Leaping into the Unknown World of Sporisorium scitamineum Candidate Effectors
Abstract
1. Introduction
2. Materials and Methods
2.1. Ethics Statement and Material Collection
2.2. Biological Material and Experimental Conditions
2.3. Selection of Candidate Effectors and Sequence Analysis
2.4. Candidate Effector Gene Expression
2.5. Vectors and Cloning Procedures for Subcellular Location Assays
2.6. Agrobacterium-Mediated Transient Expression and Confocal Microscopy
2.7. Co-Immunoprecipitation and Mass Spectrometry
2.8. Determining Sugarcane Orthologs of Nicotiana Interactors
2.9. Virulence Assay
2.10. Cloning, Bacterial Strains and Selection Conditions for EtHAn Conjugation and Infiltration
2.11. AvrB-Induced ETI Suppression Assay
3. Results
3.1. Sequence Features of the Selected Candidate Effectors
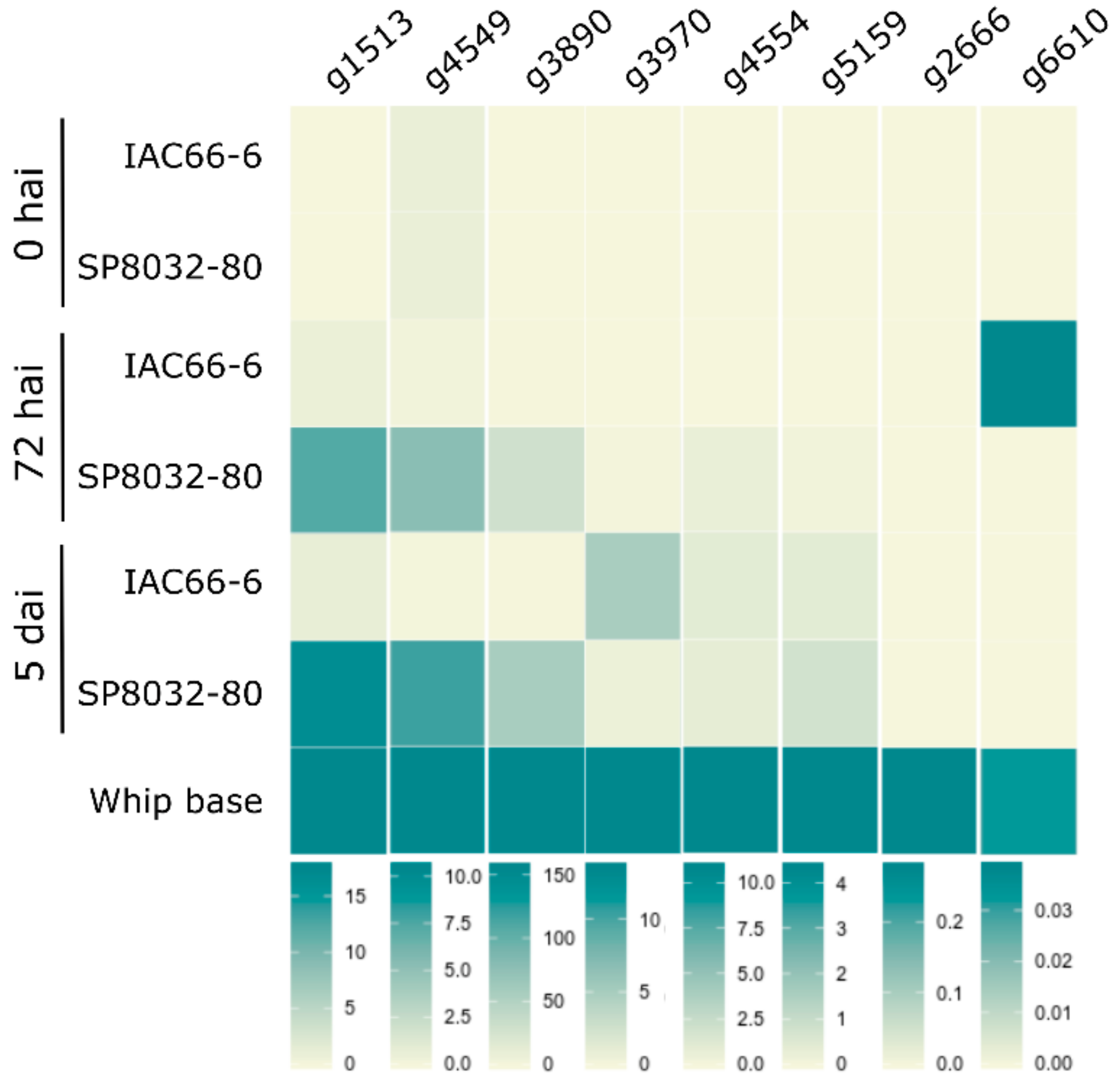
3.2. CE Gene Expression during the Interaction
3.3. Sporisorium scitamineum Candidate Effectors Accumulate in a Variety of Plant Subcellular Compartments
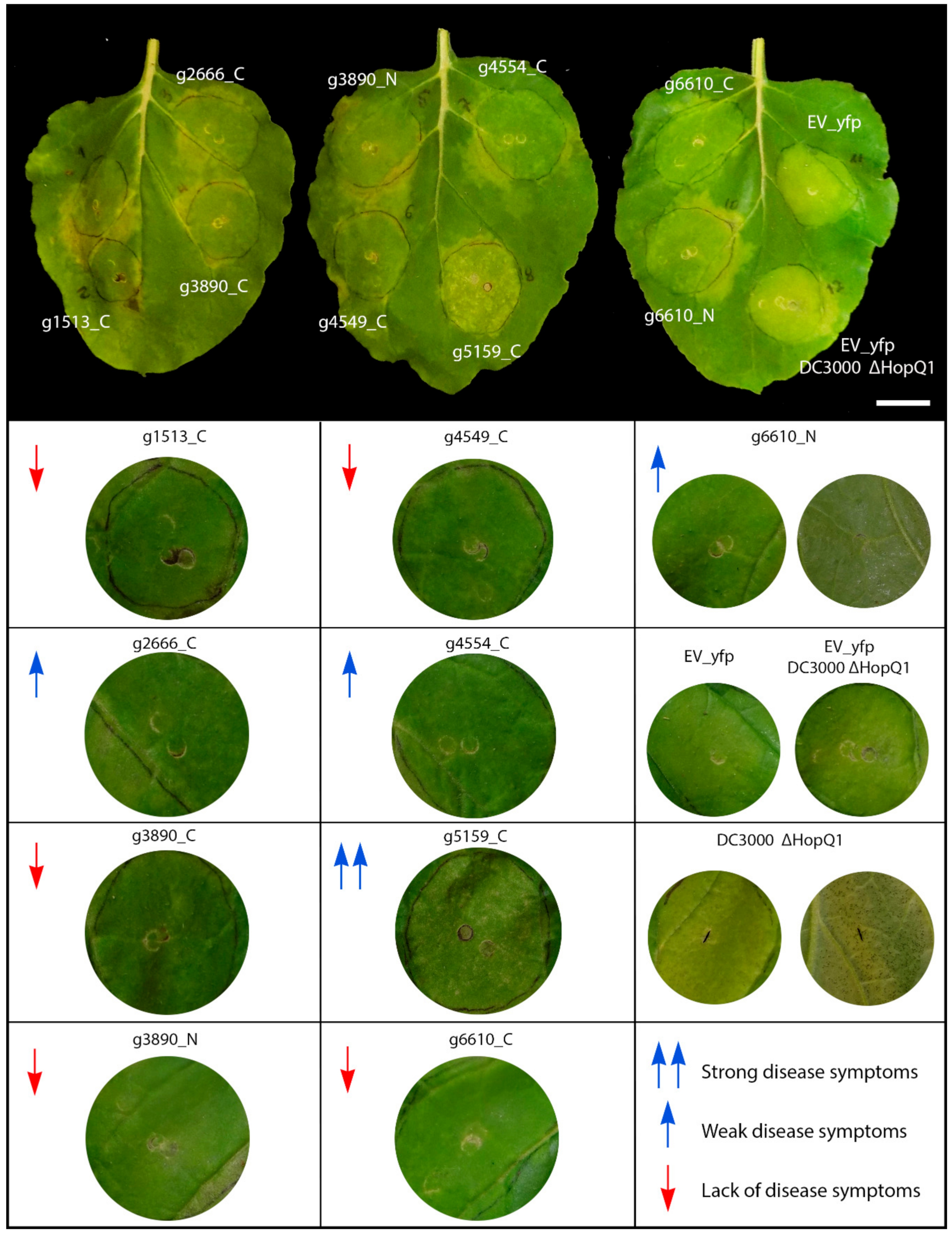
3.4. Sporisorium scitamineum Candidate Effectors Modulate Plant Immune Responses and Disease in Nicotiana benthamiana
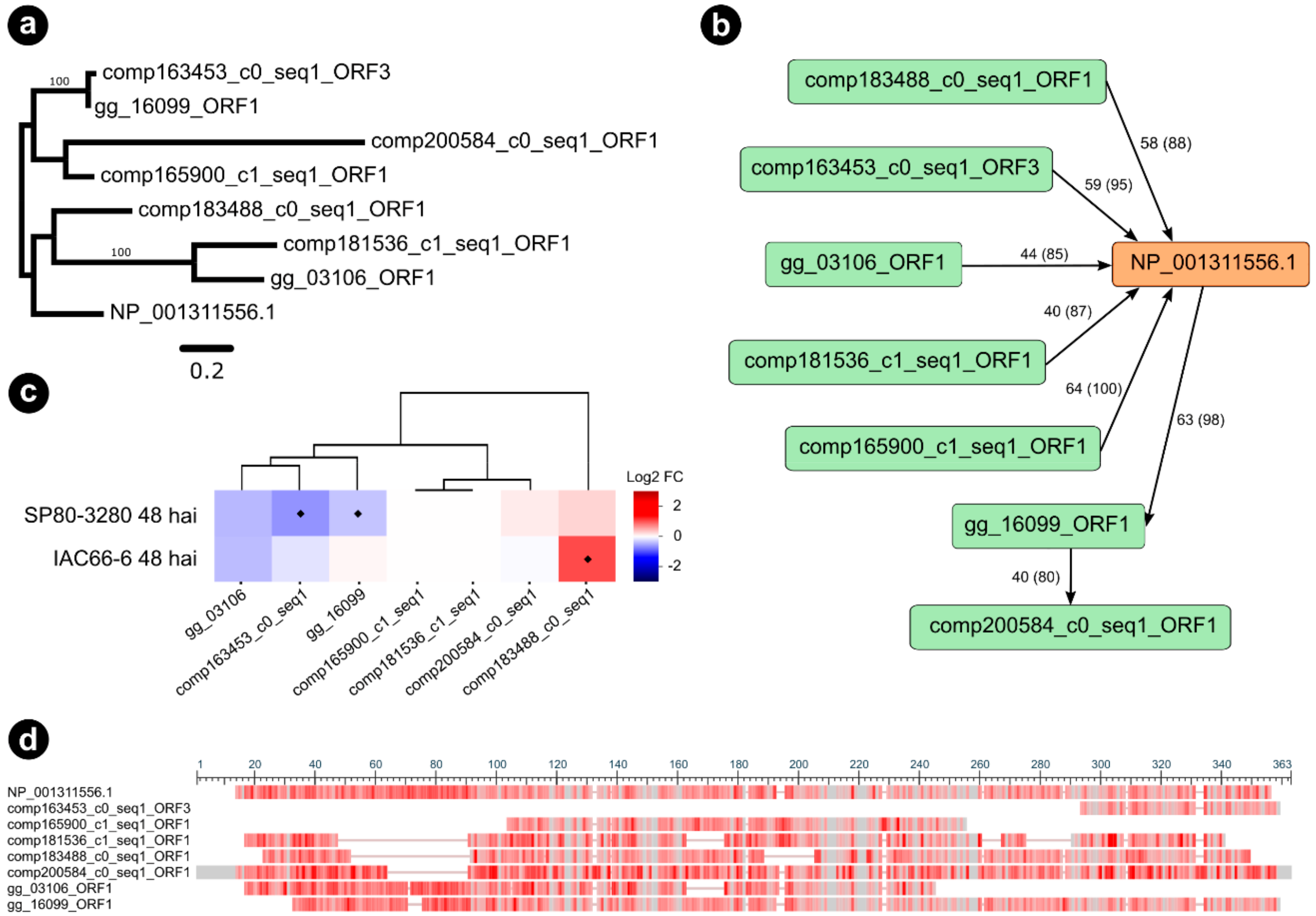
3.5. Interactors of S. scitamineum Effector Candidates
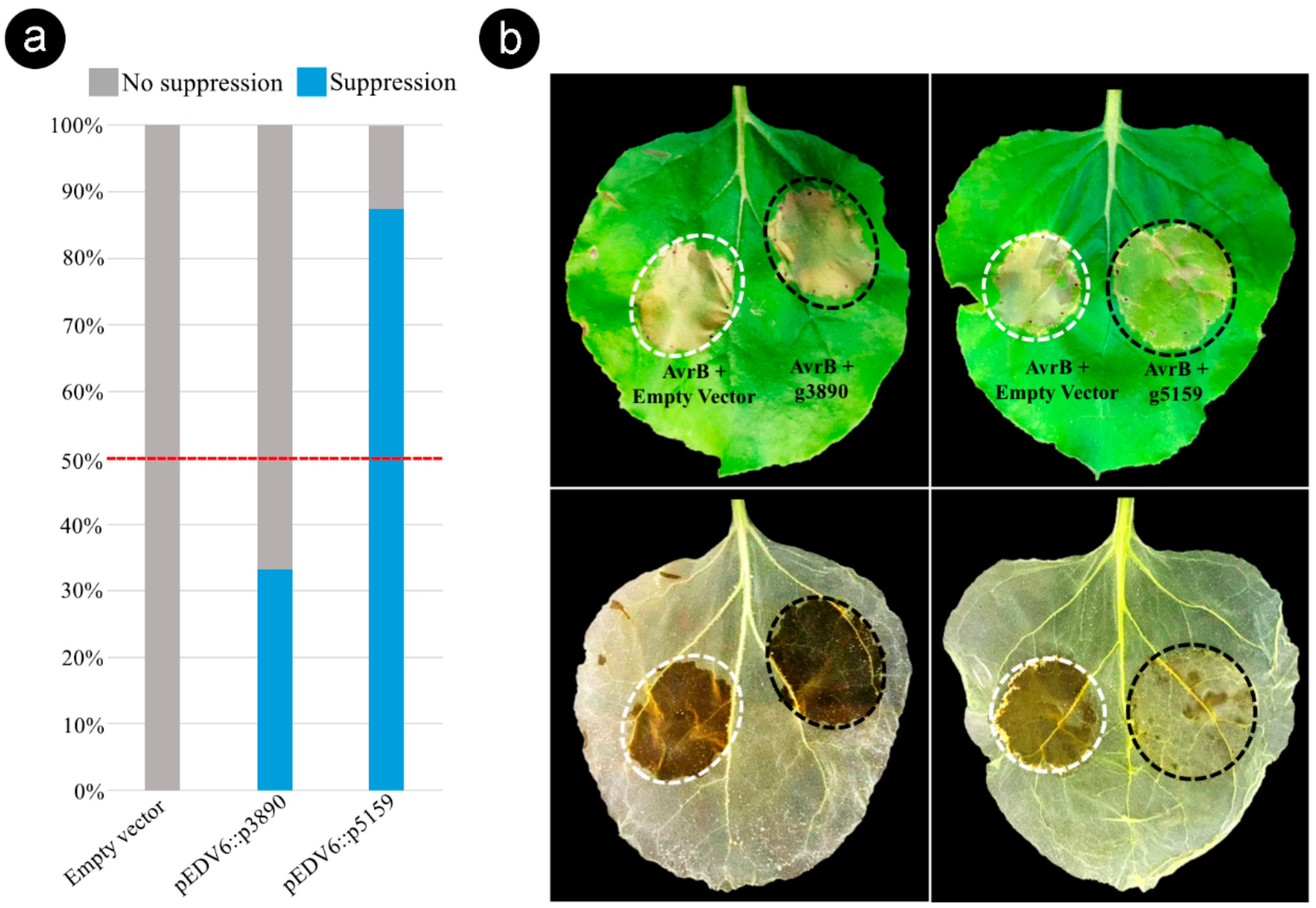
3.6. Bacterial Delivery of the Effector Candidate g5159 from S. scitamineum Suppresses AvrB-Induced ETI in Nicotiana benthamiana
4. Discussion
4.1. General Aspects of the Candidate Effectors
4.2. The Expression of S. scitamineum Effector Genes Is Dependent on the Host Genotype
