Investigating the Smuts: Common Cues, Signaling Pathways, and the Role of MAT in Dimorphic Switching and Pathogenesis
Abstract
1. Introduction
2. Physiological Aspect of Fungal Dimorphism
2.1. External Factors Affecting Fungal Dimorphism
| Lineages/Species | Life Strategy | Environmental Cues * | References |
|---|---|---|---|
| ASCOMYCOTA Saccharomycotina Saccharomyces cerevisiae | Saprobe | Nutrient limitation (carbon, nitrogen) | [14] |
| Candida albicans | Opportunistic human pathogen | Temperature, serum, CO2, pH, farnesol, GlcNAc | [16] |
| Holleya sinecauda | Plant pathogen | Media solidity | [34] |
| Yarrowia lipolytica | Saprobe | Nitrogen source, GlcNAc, serum, citrate, pH, anaerobic | [21,24,29] |
| Taphrinomycotina Taphrina deformans | Plant pathogen | Unknown cue from host leaves | [36,37] |
| Schizosaccharomyces pombe | Saprobe | Nitrogen starvation | [15] |
| Eurotiomycetes Blastomyces dermatidis | Human pathogen | Temperature, GlcNAc | [6,30] |
| Coccidioides immitis | Human pathogen | Temperature | [6] |
| Talaromyces marneffii | Human pathogen | Temperature | [6] |
| Histoplasma capsulatum | Human pathogen | Temperature, GlcNAc | [6,30,38] |
| Sordariomycetes Sporothrix schenckii | Human pathogen | Temperature | [6] |
| Ophiostoma ulmi & O. novo-ulmi | Plant pathogen | Nitrogen source, inoculum density, quorum-sensing, linoleic acid | [23,39,40,41] |
| Verticillium albo-atrum | Plant pathogen | Culture agitation, inoculum density | [35] |
| Metarhizium rileyi | Insect pathogen | Host hemolymph, quorum-sensing | [42] |
| Dothideomycetes Zymoseptoria tritici | Plant pathogen | Nitrogen starvation | [19] |
| Aureobasidium pullulans | Saprobe | Nitrogen source, quorum-sensing, Zn2+ | [22,33] |
| Hortaea werneckii ** | Saprobe, opportunistic human pathogen | Temperature, CO2, cysteine, inoculum size, agitation | [43,44] |
| BASIDIOMYCOTA Ustilaginomycotina Ustilago maydis | Plant pathogen | Mating, lipid, hydrophobicity, acidic pH, nitrogen starvation | [18,31,45,46] |
| Malassezia spp. | Opportunistic human pathogen | L-DOPA, lipid on mammal skin, high CO2 tension | [47,48,49] |
| Pucciniomycotina Microbotryum lychnidis-dioicae | Plant pathogen | Unknown | [50] |
| Agaricomycotina Cryptococcus neoformans | Human pathogen | Mating, nitrogen starvation, temperature, CO2 | [17] |
| Trichosporon cutaneum | Saprobe | Nitrogen source, pH, temperature | [20] |
| Tremella spp. | Saprobe, mycoparasite | Mating, ploidy status, carbon sources, nitrogen sources | [25,26] |
| MUCOROMYCOTA Mucor spp. | Saprobe, opportunistic human pathogen | Carbon source, CO2 | [27] |
2.2. Comparative Physiological Studies in Ustilaginomycotina
3. Mating and Fungal Dimorphism
3.1. Cellular Communication in Dimorphic Fungi
3.2. Mating (MAT) Loci in U. maydis
3.3. Comparative MAT Loci Analyses in Ustilaginomycotina
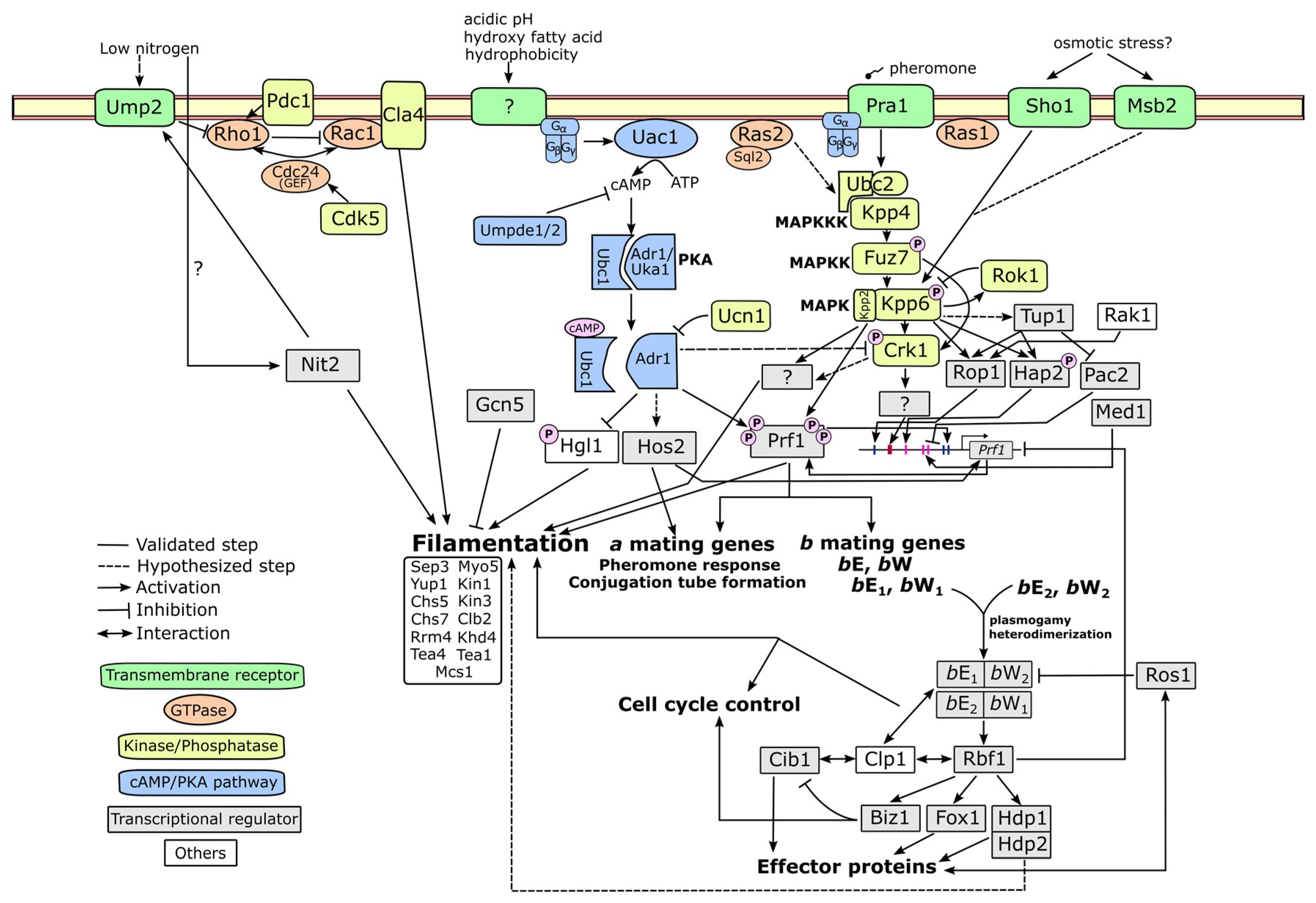
4. Molecular Mechanism Related to Fungal Dimorphism
4.1. cAMP-PKA Pathway
4.2. MAPK Pathway
4.3. Alternative Pathways
4.4. Upstream and Downstream Molecular Players
4.5. Comparative Genomics of Fungal Dimorphism Genes in Ustilaginomycotina
5. Benefits of Fungal Dimorphism
6. Conclusions and Perspectives
7. Materials and Methods
7.1. Physiological Study
7.2. MAT Loci Studies
7.3. Comparative Genomic Studies
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Zhao, Y.; Lin, J.; Fan, Y.; Lin, X. Life cycle of Cryptococcus neoformans. Annu. Rev. Microbiol. 2019, 73, 17–42. [Google Scholar] [CrossRef] [PubMed]
- Riquelme, M.; Aguirre, J.; Bartnicki-García, S.; Braus, G.H.; Feldbrügge, M.; Fleig, U.; Hansberg, W.; Herrera-Estrella, A.; Kämper, J.; Kück, U.; et al. Fungal Morphogenesis, from the Polarized Growth of Hyphae to Complex Reproduction and Infection Structures. Microbiol. Mol. Biol. Rev. 2018, 82, 1–47. [Google Scholar] [CrossRef] [PubMed]
- Kurtzman, C.P.; Sugiyama, J. Saccharomycotina and Taphrinomycotina: The Yeasts and Yeast-like Fungi of the Ascomycota. In The Mycota VII Systematics and Evolution Part B; McLaughlin, D.J., Spatafora, J.W., Eds.; Springer: Berlin/Heidelberg, Germany; New York, NY, USA, 2015; pp. 3–34. [Google Scholar]
- Berkeley, M.J. On a confervoid state of Mucor clavatus, Lk. Mag. Zool. Bot. 1838, 2, 340–343. [Google Scholar]
- Naranjo-Ortiz, M.A.; Gabaldón, T. Fungal evolution: Major ecological adaptations and evolutionary transitions. Biol. Rev. 2019. [Google Scholar] [CrossRef] [PubMed]
- Sil, A.; Andrianopoulos, A. Thermally dimorphic human fungal pathogens—Polyphyletic pathogens with a convergent pathogenicity trait. Cold Spring Harb. Perspect. Med. 2015, 5, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Begerow, D.; Schaffer, A.M.; Kellner, R.; Youkov, A.; Kemler, M.; Oberwinkler, F.; Bauer, R. Ustilaginomycotina. In Mycota VII Systematics and Evolution Part A; McLaughlin, D.J., Spatafora, J.W., Eds.; Springer: Berlin/Heidelberg, Germany; New York, NY, USA, 2014; pp. 295–329. [Google Scholar]
- Weiss, M.; Bauer, R.; Sampaio, J.P.; Oberwinkler, F. Tremellomycetes and Related Groups. In Mycota VII Systematics and Evolution Part A; McLaughlin, D.J., Spatafora, J.W., Eds.; Springer: Berlin/Heidelberg, Germany; New York, NY, USA, 2014; pp. 331–355. [Google Scholar]
- Oberwinkler, F. Yeasts in Pucciniomycotina. Mycol. Prog. 2017, 16, 831–856. [Google Scholar] [CrossRef]
- Vollmeister, E.; Schipper, K.; Baumann, S.; Haag, C.; Pohlmann, T.; Stock, J.; Feldbrügge, M. Fungal development of the plant pathogen Ustilago maydis. FEMS Microbiol. Rev. 2012, 36, 59–77. [Google Scholar] [CrossRef]
