velebny, V.; Papezikova, I.; Kubala, L. Photoprotective effects of glucomannan isolated from Candida utilis. Carbohydr. Res. 2008, 343, 501–511. [Google Scholar] [CrossRef]
- Tamakawa, H.; Ikushima, S.; Yoshida, S. Ethanol production from xylose by a recombinant Candida utilis strain expressing protein-engineered xylose reductase and xylitol dehydrogenase. Biosci. Biotechnol. Biochem. 2011, 75, 1994–2000. [Google Scholar] [CrossRef][Green Version]
- Hui, Y.H.; Khachatourians, G. Food Biotechnology: Microorganisms; Wiley-Interscience: New York, NY, USA, 1994. [Google Scholar]
- Maugeri-Filho, F.; Goma, G. SCP production from organic acids with Candida utilis. Rev. Microbiol. 1988, 19, 446–452. [Google Scholar]
- Huang, L.; Zhang, Y.; Zhang, L.; Guo, Z.; Zeng, S.; Tian, Y.; Zheng, B. Application of Candida utilis to Loquat Wine Making and Making Method for Loquat Wine. CN Patent Application Number 201210131375.2, 28 April 2012. [Google Scholar]
- Xie, B.; He, B.; Hu, J.; Yu, Q.; Huang, Q.; Zhang, Y. Dry Loquat Wine and Making Method Thereof. CN Patent Application Number 201410808872.0, 1 April 2015. [Google Scholar]
- Cai, Y.; Jiang, X.; Chen, Y.; Chen, Q.; Cai, Y.; Cai, Y. Preparation Method of Alcohol-Free Fruit Wine Being Rich in Lovastatin. CN Patent Application Number 201810869936.6, 29 January 2019. [Google Scholar]
- Ma, X.; Liu, L. Body Lotion Containing Candida utilis Beta-D-glucan. CN Patent Application Number 201810110524.4, 8 June 2018. [Google Scholar]
- Ma, X.; Liu, L. Sunscreen Cream with Candida utilis Beta-D-glucan. CN Patent Application Number 201810110525.9, 12 June 2018. [Google Scholar]
- Ma, X.; Liu, L. Facial Cleanser Containing Candida utilis Beta-D-glucan. CN Patent Application Number 201810113250.4, 8 June 2018. [Google Scholar]
- Ma, X.; Liu, L. Toner Containing Beta-D-glucan of Candida utilis. CN Patent Application Number 201810113263.1, 22 June 2018. [Google Scholar]
- Ma, X.; Liu, L. Eye Cream Containing Beta-D-glucan of Candida utilis. CN Patent Application Number 201810113248.7, 22 June 2018. [Google Scholar]
- Ma, X.; Liu, L. Shampoo Containing Candida utilis Beta-D-glucan. CN Patent Application Number 201810110923.0, 14 August 2018. [Google Scholar]
- Ma, X.; Liu, L. Body Wash with Candida utilis Beta-D-glucan. CN Patent Application Number 201810110925.X, 15 June 2018. [Google Scholar]
- Ma, X.; Liu, L.; Shao, L.; He, Y. Hand Care Cream with Candida utilis Beta-D-glucan. CN Patent Application Number 201810110885.9, 15 June 2018. [Google Scholar]
- Li, S.; Pang, H.; Liu, S. Preparation Method of Recombinant Uricase. CN Patent Application Number 201410535887.4, 11 February 2015. [Google Scholar]
- Liu, J.; Gao, X.; Shang, F.; Liu, P.; Zhu, M. Candida utilis Reduction Method for Producing Methyl Fluorophenyl Methyl Propionate. CN Patent Application Number 201410518177.0, 14 January 2015. [Google Scholar]
- Firgatovich, B.F.; Gennadevich, G.Y.; Yurevna, G.E.; Mikhajlovna, K.E. Organomineral Granular Fertilizer. Russian Patent Application Number 2019143533, 25 June 2020. [Google Scholar]
- Bai, L.; Liu, X.; Jiang, H.; Liu, H.; Deng, Y. Application of Microorganism Bacterium Agent Capable of Efficiently Converting Form of Cadmium in Contaminated Soil Biologically. CN Patent Application Number 201710284861.0, 18 August 2017. [Google Scholar]
- Cui, G.; Gu, Y.; Cui, C.; Zhou, Y. Microbial Soil Conditioner for Lithified Soil. CN Patent Application Number 201310638420.8, 3 June 2015. [Google Scholar]