4.3. S. scitamineum Effector Proteins Target Multiple Cell Compartments
4.4. g5159 Interactors and Suppression of ETI
4.5. g3890 Targets Multicellular Compartments Potentially Having Multiple Partners
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Comstock, J. Smut. In A Guide to Sugarcane Diseases; Rott, P., Bailey, R., Comstock, J., Eds.; Cirad: Montpellier, France, 2000; pp. 181–185. [Google Scholar]
- Sundar, A.; Barnabas, E.; Malathi, P.; Viswanathan, R. A mini-review on smut disease of sugarcane caused by Sporisorium scitamineum. In Botany; Mworia, J., Ed.; InTech: London, UK, 2014; p. 226. [Google Scholar]
- Monteiro-Vitorello, C.B.; Schaker, P.D.C.; Benevenuto, J.; Teixeira-Silva, N.S.; Almeida, S.S. Progress in understanding fungal diseases affecting sugarcane: Smut. In Achieving Sustainable Cultivation of Sugarcane; Rott, P., Ed.; Burleigh Dodds Science Publishing: Cambridge, UK, 2018. [Google Scholar]
- Taniguti, L.M.; Schaker, P.D.C.; Benevenuto, J.; Peters, L.P.; Carvalho, G.; Palhares, A.; Quecine, M.C.; Nunes, F.R.S.; Kmit, M.C.P.; Alvan, W.; et al. Complete genome sequence of Sporisorium scitamineum and biotrophic interaction transcriptome with sugarcane. PLoS ONE 2015, 10, e0129318. [Google Scholar] [CrossRef]
- Schaker, P.D.C.; Palhares, A.C.; Taniguti, L.M.; Peters, L.P.; Creste, S.; Aitken, K.S.; VanSluys, M.-A.; Kitajima, J.P.; Vieira, M.L.C.V.; Monteiro-Vitorello, C.B. RNAseq transcriptional profiling following whip development in sugarcane smut disease. PLoS ONE 2016, 11, e0162237. [Google Scholar] [CrossRef]