- Hibbett, D.S.; Stajich, J.E.; Spatafora, J.W. Toward genome-enabled mycology. Mycologia 2013, 105, 1339–1349. [Google Scholar] [CrossRef]
- Kijpornyongpan, T. Comparative Studies of Fungal Dimorphism in Dikarya; Purdue University: West Lafayette, IN, USA, 2019. [Google Scholar]
- Kijpornyongpan, T.; Mondo, S.J.; Barry, K.; Sandor, L.; Lee, J.; Lipzen, A.; Pangilinan, J.; LaButti, K.; Hainaut, M.; Henrissat, B.; et al. Broad Genomic Sampling Reveals a Smut Pathogenic Ancestry of the Fungal Clade Ustilaginomycotina. Mol. Biol. Evol. 2018, 35, 1840–1854. [Google Scholar] [CrossRef]
- Cullen, P.J.; Sprague, G.F. The regulation of filamentous growth in yeast. Genetics 2012, 190, 23–49. [Google Scholar] [CrossRef]
- Amoah-Buahin, E.; Bone, N.; Armstrong, J. Hyphal Growth in the Fission Yeast. Microbiology 2005, 4, 1287–1297. [Google Scholar] [CrossRef]
- Sudbery, P.E. Growth of Candida albicans hyphae. Nat. Rev. Microbiol. 2011, 9, 737–748. [Google Scholar] [CrossRef] [PubMed]
- Lin, X. Cryptococcus neoformans: Morphogenesis, infection, and evolution. Infect. Genet. Evol. 2009, 9, 401–416. [Google Scholar] [CrossRef]
- Paul, J.A.; Barati, M.T.; Cooper, M.; Perlin, H. Physical and Genetic Interaction between Ammonium Transporters and the Signaling Protein Rho1 in the Plant Pathogen Ustilago maydis. Eukaryot. Cell 2014, 13, 1328–1336. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Yemelin, A.; Brauchler, A.; Jacob, S.; Laufer, J.; Heck, L.; Foster, A.J.; Antelo, L.; Andresen, K.; Thines, E. Identification of factors involved in dimorphism and pathogenicity of Zymoseptoria tritici. PLoS ONE 2017, 12, e0183065. [Google Scholar] [CrossRef] [PubMed]
- Zhu, L.B.; Wang, Y.; Zhang, Z.B.; Yang, H.L.; Yan, R.M.; Zhu, D. Influence of environmental and nutritional conditions on yeast—Mycelial dimorphic transition in Trichosporon cutaneum. Biotechnol. Biotechnol. Equip. 2017, 2818. [Google Scholar] [CrossRef]
- Szabo, R. Dimorphism in Yarrowia lipolytica: Filament formation is suppressed by nitrogen starvation and inhibition of respiration. Folia Microbiol. (Praha) 1999, 44, 19–24. [Google Scholar] [CrossRef]
- Park, D. Population density and yeast mycelial dimorphism in Aureobasidium pullulans. Trans. Br. Mycol. Soc. 1984, 82, 39–44. [Google Scholar] [CrossRef]
- Kulkarni, R.K.; Nickerson, K.W. Nutritional control of dimorphism in Ceratocystis ulmi. Exp. Mycol. 1981, 5, 148–154. [Google Scholar] [CrossRef]
- Ruiz-Herrera, J.; Sentandreu, R. Different effectors of dimorphism in Yarrowia lipolytica. Arch. Microbiol. 2002, 178, 477–483. [Google Scholar] [CrossRef]
- Hou, L.; Chen, Y.; Ma, C.; Liu, J.; Chen, L.; Ma, A. Effects of environmental factors on dimorphic transition of the jelly mushroom Tremella fuciformis. Cryptogam. Mycol. 2011, 32, 421–428. [Google Scholar] [CrossRef]
- Pippola, E.; Kyroviita, M.-M. Growth and dimorphism of the mycoparasite Tremella encephala as affected by different nitrogen and carbon sources and the host presence. Cryptogamie 2009, 30, 3. [Google Scholar]
- Orlowski, M. Mucor Dimorphism. Microbiol. Rev. 1991, 55, 234–258. [Google Scholar] [CrossRef] [PubMed]
- Cullen, P.J.; Sprague, G.F. Glucose depletion causes haploid invasive growth in yeast. Proc. Natl. Acad. Sci. USA 2000, 97, 13619–13624. [Google Scholar] [CrossRef]
- Kim, J.; Cheon, S.A.; Park, S.; Song, Y.; Kim, J.Y. Serum-induced hypha formation in the dimorphic yeast Yarrowia lipolytica. FEMS Microbiol. Lett. 2000, 190, 9–12. [Google Scholar] [CrossRef]
- Gilmore, S.A.; Naseem, S.; Konopka, J.B.; Sil, A. N-acetylglucosamine (GlcNAc) Triggers a Rapid, Temperature-Responsive Morphogenetic Program in Thermally Dimorphic Fungi. PLoS Genet. 2013, 9. [Google Scholar] [CrossRef]
- Ruiz-Herrera, J.; Guevara-Olvera, C.G.L.L.; Cdrabez-Trejo, A. Yeast-mycelial dimorphism of haploid and diploid strains of Ustiago maydis. Microbiology 1995, 695–703. [Google Scholar] [CrossRef]
- Buffo, J.; Herman, M.A.; Soll, D.R. A characterization of pH-regulated dimorphism in Candida albicans. Mycopathologia 1984, 85, 21–30. [Google Scholar] [CrossRef]
- Reeslev, M.; Jorgensen, B.B.; Jorgensen, O.B. Influence of Zn2+ on yeast-mycelium dimorphism and exopolysaccharide production by the fungus Aureobasidium pullulans grown in a defined medium in continuous culture. J. Gen. Microbiol. 1993, 139, 3065–3070. [Google Scholar] [CrossRef][Green Version]
- Schade, D.; Walther, A.; Wendland, J. The development of a transformation system for the dimorphic plant pathogen Holleya sinecauda based on Ashbya gossypii DNA elements. Fungal Genet. Biol. 2003, 40, 65–71. [Google Scholar] [CrossRef]
- Keen, N.T.; Wang, M.C.; Long, M.; Erwin, D.C. Dimorphism in Verticillium albo-atrum as affected by initial spore concentration and antisporulant chemicals. Phytopathology 1971, 61, 1266–1269. [Google Scholar] [CrossRef]
- Rodrigues, M.G.; Fonseca, A. Molecular systematics of the dimorphic ascomycete genus Taphrina. Int. J. Syst. Evol. Microbiol. 2003, 53, 607–616. [Google Scholar] [CrossRef] [PubMed]
- Svetaz, L.A.; Bustamante, C.A.; Goldy, C.; Rivero, N.; Müller, G.L.; Valentini, G.H.; Fernie, A.R.; Drincovich, M.F.; Lara, M.V. Unravelling early events in the Taphrina deformans–Prunus persica interaction: An insight into the differential responses in resistant and susceptible genotypes. Plant Cell Environ. 2017, 40, 1456–1473. [Google Scholar] [CrossRef] [PubMed]