- Soares-Silva, I.; Ribas, D.; Sousa-Silva, M.; Azevedo-Silva, J.; Rendulić, T.; Casal, M. Membrane transporters in the bioproduction of organic acids: State of the art and future perspectives for industrial applications. Fems Microbiol. Lett. 2020, 367, fnaa118. [Google Scholar] [CrossRef]
- Parkin, M.; Ross, I. Uptake of copper and manganese by the yeast, Candida utilis. Microbios Lett. 1985, 29, 115–120. [Google Scholar]
- Parkin, M.; Ross, I. The specific uptake of manganese in the yeast Candida utilis. Microbiology 1986, 132, 2155–2160. [Google Scholar] [CrossRef][Green Version]
- Lawford, H.; Pik, J.; Lawford, G.; Williams, T.; Kligerman, A. Hyperaccumulation of zinc by zinc-depleted Candida utilis grown in chemostat culture. Can. J. Microbiol. 1980, 26, 71–76. [Google Scholar] [CrossRef] [PubMed]
- Failla, M.; Benedict, C.; Weinberg, E. Accumulation and storage of Zn2+ by Candida utilis. Microbiology 1976, 94, 23–36. [Google Scholar] [CrossRef] [PubMed]
- Failla, M.; Weinberg, E. Cyclic accumulation of zinc by Candida utilis during growth in batch culture. Microbiology 1977, 99, 85–97. [Google Scholar] [CrossRef] [PubMed]
- García, M.; Benitez, J.; Delgado, J.; Kotyk, A. Isolation of sulphate transport defective mutants of Candida utilis: Further evidence for a common transport system for sulphate, sulphite and thiosulphate. Folia Microbiol. 1983, 28, 1–5. [Google Scholar] [CrossRef]
- Benitez, J.; Alonso, A.; Delgado, J.; Kotyk, A. Sulphate transport in Candida utilis. Folia Microbiol. 1983, 28, 6–11. [Google Scholar] [CrossRef]
- Eddy, A.; Hopkins, P. The putative electrogenic nitrate-proton symport of the yeast Candida utilis. Comparison with the systems absorbing glucose or lactate. Biochem. J. 1985, 231, 291–297. [Google Scholar] [CrossRef]
- Ali, A.; Hipkin, C. Nitrate assimilation in Candida nitratophila and other yeasts. Arch. Microbiol. 1986, 144, 263–267. [Google Scholar] [CrossRef]
- Barnett, J. A history of research on yeasts 13. Active transport and the uptake of various metabolites. Yeast 2008, 25, 689–731. [Google Scholar] [CrossRef]
- De Koning, W.; Zwart, K.; Harder, W. A spectrophotometric method for the determination of the kinetic parameters of NH+4 uptake by yeast cells. Fems Microbiol. Lett. 1984, 23, 257–262. [Google Scholar] [CrossRef]
- Eddy, A.; Philo, R.; Earnshaw, P.; Brocklehurst, R. Some common aspects of active solute transport in yeast and mouse ascites tumour cells. In Biochemistry of Membrane Transport; Springer: Berlin/Heidelberg, Germany, 1977; pp. 250–260. [Google Scholar]
- Eddy, A.; Seaston, A.; Gardner, D.; Hacking, C. The thermodynamic efficiency of cotransport mechanisms with special reference to proton and anion transport in yeast. Ann. New York Acad. Sci. USA 1980, 341, 494–509. [Google Scholar] [CrossRef] [PubMed]
- Robins, R.; Davies, D. The role of glutathione in amino-acid absorption. Lack of correlation between glutathione turnover and amino-acid absorption by the yeast Candida utilis. Biochem. J. 1981, 194, 63–70. [Google Scholar] [CrossRef] [PubMed]
- Klein, M.; Islam, Z.; Knudsen, P.; Carrillo, M.; Swinnen, S.; Workman, M.; Nevoigt, E. The expression of glycerol facilitators from various yeast species improves growth on glycerol of Saccharomyces cerevisiae. Metab. Eng. Commun. 2016, 3, 252–257. [Google Scholar] [CrossRef] [PubMed]
- Peinado, J.; Cameira-Dos-Santos, P.; Loureiro-Dias, M. Regulation of glucose transport in Candida utilis. Microbiology 1989, 135, 195–201. [Google Scholar] [CrossRef] [PubMed]