- Marques, J.P.R.; Appezzato-da-Glória, B.; Piepenbring, M.; Massola, N.S.; Monteiro-Vitorello, C.B.; Vieira, M.L.C. Sugarcane smut: Shedding light on the development of the whip-shaped sorus. Ann. Bot. 2017, 119, 815–827. [Google Scholar] [CrossRef]
- Alexander, K.C.; Srinivasan, K.V. Sexuality in Ustilago scitaminea Syd. Curr. Sci. 1966, 35, 603–604. [Google Scholar]
- Martinez, M.; Medina, I.; Naranjo, S.; Rodriguez, C.W.; Armas, R.; Piñon, D.; Vicente, C.; Legaz, M.E. Changes of some chemical parameters, involved in sucrose recovery from sugarcane juices, related to the susceptibility or resistance of sugarcane plants to smut (Ustilago scitaminea). Int. Sugar J. 2000, 102, 445–448. [Google Scholar]
- Wada, A.C.; Anaso, A.B.; Bassey, M.S. Sugar cane whip smut (Sporisorium scitamineum Syd) caused field sucrose and juice quality losses of two sugar cane varieties in Nigeria. Int. J. Plant Soil Sci. 2016, 10, 1–11. [Google Scholar] [CrossRef]
- Hogenhout, S.; Van der Hoorn, R.L.; Terauchi, R.; Kamoun, S. Emerging concepts in effector biology of plant-associated organisms. Mol. Plant-Microbe Interact 2009, 22, 115–122. [Google Scholar] [CrossRef]
- De Jonge, R.; Bolton, M.D.; Thomma, B.P.H.J. How filamentous pathogens co-opt plants: The ins and outs of fungal effectors. Curr. Opin. Plant Biol. 2011, 14, 400–406. [Google Scholar] [CrossRef]
- Ökmen, B.; Doehlemann, G. Inside plant: Biotrophic strategies to modulate host immunity and metabolism. Curr. Opin. Plant Biol. 2014, 20, 19–25. [Google Scholar] [CrossRef]