- Maresca, B.; Lambowitz, A.M.; Kumar, V.B.; Grant, G.A.; Kobayashi, G.S.; Medoff, G. Role of cysteine in regulating morphogenesis and mitochondrial activity in the dimorphic fungus Histoplasma capsulatum. Proc. Natl. Acad. Sci. USA 1981, 78, 4596–4600. [Google Scholar] [CrossRef]
- Hornby, J.M.; Jacobitz-Kizzier, S.M.; McNeel, D.J.; Jensen, E.C.; Treves, D.S.; Nickerson, K.W. Inoculum size effect in dimorphic fungi: Extracellular control of yeast-mycelium dimorphism in Ceratocystis ulmi. Appl. Environ. Microbiol. 2004, 70, 1356–1359. [Google Scholar] [CrossRef]
- Naruzawa, E.S.; Bernier, L. Control of yeast-mycelium dimorphism invitro in Dutch elm disease fungi by manipulation of specific external stimuli. Fungal Biol. 2014, 118, 872–884. [Google Scholar] [CrossRef]
- Naruzawa, E.S.; Malagnac, F.; Bernier, L. Effect of linoleic acid on reproduction and yeast-mycelium dimorphism in the Dutch elm disease pathogens. Botany 2016, 96, 31–39. [Google Scholar] [CrossRef]
- Boucias, D.; Liu, S.; Meagher, R.; Baniszewski, J. Fungal dimorphism in the entomopathogenic fungus Metarhizium rileyi: Detection of an in vivo quorum-sensing system. J. Invertebr. Pathol. 2016, 136, 100–108. [Google Scholar] [CrossRef]
- Hardcastle, R.V.; Szaniszlo, P.J. Characterization of dimorphism in Cladosporium werneckii. J. Bacteriol. 1974, 119, 294–302. [Google Scholar] [CrossRef]
- Houston, M.R.; Meyer, K.H.; Thomas, N.; Wolf, F.T. Dimorphism in Cladosporium werneckii. Sabouraudia J. Med. Vet. Mycol. 1969, 7, 195–198. [Google Scholar] [CrossRef]
- Brefort, T.; Doehlemann, G.; Mendoza-mendoza, A.; Reissmann, S.; Djamei, A.; Kahmann, R. Ustilago maydis as a Pathogen. Annu. Rev. Phytopathol. 2009, 47, 423–445. [Google Scholar] [CrossRef] [PubMed]
- Mendoza-Mendoza, A.; Berndt, P.; Djamei, A.; Weise, C.; Linne, U.; Marahiel, M.; Vraneš, M.; Kämper, J.; Kahmann, R. Physical-chemical plant-derived signals induce differentiation in Ustilago maydis. Mol. Microbiol. 2009, 71, 895–911. [Google Scholar] [CrossRef] [PubMed]
- Youngchim, S.; Nosanchuk, J.D.; Pornsuwan, S.; Kajiwara, S.; Vanittanakom, N. The role of L-DOPA on melanization and mycelial production in Malassezia furfur. PLoS ONE 2013, 8, e63764. [Google Scholar] [CrossRef] [PubMed]
- Faergemann, J.; Bernander, S. Micro-aerophilic and anaerobic growth of Pityrosporum species. Med. Mycol. 1981, 19, 117–121. [Google Scholar] [CrossRef]
- Faergemann, J. A new model for growth and filament production of Pityrosporum ovale (orbiculare) on human stratum corneum in vitro. J. Investig. Dermatol. 1989, 92, 117–119. [Google Scholar] [CrossRef]
- Schäfer, A.M.; Kemler, M.; Bauer, R.; Begerow, D. The illustrated life cycle of Microbotryum on the host plant Silene latifolia. Botany 2010, 88, 875–885. [Google Scholar] [CrossRef]
- Morrow, C.A.; Fraser, J.A. Sexual reproduction and dimorphism in the pathogenic basidiomycetes. FEMS Yeast Res. 2009, 9, 161–177. [Google Scholar] [CrossRef]
- Lanver, D.; Mendoza-Mendoza, A.; Brachmann, A.; Kahmann, R. Sho1 and Msb2-related proteins regulate appressorium development in the smut fungus Ustilago maydis. Plant Cell 2010, 22, 2085–2101. [Google Scholar] [CrossRef]
- Domínguez, E.; Cuartero, J.; Heredia, A. An overview on plant cuticle biomechanics. Plant Sci. 2011, 181, 77–84. [Google Scholar] [CrossRef]
- Klose, J.; De Sá, M.M.; Kronstad, J.W. Lipid-induced filamentous growth in Ustilago maydis. Mol. Microbiol. 2004, 52, 823–835. [Google Scholar] [CrossRef]
- Rush, T.A.; Aime, M.C. The genus Meira: Phylogenetic placement and description of a new species. Antonie Van Leeuwenhoek 2013, 103, 1097–1106. [Google Scholar] [CrossRef] [PubMed]
- Kijpornyongpan, T.; Aime, M.C. Taxonomic revisions in the Microstromatales: Two new yeast species, two new genera, and validation of Jaminaea and two Sympodiomycopsis species. Mycol. Prog. 2017, 16, 495–505. [Google Scholar] [CrossRef]
- Albu, S.; Toome, M.; Aime, M.C. Violaceomyces palustris gen. et sp. nov. and a new monotypic lineage, Violaceomycetales ord. nov. in Ustilaginomycetes. Mycologia 2015, 107, 1193–1204. [Google Scholar] [CrossRef] [PubMed]
- Boekhout, T.; Gildemacher, P.; Theelen, B.; Müller, W.H.; Heijne, B.; Lutz, M. Extensive colonization of apples by smut anamorphs causes a new postharvest disorder. FEMS Yeast Res. 2006, 6, 63–76. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.; Zhou, X.; Li, G.; Li, L.; Kong, L.; Wang, C.; Zhang, H.; Xu, J.R. Multiple plant surface signals are sensed by different mechanisms in the rice blast fungus for appressorium formation. PLoS Pathog. 2011, 7. [Google Scholar] [CrossRef] [PubMed]
- Zabka, V.; Stangl, M.; Bringmann, G.; Vogg, G.; Riederer, M.; Hildebrandt, U. Host surface properties affect prepenetration processes in the barley powdery mildew fungus. New Phytol. 2008, 177, 251–263. [Google Scholar] [CrossRef]
- Borges-Walmsley, M.I.; Walmsley, A.R. cAMP signalling in pathogenic fungi: Control of dimorphic switching and pathogenicity. Trends Microbiol. 2000, 8, 133–141. [Google Scholar] [CrossRef]
- Wongsuk, T.; Pumeesat, P.; Luplertlop, N. Fungal quorum sensing molecules: Role in fungal morphogenesis and pathogenicity. J. Basic Microbiol. 2016, 56, 440–447. [Google Scholar] [CrossRef]
- Boekhout, T.; Fonseca, A.; Sampaio, J.P.; Bandoni, R.J.; Fell, J.W.; Kwon-Chung, K.J. Discussion of Teleomorphic and Anamorphic Basidiomycetous Yeasts. In The Yeasts, a Taxonomic Study Volume 3; Kurtzman, C.P., Fell, J.W., Boekhout, T., Eds.; Elseiver: Burlington, MA, USA, 2011; pp. 1339–1371. [Google Scholar]