- Barnett, J.; Sims, A. A note on the kinetics of uptake of d-glucose by the food yeast, Candida utilis. Arch. Microbiol. 1976, 111, 193–194. [Google Scholar] [CrossRef]
- Flores, C.; Rodríguez, C.; Petit, T.; Gancedo, C. Carbohydrate and energy-yielding metabolism in non-conventional yeasts. Fems Microbiol. Rev. 2000, 24, 507–529. [Google Scholar] [CrossRef]
- Broek, P.; Gompel, A.; Luttik, M.; Pronk, J.; Leeuwen, C. Mechanism of glucose and maltose transport in plasma-membrane vesicles from the yeast Candida utilis. Biochem. J. 1997, 321, 487–495. [Google Scholar] [CrossRef]
- Roush, A.; Domnas, A. Induced biosynthesis of uricase in yeast. Science 1956, 124, 125–126. [Google Scholar] [CrossRef]
- Kilian, S.; Prior, B.; Du Preez, J. The kinetics and regulation of M-xylose transport in Candida utilis. World J. Microbiol. Biotechnol. 1993, 9, 357–360. [Google Scholar] [CrossRef]
- Saayman, M.; Van Vuuren, H.; Van Zyl, W.; Viljoen-Bloom, M. Differential uptake of fumarate by Candida utilis and Schizosaccharomyces pombe. Appl. Microbiol. Biotechnol. 2000, 54, 792–798. [Google Scholar] [CrossRef]
- Roush, A.; Questiaux, L.; Domnas, A. The active transport and metabolism of purines in the yeast, Candida utilis. J. Cell. Comp. Physiol. 1959, 54, 275–286. [Google Scholar] [CrossRef] [PubMed]
- Casal, M.; Paiva, S.; Queirós, O.; Soares-Silva, I. Transport of carboxylic acids in yeasts. Fems Microbiol. Rev. 2008, 32, 974–994. [Google Scholar] [CrossRef] [PubMed]
- Paiva, S.; Devaux, F.; Barbosa, S.; Jacq, C.; Casal, M. Ady2p is essential for the acetate permease activity in the yeast Saccharomyces cerevisiae. Yeast 2004, 21, 201–210. [Google Scholar] [CrossRef]
- Casal, M.; Paiva, S.; Andrade, R.; Gancedo, C.; Leão, C. The lactate-proton symport of Saccharomyces cerevisiae is encoded by JEN1. J. Bacteriol. 1999, 181, 2620–2623. [Google Scholar] [CrossRef] [PubMed]
- Soares-Silva, I.; Schuller, D.; Andrade, R.; Baltazar, F.; Cássio, F.; Casal, M. Functional expression of the lactate permease Jen1p of Saccharomyces cerevisiae in Pichia pastoris. Biochem. J. 2003, 376, 781–787. [Google Scholar] [CrossRef]
- Queirós, O.; Pereira, L.; Paiva, S.; Moradas-Ferreira, P.; Casal, M. Functional analysis of Kluyveromyces lactis carboxylic acids permeases: Heterologous expression of KlJEN1 and KlJEN2 genes. Curr. Genet. 2007, 51, 161–169. [Google Scholar] [CrossRef]
- Soares-Silva, I.; Ribas, D.; Foskolou, I.; Barata, B.; Bessa, D.; Paiva, S.; Queirós, O.; Casal, M. The Debaryomyces hansenii carboxylate transporters Jen1 homologues are functional in Saccharomyces cerevisiae. FEMS Yeast Res. 2015, 15, fov094. [Google Scholar] [CrossRef][Green Version]
- Sá-Pessoa, J.; Paiva, S.; Ribas, D.; Silva, I.; Viegas, S.; Arraiano, C.; Casal, M. SATP (YaaH), a succinate–acetate transporter protein in Escherichia coli. Biochem. J. 2013, 454, 585–595. [Google Scholar] [CrossRef]
- Sá-Pessoa, J.; Amillis, S.; Casal, M.; Diallinas, G. Expression and specificity profile of the major acetate transporter AcpA in Aspergillus nidulans. Fungal Genet. Biol. 2015, 76, 93–103. [Google Scholar] [CrossRef]
- Ribas, D.; Soares-Silva, I.; Vieira, D.; Sousa-Silva, M.; Sá-Pessoa, J.; Azevedo-Silva, J.; Viegas, S.; Arraiano, C.; Diallinas, G.; Paiva, S.; et al. The acetate uptake transporter family motif “NPAPLGL (M/S)” is essential for substrate uptake. Fungal Genet. Biol. 2019, 122, 1–10. [Google Scholar] [CrossRef]