- Takahara, H.; Hacquard, S.; Kombrink, A.; Hughes, H.D.; Robin, G.P.; Hiruma, K.; Neumann, U.; Shinya, T.; Kombrik, E.; Shibuya, N.; et al. Colletotrichum higginsianum extracellular LysM proteins play dual roles in appressorial function and suppression of chitin-triggered plant immunity. New Phytol. 2016, 211, 1323–1337. [Google Scholar] [CrossRef]
- Hemetsberger, C.; Mueller, A.N.; Matei, A.; Herrberger, C.; Hensel, G.; Kumlehn, J.; Mishra, B.; Sharma, R.; Thines, M.; Hückelhoven, R.; et al. The fungal core effector Pep1 is conserved across smuts of dicots and monocots. New Phytol. 2015, 206, 1116–1126. [Google Scholar] [CrossRef]
- Houterman, P.M.; Cornelissen, B.J.C.; Rep, M. Suppression of plant resistance gene-based immunity by a fungal effector. PLoS Pathog. 2008, 4, 1–6. [Google Scholar] [CrossRef]
- Wahl, R.; Wippel, K.; Goos, S.; Kämper, J.; Sauer, N. A novel high-affinity sucrose transporter is required for virulence of the plant pathogen Ustilago maydis. PLoS Biol. 2010, 8, e1000303. [Google Scholar] [CrossRef]
- Caillaud, M.-C.; Asai, S.; Rallapalli, G.; Piquerez, S.; Fabro, G.; Jones, J.D.G. A downy mildew effector attenuates salicylic acid–triggered immunity in Arabidopsis by interacting with the host mediator complex. PLoS Biol. 2013, 11, e1001732. [Google Scholar] [CrossRef]
- Tanaka, S.; Brefort, T.; Neidig, N.; Djamei, A.; Kahnt, J.; Vermerris, W.; Koenig, S.; Feussner, K.; Feussner, I.; Kahmann, R. A secreted Ustilago maydis effector promotes virulence by targeting anthocyanin biosynthesis in maize. eLife 2014, 3, e01355. [Google Scholar] [CrossRef]