- Spellig, T.; Bölker, M.; Lottspeich, F.; Frank, R.W.; Kahmann, R. Pheromones trigger filamentous growth in Ustilago maydis. EMBO J. 1994, 13, 1620–1627. [Google Scholar] [CrossRef]
- Bölker, M.; Urban, M.; Kahmann, R. The a mating type locus of U. maydis specifies cell signaling components. Cell 1992, 68, 441–450. [Google Scholar] [CrossRef]
- Hartmann, H.A.; Kahmann, R.; Bolker, M. The pheromone response factor coordinates filamentous growth and pathogenicity in Ustilago maydis. Embo J. 1996, 15, 1632–1641. [Google Scholar] [CrossRef]
- Schirawski, J.; Heinze, B.; Wagenknecht, M.; Kahmann, R. Mating type loci of Sporisorium reilianum: Novel pattern with three a and multiple b specificities. Eukaryot. Cell 2005, 4, 1317–1327. [Google Scholar] [CrossRef]
- Yan, M.; Zhu, G.; Lin, S.; Xian, X.; Chang, C.; Xi, P.; Shen, W.; Huang, W.; Cai, E.; Jiang, Z.; et al. The mating-type locus b of the sugarcane smut Sporisorium scitamineum is essential for mating, filamentous growth and pathogenicity. Fungal Genet. Biol. 2016, 86, 1–8. [Google Scholar] [CrossRef]
- Kües, U.; James, T.; Heitman, J. Mating Type in Basidiomycetes: Unipolar, Bipolar, and Tetrapolar Patterns of Sexuality. Evol. Fungi Fungal-Like Org. 2011, 14, 97–160. [Google Scholar] [CrossRef]
- Sabbagh, S.K.; Diagne-Lèye, G.; Naudan, M.; Roux, C.P. Solopathogenic strain formation strongly differs among Ustilaginaceae species. FEMS Microbiol. Lett. 2010, 305, 121–127. [Google Scholar] [CrossRef][Green Version]
- Holliday, R. Ustilago maydis . In Handbook of Genetics, Vol. 1. Bacteria, Bacteriophages and Fungi; King, R.C., Ed.; Springer: Plenum, NY, USA, 1997; pp. 575–595. [Google Scholar]
- Bölker, M.; Genin, S.; Lehmler, C.; Kahmann, R. Genetic regulation of mating and dimorphism in Ustilago maydis. Can. J. Bot. 1995, 73, 320–325. [Google Scholar] [CrossRef]
- Billiard, S.; López-Villavicencio, M.; Devier, B.; Hood, M.E.; Fairhead, C.; Giraud, T. Having sex, yes, but with whom? Inferences from fungi on the evolution of anisogamy and mating types. Biol. Rev. 2011, 86, 421–442. [Google Scholar] [CrossRef]
- Kellner, R.; Vollmeister, E.; Feldbrügge, M.; Begerow, D. Interspecific sex in grass smuts and the genetic diversity of their pheromone-receptor system. PLoS Genet. 2011, 7. [Google Scholar] [CrossRef]
- Kämper, J.; Kahmann, R.; Bölker, M.; Ma, L.-J.; Brefort, T.; Saville, B.J.; Banuett, F.; Kronstad, J.W.; Gold, S.E.; Müller, O.; et al. Insights from the genome of the biotrophic fungal plant pathogen Ustilago maydis. Nature 2006, 444, 97–101. [Google Scholar] [CrossRef]
- Schirawski, J.; Mannhaupt, G.; Münch, K.; Brefort, T.; Schipper, K.; Doehlemann, G.; Di Stasio, M.; Rössel, N.; Mendoza-Mendoza, A.; Pester, D.; et al. Pathogenicity determinants in smut fungi revealed by genome comparison. Science 2010, 330, 1546–1548. [Google Scholar] [CrossRef]
- Konishi, M.; Hatada, Y.; Horiuchi, J.-I. Draft Genome Sequence of the Basidiomycetous Yeast-Like Fungus Pseudozyma hubeiensis SY62, Which Produces an Abundant Amount of the Biosurfactant Mannosylerythritol Lipids. Genome Announc. 2013, 1, e00409-13. [Google Scholar] [CrossRef]
- Morita, T.; Koike, H.; Koyama, Y.; Hagiwara, H.; Ito, E.; Fukuoka, T.; Imura, T.; Machida, M.; Kitamoto, D. Genome sequence of the basidiomycetous yeast Pseudozyma antarctica T-34, a producer of the glycolipid biosurfactants mannosylerythritol lipids. Genome Announc. 2013, 1, e00064-13. [Google Scholar] [CrossRef]
- Xu, J.; Saunders, C.W.; Hu, P.; Grant, R.A.; Boekhout, T.; Kuramae, E.E.; Kronstad, J.W.; Deangelis, Y.M.; Reeder, N.L.; Johnstone, K.R.; et al. Dandruff-associated Malassezia genomes reveal convergent and divergent virulence traits shared with plant and human fungal pathogens. Proc. Natl. Acad. Sci. USA 2007, 104, 18730–18735. [Google Scholar] [CrossRef]
- Gioti, A.; Nystedt, B.; Li, W. Genomic Insights into the Atopic Eczema-Associated Skin Commensal Yeast Malassezia sympodialis. MBio 2013, 4, 1–16. [Google Scholar] [CrossRef]
- Toome, M.; Kuo, A.; Henrissat, B.; Lipzen, A.; Tritt, A.; Yoshinaga, Y.; Zane, M.; Barry, K.; Grigoriev, I.V.; Spatafora, J.W.; et al. Draft Genome Sequence of a Rare Smut Relative, Tilletiaria anomala UBC 951. Genome Announc. 2014, 2, 2–3. [Google Scholar] [CrossRef]
- Hamel, L.-P.; Nicole, M.-C.; Duplessis, S.; Ellis, B.E. Mitogen-Activated Protein Kinase Signaling in Plant-Interacting Fungi: Distinct Messages from Conserved Messengers. Plant Cell 2012, 24, 1327–1351. [Google Scholar] [CrossRef]
- Fuller, K.K.; Rhodes, J.C. Protein kinase A and fungal virulence A sinister side to a conserved nutrient sensing pathway. Virulence 2012, 3, 109–121. [Google Scholar] [CrossRef]
- Regenfelder, E.; Spellig, T.; Hartmann, A.; Lauenstein, S.; Bölker, M.; Kahmann, R. G proteins in Ustilago maydis: Transmission of multiple signals? EMBO J. 1997, 16, 1934–1942. [Google Scholar] [CrossRef]
- Müller, P.; Leibbrandt, A.; Teunissen, H.; Cubasch, S.; Aichinger, C.; Kahmann, R. The Gβ-subunit-encoding gene bpp1 controls cyclic-AMP signaling in Ustilago maydis. Eukaryot. Cell 2004, 3, 806–814. [Google Scholar] [CrossRef]
- Gold, S.; Duncan, G.; Barrett, K.; Kronstad, J. cAMP regulates morphogenesis in the fungal pathogen Ustilago maydis. Genes Dev. 1994, 8, 2805–2816. [Google Scholar] [CrossRef]
- Gold, S.E.; Brogdon, S.M.; Mayorga, M.E.; Kronstad, J.W. The Ustilago maydis regulatory subunit of a cAMP-dependent protein kinase is required for gall formation in maize. Plant Cell 1997, 9, 1585–1594. [Google Scholar]