- Ribas, D.; Sá-Pessoa, J.; Soares-Silva, I.; Paiva, S.; Nygard, Y.; Ruohonen, L.; Penttila, M.; Casal, M. Yeast as a tool to express sugar acid transporters with biotechnological interest. Fems Yeast Res. 2017, 17. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Lin, M.; Xu, M.; Yang, S. Anaerobic fermentation for production of carboxylic acids as bulk chemicals from renewable biomass. In Anaerobes in Biotechnology; Springer: Berlin/Heidelberg, Germany, 2016; pp. 323–361. [Google Scholar]
- Kell, D.; Swainston, N.; Pir, P.; Oliver, S. Membrane transporter engineering in industrial biotechnology and whole cell biocatalysis. Trends Biotechnol. 2015, 33, 237–246. [Google Scholar] [CrossRef] [PubMed]
| Nomenclature in Literature | Current Nomenclature | Isolation Source | Reference |
|---|---|---|---|
| Candida utilis NBRC 0988 | C. jadinii ATCC 9950; CBS 5609; DSM 2361; NBRC 0988; NCYC 707; NRRL Y-900 | Yeast factory in Germany | [24] |
| C. utilis ATCC 9256 a | C. jadinii NRRL Y1084; CBS 841; CCRC 20334; DSM 70167; NCYC 359; VKM Y768; VTT C79091 | Unknown | [25,26] |
| C. utilis ATCC 9226 a NBRC 1086 | C. jadinii VTT C-71015; FMJ 4026; NBRC 1086 | Unknown | [25,27,28] |
| C. utilis IGC 3092 | C. jadinii PYCC 3092; CBS 890; VKM Y-33 | Unknown | [29,30,31] |
| C. utilis CCY 39-38-18 | C. jadinii CCY 029-38-18 b | Unknown | [32] |
| C. utilis NCYC 708 | C. jadinii NCYC 708; ATCC 42181; CBS 5947; VTT C-84157 | Unknown | [33] |
| C. utilis CBS 4885 NRRL Y-6756 | C. jadinii CBS 4885; NRRL Y-6756; NBRC 10708 | Cow with mastitis | [34] |
| C. utilis CBS 567 NRRL Y-1509 | C. jadinii CBS 567; NRRL Y-1509 | Yeast deposit in distillery | [34] |
| C. utilis CBS 2160 | C. jadinii CBS 2160 | Flower of Taraxacum sp. | [34] |
| C. utilis CBS 621 | C. jadinii CBS 621; NRRL Y-7586; ATCC 22023; PYCC 4182 | Yeast factories | [35] |
| C. utilis CBS 1600 | C. jadinii CBS 1600; NRRL Y-1542; ATCC 18201 | Pus of a woman abscess | [16] |
| Components | Cyberlindnera jadinii Strains | Saccharomyces cerevisiae S288c | ||
|---|---|---|---|---|
| NBRC 0988 | CBS 1600 | NRRL Y-1542 | ||
| NCBI assembly reference | GCA_000328385.1 | GCA_001245095.1 | GCA_001661405.1 | GCA_000146045.2 |
| Assembly level | Chromosomes | Scaffold | Scaffold | Complete genome |
| Genome size | 14.3 Mb | 12.7 Mb | 13.0 Mb | 12.2 Mb |
| Genes a | 8864 | 5566 | 6184 | 6002 |
| No of scaffolds b | 1002 | 7 | 76 | 17 |
| Scaffold N50 b | 189,765 | 2,123,196 | 700,888 | 924,431 |
| No. of contigs b | 1163 | 91 | 392 | 17 |
| Contig N50 (bp) b | 158,681 | 287,918 | 111,555 | 924,431 |
| No. of chromosomes | 13 | – | – | 16 |
| GC-content (%) | 44.7 | 44.5 | 44.6 | 38.3 |
| Total of CDS a | 8646 | 5057 | 6032 | 5771 |
| Gene annotation | [24] | [16] | [37] | [47,48] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sousa-Silva, M.; Vieira, D.; Soares, P.; Casal, M.; Soares-Silva, I. Expanding the Knowledge on the Skillful Yeast Cyberlindnera jadinii. J. Fungi 2021, 7, 36. https://doi.org/10.3390/jof7010036
Sousa-Silva M, Vieira D, Soares P, Casal M, Soares-Silva I. Expanding the Knowledge on the Skillful Yeast Cyberlindnera jadinii. Journal of Fungi. 2021; 7(1):36. https://doi.org/10.3390/jof7010036
Chicago/Turabian StyleSousa-Silva, Maria, Daniel Vieira, Pedro Soares, Margarida Casal, and Isabel Soares-Silva. 2021. "Expanding the Knowledge on the Skillful Yeast Cyberlindnera jadinii" Journal of Fungi 7, no. 1: 36. https://doi.org/10.3390/jof7010036
APA StyleSousa-Silva, M., Vieira, D., Soares, P., Casal, M., & Soares-Silva, I. (2021). Expanding the Knowledge on the Skillful Yeast Cyberlindnera jadinii. Journal of Fungi, 7(1), 36. https://doi.org/10.3390/jof7010036