- Ahmed, M.B.; dos Santos, K.C.G.; Sanchez, I.B.; Petre, B.; Lorrains, C.; Plourde, M.B.; Duplessis, S.; Desgagné-Penis, I.; Germain, H. A rust fungal effector binds plant DNA and modulates transcription. Sci. Rep. 2018, 8, 14718. [Google Scholar] [CrossRef]
- Bhadauria, V.; MacLachlan, R.; Pozniak, C.; Banniza, S. Candidate effectors contribute to race differentiation and virulence of the lentil anthracnose pathogen Colletotrichum lentis. BMC Genom. 2015, 16, 628. [Google Scholar] [CrossRef]
- Lorrain, C.; Petre, B.; Duplessis, S. Show me the way: Rust effector targets in heterologous plant systems. Curr. Opin. Microbiol. 2018, 46, 19–25. [Google Scholar] [CrossRef]
- Petit-Houdenot, Y.; Fudal, I. Complex interactions between fungal avirulence genes and their corresponding plant resistance genes and consequences for disease resistance management. Front. Plant Sci. 2017, 8, 1072. [Google Scholar] [CrossRef]
- Thirugnanasambandam, P.P.; Hoang, N.V.; Henry, R.J. The challenge of analyzing the sugarcane genome. Front. Plant Sci. 2018, 9, 616. [Google Scholar] [CrossRef]
- Carvalho, G.; Quecine, M.C.; Longatto, D.P.; Peters, L.P.; Almeida, J.R.; Shyton, T.G.; Silva, M.M.L.; Crestana, G.S.; Creste, S.; Monteiro-Vitorello, C.B. Sporisorium scitamineum colonisation of sugarcane genotypes susceptible and resistant to smut revealed by GFP-tagged. Ann. Appl. Biol. 2016, 169, 329–341. [Google Scholar] [CrossRef]
- Peters, L.; Carvalho, G.; Vilhena, M.B.; Creste, S.; Azevedo, R.A.; Monteiro-Vitorello, C.B. Functional analysis of oxidative burst in sugarcane smut-resistant and -susceptible genotypes. Planta 2017, 245, 749–764. [Google Scholar] [CrossRef]