- Dürrenberger, F.; Wong, K.; Kronstad, J.W. Identification of a cAMP-dependent protein kinase catalytic subunit required for virulence and morphogenesis in Ustilago maydis. Proc. Natl. Acad. Sci. USA 1998, 95, 5684–5689. [Google Scholar] [CrossRef]
- Agarwal, C.; Aulakh, K.B.; Edelen, K.; Cooper, M.; Wallen, R.M.; Adams, S.; Schultz, D.J.; Perlin, M.H. Ustilago maydis phosphodiesterases play a role in the dimorphic switch and in pathogenicity. Microbiology 2013, 159, 857–868. [Google Scholar] [CrossRef][Green Version]
- Krüger, J.; Loubradou, G.; Regenfelder, E.; Hartmann, A.; Kahmann, R. Crosstalk between cAMP and pheromone signalling pathways in Ustilago maydis. Mol. Gen. Genet. 1998, 260, 193–198. [Google Scholar] [CrossRef]
- Martínez-Espinoza, A.D.; Ruiz-Herrera, J.; León-Ramírez, C.G.; Gold, S.E. MAP kinase and cAMP signaling pathways modulate the pH-induced yeast-to-mycelium dimorphic transition in the corn smut fungus Ustilago maydis. Curr. Microbiol. 2004, 49, 274–281. [Google Scholar] [CrossRef]
- Müller, P.; Weinzierl, G.; Brachmann, A.; Feldbrügge, M.; Kahmann, R. Mating and pathogenic development of the smut fungus Ustilago maydis are regulated by one mitogen-activated protein kinase cascade. Eukaryot. Cell 2003, 2, 1187–1199. [Google Scholar] [CrossRef]
- Andrews, D.L.; Egan, J.D.; Mayorga, M.E.; Gold, S.E. The Ustilago maydis ubc4 and ubc5 genes encode members of a MAP kinase cascade required for filamentous growth. Mol. Plant-Microbe Interact. 2000, 13, 781–786. [Google Scholar] [CrossRef]
- Banuett, F.; Herskowitz, I. Identification of fuz7, a Ustilago maydis MEK/MAPKK homolog required for a-locus-dependent and-independent steps in the fungal life cycle. Genes Dev. 1994, 8, 1367–1378. [Google Scholar] [CrossRef] [PubMed]
- Müller, P.; Aichinger, C.; Feldbrügge, M.; Kahmann, R. The MAP kinase kpp2 regulates mating and pathogenic development in Ustilago maydis. Mol. Microbiol. 1999, 34, 1007–1017. [Google Scholar] [CrossRef]
- Mayorga, M.E.; Gold, S.E. A MAP kinase encoded by the ubc3 gene of Ustilago maydis is required for filamentous growth and full virulence. Mol. Microbiol. 1999, 34, 485–497. [Google Scholar] [CrossRef]
- Brachmann, A.; Schirawski, J.; Müller, P.; Kahmann, R. An unusual MAP kinase is required for efficient penetration of the plant surface by Ustilago maydis. EMBO J. 2003, 22, 2199–2210. [Google Scholar] [CrossRef] [PubMed]
- Garrido, E.; Voß, U.; Müller, P.; Castillo-lluva, S.; Kahmann, R. The induction of sexual development and virulence in the smut fungus Ustilago maydis depends on Crk1, a novel MAPK protein. Genes Dev. 2004, 7, 3117–3130. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Klosterman, S.J.; Martinez-Espinoza, A.D.; Andrews, D.L.; Seay, J.R.; Gold, S.E. Ubc2, an ortholog of the yeast Ste50p adaptor, possesses a basidiomycete-specific carboxy terminal extension essential for pathogenicity independent of pheromone response. Mol. Plant-Microbe Interact. 2008, 21, 110–121. [Google Scholar] [CrossRef] [PubMed]
- Mayorga, M.E.; Gold, S.E. The ubc2 gene of Ustilago maydis encodes a putative novel adaptor protein required for filamentous growth, pheromone response and virulence. Mol. Microbiol. 2001, 41, 1365–1379. [Google Scholar] [CrossRef]
- Di Stasio, M.; Brefort, T.; Mendoza-Mendoza, A.; Münch, K.; Kahmann, R. The dual specificity phosphatase Rok1 negatively regulates mating and pathogenicity in Ustilago maydis. Mol. Microbiol. 2009, 73, 73–88. [Google Scholar] [CrossRef]
- Garrido, E.; Pérez-Martín, J. The crk1 gene encodes an ime2-related protein that is required for morphogenesis in the plant pathogen Ustilago maydis. Mol. Microbiol. 2003, 47, 729–743. [Google Scholar] [CrossRef]
- Strudwick, N.; Brown, M.; Parmar, V.M.; Schroder, M. Ime1 and Ime2 Are Required for Pseudohyphal Growth of Saccharomyces cerevisiae on Nonfermentable Carbon Sources. Mol. Cell. Biol. 2010, 30, 5514–5530. [Google Scholar] [CrossRef]
- Lee, N.; Kronstad, J.W. ras2 controls morphogenesis, pheromone response, and pathogenicity in the fungal pathogen Ustilago maydis. Eukaryot. Cell 2002, 1, 954–966. [Google Scholar] [CrossRef]
- Müller, P.; Katzenberger, D.; Loubradou, G.; Kahmann, R. Guanyl Nucleotide Exchange Factor Sql2 and Ras2 Regulate Filamentous Growth in Ustilago maydis. Eukaryot. Cell 2003, 2, 609–617. [Google Scholar] [CrossRef]
- Mahlert, M.; Leveleki, L.; Hlubek, A.; Sandrock, B.; Bölker, M. Rac1 and Cdc42 regulate hyphal growth and cytokinesis in the dimorphic fungus Ustilago maydis. Mol. Microbiol. 2006, 59, 567–578. [Google Scholar] [CrossRef]
- Pham, C.D.; Yu, Z.; Bolker, M.; Gold, S.E.; Perlin, M.H. Ustilago maydis Rho1 and 14-3-3 Homologues Participate in Pathways Controlling Cell Separation and Cell Polarity. Eukaryot. Cell 2009, 8, 977–989. [Google Scholar] [CrossRef] [PubMed]
- Leveleki, L.; Mahlert, M.; Sandrock, B.; Bölker, M. The PAK family kinase Cla4 is required for budding and morphogenesis in Ustilago maydis. Mol. Microbiol. 2004, 54, 396–406. [Google Scholar] [CrossRef] [PubMed]
- Castillo-Lluva, S.; Alvarez-Tabares, I.; Weber, I.; Steinberg, G.; Perez-Martin, J. Sustained cell polarity and virulence in the phytopathogenic fungus Ustilago maydis depends on an essential cyclin-dependent kinase from the Cdk5/Pho85 family. J. Cell Sci. 2007, 120, 1584–1595. [Google Scholar] [CrossRef] [PubMed]
- Boyce, K.J.; Andrianopoulos, A. Fungal dimorphism: The switch from hyphae to yeast is a specialized morphogenetic adaptation allowing colonization of a host. FEMS Microbiol. Rev. 2015, 39, 797–811. [Google Scholar] [CrossRef] [PubMed]