- Peters, L.P.; Teixeira-Silva, N.S.; Bini, A.P.; Silva, M.M.L.; Crestana, G.S.; Creste, S.; Azevedo, R.A.; Carvalho, G.; Monteiro-Vitorello, C.B. Differential responses of genes and enzymes associated with ROS protective responses in the sugarcane smut fungus. Fungal Biol. 2020. [Google Scholar] [CrossRef]
- Benevenuto, J.; Teixeira-Silva, N.S.; Kuramae, E.E.; Croll, D.; Monteiro-Vitorello, C.B. Comparative genomics of smut pathogens: Insights from orphans and positively selected genes into host specialization. Front. Microbiol. 2018, 9, 660. [Google Scholar] [CrossRef]
- Jones, P.; Binns, D.; Chang, H.-Y.; Fraser, M.; Li, W.; McAnulla, C.; McWilliam, H.; Maslen, J.; Mitchell, A.; Nuka, G. InterProScan 5: Genome-scale protein function classification. Bioinformatics 2014, 30, 1236–1240. [Google Scholar] [CrossRef]
- Petersen, T.N.; Brunak, S.; von Heijne, G.; Nielsen, H. SignalP 4.0: Discriminating signal peptides from transmembrane regions. Nat. Methods 2011, 8, 785–786. [Google Scholar] [CrossRef]
- Gasteiger, E.; Hoogland, C.; Gattiker, A.; Duvaud, S.; Wilkins, M.; Appel, R.O.; Bairoch, A. Protein identification and analysis tools on the ExPASy server. In The Proteomics Protocols Handbook; Humana Press: Totowa, NJ, USA, 2005; pp. 571–607. [Google Scholar]
- Kosugi, S.; Hasebe, M.; Matsumura, N.; Takashima, H.; Miyamoto-Sato, E.; Tomita, M.; Yanagawa, H. Six classes of nuclear localization signals specific to different binding grooves of importin alpha. J. Biol. Chem. 2009, 284, 478–485. [Google Scholar] [CrossRef]
- Heger, A.; Holm, L. Rapid automatic detection and alignment of repeats in protein sequences. Proteins Struct. Funct. Genet 2000, 41, 224–237. [Google Scholar] [CrossRef]
- Sperschneider, J.; Dodds, P.N.; Singh, K.B.; Taylor, J.M. ApoplastP: Prediction of effectors and plant proteins in the apoplast using machine learning. New Phytol. 2018, 217, 1764–1778. [Google Scholar] [CrossRef]
- Ramakers, C.; Ruijter, J.M.; Lekanne Deprez, R.H.; Moorman, A.F.M. Assumption-free analysis of quantitative real-time polymerase chain reaction (PCR) data. Neurosci. Lett. 2003, 339, 62–66. [Google Scholar] [CrossRef]
- Petre, B.; Saunders, D.G.; Sklenar, J.; Lorrain, C.; Win, J.; Duplessis, S.; Kamoun, S. Candidate effector proteins of the rust pathogen Melampsora larici-populina target diverse plant cell compartments. Mol. Plant-Microbe Interact. 2015, 28, 689–700. [Google Scholar] [CrossRef] [PubMed]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Cardoso-Silva, C.B.; Costa, E.A.; Mancini, M.C.; Balsalobre, T.W.A.; Canesin, L.E.C.; Pinto, L.R.; Carneiro, M.S.; Garcia, A.A.F.; Souza, A.P.; Vicentini, R. De Novo assembly and transcriptome analysis of contrasting sugarcane varieties. PLoS ONE 2014, 9, e88462. [Google Scholar] [CrossRef] [PubMed]
- Hagberg, A.; Swart, P.; Chult, D.S. Exploring Network Structure, Dynamics, and Function Using NetworkX; Los Alamos National Lab. (LANL): Los Alamos, NM, USA, 2008; pp. 11–15. [Google Scholar]