- Campos, C.B.L.; Di Benedette, J.P.T.; Morais, F.V.; Ovalle, R.; Nobrega, M.P. Evidence for the role of calcineurin in morphogenesis and calcium homeostasis during mycelium-to-yeast dimorphism of Paracoccidioides brasiliensis. Eukaryot. Cell 2008, 7, 1856–1864. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.C.; Li, A.; Calo, S.; Heitman, J. Calcineurin Plays Key Roles in the Dimorphic Transition and Virulence of the Human Pathogenic Zygomycete Mucor circinelloides. PLoS Pathog. 2013, 9. [Google Scholar] [CrossRef]
- Lee, S.C.; Li, A.; Calo, S.; Inoue, M.; Tonthat, N.K.; Bain, J.M.; Louw, J.; Shinohara, M.L.; Erwig, L.P.; Schumacher, M.A.; et al. Calcineurin orchestrates dimorphic transitions, antifungal drug responses and host—Pathogen interactions of the pathogenic mucoralean fungus Mucor circinelloides. Mol. Microbiol. 2015, 97, 844–865. [Google Scholar] [CrossRef]
- Egan, J.D.; García-pedrajas, M.D.; Andrews, D.L.; Gold, S.E. Calcineurin Is an Antagonist to PKA Protein Phosphorylation Required for Postmating Filamentation and Virulence, While PP2A Is Required for Viability in Ustilago maydis. Mol. Plant-Microbe Interact. 2009, 22, 1293–1301. [Google Scholar] [CrossRef]
- Paul, J.A.; Wallen, R.M.; Zhao, C.; Shi, T.; Perlin, M.H. Coordinate regulation of Ustilago maydis ammonium transporters and genes involved in mating and pathogenicity. Fungal Biol. 2018, 122, 639–650. [Google Scholar] [CrossRef]
- Brefort, T.; Müller, P.; Kahmann, R. The high-mobility-group domain transcription factor Rop1 is a direct regulator of prf1 in Ustilago maydis. Eukaryot. Cell 2005, 4, 379–391. [Google Scholar] [CrossRef][Green Version]
- Hartmann, H.A.; Krüger, J.; Lottspeich, F.; Kahmann, R. Environmental signals controlling sexual development of the corn smut fungus Ustilago maydis through the transcriptional regulator Prf1. Plant Cell 1999, 11, 1293–1305. [Google Scholar] [CrossRef]
- Elías-Villalobos, A.; Fernández-Álvarez, A.; Ibeas, J.I. The general transcriptional repressor Tup1 is required for dimorphism and virulence in a fungal plant pathogen. PLoS Pathog. 2011, 7, e1002235. [Google Scholar] [CrossRef]
- Chacko, N.; Gold, S. Deletion of the Ustilago maydis ortholog of the Aspergillus sporulation regulator medA affects mating and virulence through pheromone response. Fungal Genet. Biol. 2012, 49, 426–432. [Google Scholar] [CrossRef]
- Wang, L.; Berndt, P.; Xia, X.; Kahnt, J.; Kahmann, R. A seven-WD40 protein related to human RACK1 regulates mating and virulence in Ustilago maydis. Mol. Microbiol. 2011, 81, 1484–1498. [Google Scholar] [CrossRef]
- Elías-Villalobos, A.; Fernández-Alvarez, A.; Moreno-Sánchez, I. The Hos2 Histone Deacetylase Controls Ustilago maydis Virulence through Direct Regulation of Mating-Type Genes. PLoS Pathog. 2015, 11, e1005134. [Google Scholar] [CrossRef]
- Kaffarnik, F.; Müller, P.; Leibundgut, M.; Kahmann, R.; Feldbrügge, M. PKA and MAPK phosphorylation of Prf1 allows promoter discrimination in Ustilago maydis. EMBO J. 2003, 22, 5817–5826. [Google Scholar] [CrossRef]
- Brachmann, A.; Weinzierl, G. Identification of genes in the bW/bE regulatory cascade in Ustilago maydis. Mol. Microbiol. 2001, 42, 1047–1063. [Google Scholar] [CrossRef]
- Heimel, K.; Scherer, M.; Vranes, M.; Wahl, R.; Pothiratana, C.; Schuler, D.; Vincon, V.; Finkernagel, F.; Flor-Parra, I.; Kämper, J. The Transcription Factor Rbf1 Is the Master Regulator for b-Mating Type Controlled Pathogenic Development in Ustilago maydis. PLOS Pathog. 2010, 6, e1001035. [Google Scholar] [CrossRef]
- Tollot, M.; Assmann, D.; Becker, C.; Altmüller, J. The WOPR Protein Ros1 Is a Master Regulator of Sporogenesis and Late Effector Gene Expression in the Maize Pathogen Ustilago maydis. PLoS Pathog. 2016, 12, e1005697. [Google Scholar] [CrossRef]
- Dürrenberger, F.; Laidlaw, R.D.; Kronstad, J.W. The hgl1 gene is required for dimorphism and teliospore formation in the fungal pathogen Ustilago maydis. Mol. Microbiol. 2001, 41, 337–348. [Google Scholar] [CrossRef]
- Horst, R.J.; Zeh, C.; Saur, A.; Sonnewald, S.; Sonnewald, U.; Voll, L.M. The Ustilago maydis Nit2 Homolog Regulates Nitrogen Utilization and Is Required for Efficient Induction of Filamentous Growth. Eukaryot. Cell 2012, 11, 368–380. [Google Scholar] [CrossRef]
- González-prieto, J.M.; Rosas-quijano, R.; Domínguez, A.; Ruiz-herrera, J. The UmGcn5 gene encoding histone acetyltransferase from Ustilago maydis is involved in dimorphism and virulence. Fungal Genet. Biol. 2014, 71, 86–95. [Google Scholar] [CrossRef]
- Weber, I.; Gruber, C.; Steinberg, G. A class-V myosin required for mating, hyphal growth, and pathogenicity in the dimorphic plant pathogen Ustilago maydis. Plant Cell 2003, 15, 2826–2842. [Google Scholar] [CrossRef]
- Steinberg, G.; Schliwa, M.; Lehmler, C.; Bolker, M.; Kahmann, R.; McIntosh, J.R. Kinesin from the plant pathogenic fungus Ustilago maydis is involved in vacuole formation and cytoplasmic migration. J. Cell Sci. 1998, 111, 2235–2246. [Google Scholar]
- Schuchardt, I.; Aßmann, D.; Thines, E.; Schuberth, C.; Steinberg, G. Myosin-V, Kinesin-1, and Kinesin-3 cooperate in hyphal growth of the fungus Ustilago maydis. Mol. Biol. Cell 2005, 16, 5191–5201. [Google Scholar] [CrossRef]
- Weber, I.; Thines, E.; Steinberg, G. Polar Localizing Class V Myosin Chitin Synthases Are Essential during Early Plant Infection in the Plant Pathogenic Fungus Ustilago maydis. Plant Cell 2006, 18, 225–242. [Google Scholar] [CrossRef]
- García-Muse, T.; Steinberg, G.; Pérez-Martín, J. Characterization of B-type cyclins in the smut fungus Ustilago maydis: Roles in morphogenesis and pathogenicity. J. Cell Sci. 2004, 117, 487–506. [Google Scholar] [CrossRef]
- Boyce, K.J.; Chang, H.; D’Souza, C.A.; Kronstad, J.W. An Ustilago maydis septin is required for filamentous growth in culture and for full symptom development on maize. Eukaryot. Cell 2005, 4, 2044–2056. [Google Scholar] [CrossRef]
- Scherer, M.; Heimel, K.; Starke, V.; Kämper, J. The Clp1 Protein Is Required for Clamp Formation and Pathogenic Development of Ustilago maydis. Plant Cell 2006, 18, 2388–2401. [Google Scholar] [CrossRef]
- Wedlich-Söldner, R.; Bölker, M.; Kahmann, R.; Steinberg, G. A putative endosomal t-SNARE links exo-and endocytosis in the phytopathogenic fungus Ustilago maydis. EMBO J. 2000, 19, 1974–1986. [Google Scholar] [CrossRef]
- Valinluck, M.; Woraratanadharm, T.; Lu, C.Y.; Quintanilla, R.H.; Banuett, F. The cell end marker Tea4 regulates morphogenesis and pathogenicity in the basidiomycete fungus Ustilago maydis. Fungal Genet. Biol. 2014, 66, 54–68. [Google Scholar] [CrossRef]
- Woraratanadharm, T.; Kmosek, S.; Banuett, F. UmTea1, a Kelch and BAR domain-containing protein, acts at the cell cortex to regulate cell morphogenesis in the dimorphic fungus Ustilago maydis. Fungal Genet. Biol. 2018, 121, 10–28. [Google Scholar] [CrossRef]
- Becht, P.; Vollmeister, E.; Feldbrügge, M. Role for RNA-Binding Proteins Implicated in Pathogenic Development of Ustilago maydis. Eukaryot. Cell 2005, 4, 121–133. [Google Scholar] [CrossRef]
- Vollmeister, E.; Haag, C.; Zarnack, K.; Baumann, S.; König, J.; Mannhaupt, G.; Feldbrügge, M. Tandem KH domains of Khd4 recognize AUACCC and are essential for regulation of morphology as well as pathogenicity in Ustilago maydis. RNA 2009, 15, 2206–2218. [Google Scholar] [CrossRef]
- Becht, P.; König, J.; Feldbrügge, M. The RNA-binding protein Rrm4 is essential for polarity in Ustilago maydis and shuttles along microtubules. J. Cell Sci. 2006, 119, 4964–4973. [Google Scholar] [CrossRef]
- Lorenz, S.; Guenther, M.; Grumaz, C.; Rupp, S.; Zibek, S.; Sohn, K. Genome sequence of the basidiomycetous fungus Pseudozyma aphidis DSM70725, an efficient producer of biosurfactant mannosylerythritol lipids. Genome Announc. 2014, 2, e00053-14. [Google Scholar] [CrossRef]
- Gauthier, G.M. Dimorphism in Fungal Pathogens of Mammals, Plants, and Insects. PLoS Pathog. 2015, 11, e1004608. [Google Scholar] [CrossRef]
- Noble, S.M.; Gianetti, B.A.; Witchley, J.N. Candida albicans cell-type switching and functional plasticity in the mammalian host. Nat. Rev. Microbiol. 2017, 15, 96–108. [Google Scholar] [CrossRef]
- Joo, H.; Choi, Y.; Cho, S.; Choi, J.; Lee, D.; Kim, H.; Jo, I.; Park, Y.; Lee, K. Pseudozyma aphidis fungaemia with invasive fungal pneumonia in a patient with acute myeloid leukaemia: Case report and literature review. Mycoses 2016, 59, 56–61. [Google Scholar] [CrossRef]
- Herb, A.; Sabou, M.; Delhorme, J.-B.; Pessaux, P.; Mutter, D.; Candolfi, E.; Letscher-Bru, V. Pseudozyma aphidis fungemia after abdominal surgery: First adult case. Med. Mycol. Case Rep. 2015, 8, 37–39. [Google Scholar] [CrossRef]
- Nadal, M.; García-Pedrajas, M.D.; Gold, S.E. Dimorphism in fungal plant pathogens. FEMS Microbiol. Lett. 2008, 284, 127–134. [Google Scholar] [CrossRef] [PubMed]
- Baric, S.; Lindner, L.; Marschall, K.; Dalla Via, J. Haplotype diversity of Tilletiopsis spp. causing white haze in apple orchards in Northern Italy. Plant Pathol. 2010, 59, 535–541. [Google Scholar] [CrossRef]
- Comeau, A.M.; Dufour, J.; Bouvet, G.F.; Jacobi, V.; Nigg, M.; Henrissat, B.; Laroche, J.; Levesque, R.C.; Bernier, L. Functional annotation of the Ophiostoma novo-ulmi genome: Insights into the phytopathogenicity of the fungal agent of Dutch elm disease. Genome Biol. Evol. 2015, 7, 410–430. [Google Scholar] [CrossRef] [PubMed]
- Wanchoo, A.; Lewis, M.W.; Keyhani, N.O. Lectin mapping reveals stage-specific display of surface carbohydrates in in vitro and haemolymph- derived cells of the entomopathogenic fungus Beauveria bassiana. Microbiology 2009, 155, 3121–3133. [Google Scholar] [CrossRef]
- Manning, M.; Mitchell, T.G. Strain variation and morphogenesis of yeast- and mycelial-phase Candida albicans in low-sulfate, synthetic medium. J. Bacteriol. 1980, 142, 714–719. [Google Scholar] [CrossRef]
- Henninger, W.; Windisch, S. A New Yeast of Sterigmatomyces, S. aphidis sp. nov. Arch. Microbiol. 1975, 105, 49–50. [Google Scholar] [CrossRef]
- Boekhout, T. Pseudozyma Bandoni emend. for yeast-like anamorphs of Ustilaginales. J. Gen. Appl. Microbiol. 1995, 41, 359–366. [Google Scholar] [CrossRef]
- Doiphode, N.; Joshi, C.; Ghormade, V.; Deshpande, M. V Biotechnological Applications of Dimorphic Yeasts. In Yeast Biotechnology: Diversity and Applications; Satyanarayana, T., Kunze, G., Eds.; Springer: Berlin/Heidelberg, Germany, 2009; pp. 635–650. [Google Scholar]
- Ceccato-antonini, S.R.; Sudbery, P.E. Filamentous Growth in Saccharomyces cerevisiae. Braz. J. Microbiol. 2004, 35, 173–181. [Google Scholar] [CrossRef]
- Ruiz-herrera, J.; Campos-Góngora, E. An Introduction to Fungal Dimorphism. In Dimorphic Fungi: Their Importance as Models for Differentiation and Fungal Pathogenesis; Ruiz-herrera, J., Ed.; Bentham Science: Sharjah, UAE, 2012; pp. 3–15. [Google Scholar]
- Doëhlemann, G. Exobasidium vaccinii MPITM Genome sequencing. unpublished.
- Wickham, H. ggplot2: Elegant Graphics for Data Analysis; Springer: Berlin/Heidelberg, Germany, 2016; ISBN 3319242776. [Google Scholar]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- Emms, D.M.; Kelly, S. OrthoFinder: Solving fundamental biases in whole genome comparisons dramatically improves orthogroup inference accuracy. Genome Biol. 2015, 16, 157. [Google Scholar] [CrossRef]

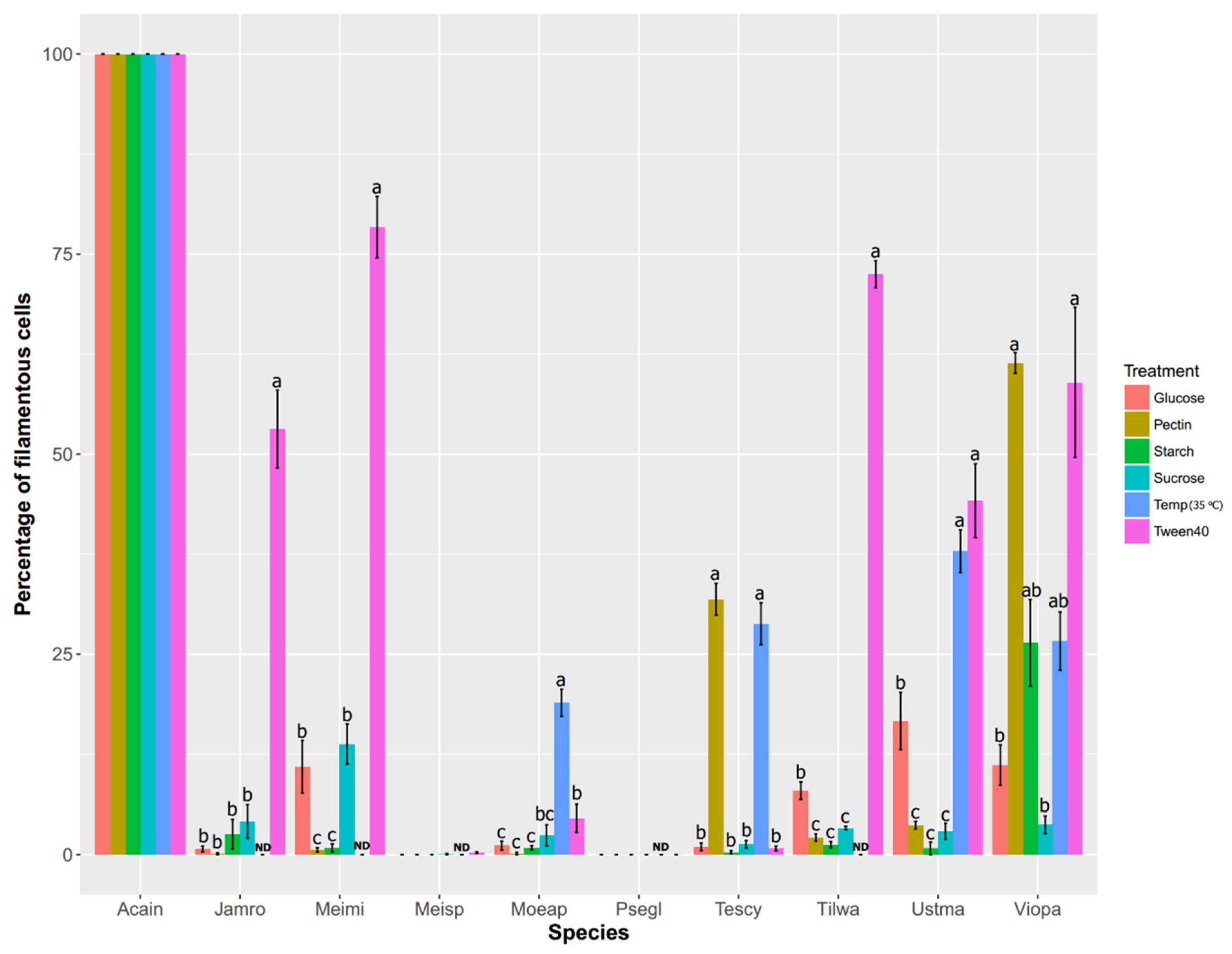

| Gene Name | Ustma GeneID | Acain | Cergu | Exova | Jamro | Malgl | Meimi | Moeap | Psean | Psehu | Psegl | Spore | Tescy | Tilan | Tilwa | Ustma | Viopa |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Msb2 | UMAG_00480 | ||||||||||||||||
| Pra1 | UMAG_02383 | ||||||||||||||||
| Sho1 | UMAG_03156 | ||||||||||||||||
| Ump2 | UMAG_05889 | ||||||||||||||||
| Adr1 | UMAG_04456 | ||||||||||||||||
| Bpp1 | UMAG_00703 | ||||||||||||||||
| Gpa3 | UMAG_04474 | ||||||||||||||||
| Uac1 | UMAG_05232 | ||||||||||||||||
| Ubc1 | UMAG_00525 | ||||||||||||||||
| Ucn1 | UMAG_00936 | ||||||||||||||||
| Uka1 | UMAG_11860 | ||||||||||||||||
| Umpde1 | UMAG_02531 | ||||||||||||||||
| Umpde2 | UMAG_10895 | ||||||||||||||||
| Crk1 | UMAG_11410 | ||||||||||||||||
| Fuz7/Ubc5 | UMAG_01514 | ||||||||||||||||
| Kpp2/Ubc3 | UMAG_03305 | ||||||||||||||||
| Kpp4/Ubc4 | UMAG_04258 | ||||||||||||||||
| Kpp6 | UMAG_02331 | ||||||||||||||||
| Rok1 | UMAG_03701 | ||||||||||||||||
| Ubc2 | UMAG_05261 | ||||||||||||||||
| Cla4 | UMAG_10145 | ||||||||||||||||
| Pdc1 | UMAG_01366 | ||||||||||||||||
| Ras1 | UMAG_01643 | ||||||||||||||||
| Ras2 | UMAG_03172 | ||||||||||||||||
| Rho1 | UMAG_05734 | ||||||||||||||||
| Sql2 | UMAG_10803 | ||||||||||||||||
| Biz1 | UMAG_02549 | ||||||||||||||||
| Chs5 | UMAG_10277 | ||||||||||||||||
| Chs7 | UMAG_05480 | ||||||||||||||||
| Cib1 | UMAG_11782 | ||||||||||||||||
| Clb2 | UMAG_10279 | ||||||||||||||||
| Clp1 | UMAG_02438 | ||||||||||||||||
| Gcn5 | UMAG_10190 | ||||||||||||||||
| Hap2 | UMAG_01597 | ||||||||||||||||
| Hgl1 | UMAG_11450 | ||||||||||||||||
| Hos2 | UMAG_11828 | ||||||||||||||||
| Khd4 | UMAG_03837 | ||||||||||||||||
| Kin1 | UMAG_04218 | ||||||||||||||||
| Kin3 | UMAG_06251 | ||||||||||||||||
| Mcs1 | UMAG_03204 | ||||||||||||||||
| Med1 | UMAG_03588 | ||||||||||||||||
| Myo5 | UMAG_04555 | ||||||||||||||||
| Nit2 | UMAG_10417 | ||||||||||||||||
| Pac2 | UMAG_15096 | ||||||||||||||||
| Prf1 | UMAG_02713 | ||||||||||||||||
| Rac1 | UMAG_00774 | ||||||||||||||||
| Rak1 | UMAG_10146 | ||||||||||||||||
| Rbf1 | UMAG_03167 | ||||||||||||||||
| Rop1 | UMAG_12033 | ||||||||||||||||
| Ros1 | UMAG_05853 | ||||||||||||||||
| Rrm4 | UMAG_03494 | ||||||||||||||||
| Sep3 | UMAG_03449 | ||||||||||||||||
| Tea1 | UMAG_15019 | ||||||||||||||||
| Tea4 | UMAG_01012 | ||||||||||||||||
| Tup1 | UMAG_03280 | ||||||||||||||||
| Yup1 | UMAG_05406 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kijpornyongpan, T.; Aime, M.C. Investigating the Smuts: Common Cues, Signaling Pathways, and the Role of MAT in Dimorphic Switching and Pathogenesis. J. Fungi 2020, 6, 368. https://doi.org/10.3390/jof6040368
Kijpornyongpan T, Aime MC. Investigating the Smuts: Common Cues, Signaling Pathways, and the Role of MAT in Dimorphic Switching and Pathogenesis. Journal of Fungi. 2020; 6(4):368. https://doi.org/10.3390/jof6040368
Chicago/Turabian StyleKijpornyongpan, Teeratas, and M. Catherine Aime. 2020. "Investigating the Smuts: Common Cues, Signaling Pathways, and the Role of MAT in Dimorphic Switching and Pathogenesis" Journal of Fungi 6, no. 4: 368. https://doi.org/10.3390/jof6040368
APA StyleKijpornyongpan, T., & Aime, M. C. (2020). Investigating the Smuts: Common Cues, Signaling Pathways, and the Role of MAT in Dimorphic Switching and Pathogenesis. Journal of Fungi, 6(4), 368. https://doi.org/10.3390/jof6040368