- Nguyen, L.T.; Schmidt, H.A.; Von Haeseler, A.; Minh, B.Q. IQ-TREE: A fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 2015, 32, 268–274. [Google Scholar] [CrossRef] [PubMed]
- Edgar, R.C. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004, 32, 1792–1797. [Google Scholar] [CrossRef] [PubMed]
- Kalyaanamoorthy, S.; Minh, B.Q.; Wong, T.K.F.; von Haeseler, A.; Jermiin, L.S. ModelFinder: Fast model selection for accurate phylogenetic estimates. Nat. Methods 2017, 14, 587–589. [Google Scholar] [CrossRef]
- Hoang, D.T.; Chernomor, O.; Von Haeseler, A.; Minh, B.Q.; Vin, L.S. UFBoot2: Improving the ultrafast bootstrap approximation. Mol. Biol. Evol. 2018, 35, 518–522. [Google Scholar] [CrossRef]
- Rody, H.V.S.; Bombardelli, R.G.H.; Creste, S.; Camargo, L.E.A.; VanSluys, M.-A.; Monteiro-Vitorello, C.B. Genome survey of resistance gene analogs in sugarcane: Genomic features and differential expression of the innate immune system from a smut-resistant genotype. BMC Genom. 2019, 20, 809. [Google Scholar] [CrossRef]
- Maia, T.; Badel, J.L.; Marin-Ramirez, G.; Rocha, C.M.; Fernandes, M.B.; Silva, J.C.F.; Azevedo-Junior, G.M.; Brommonschenkel, S.H. The Hemileia vastatrix effector HvEC-016 suppresses bacterial blight symptoms in coffee genotypes with the SH1 rust resistance gene. New Phytol. 2017, 213, 1315–1329. [Google Scholar] [CrossRef]
- Innes, R.W.; Bisgrove, S.R.; Smith, N.M.; Bent, A.F.; Staskawicz, B.J.; Liu, Y.C. Identification of a disease resistance locus in Arabidopsis that is functionally homologous to the RPG1 locus of soybean. Plant J. 1993, 4, 813–820. [Google Scholar] [CrossRef]
- Ma, L.-S.; Wang, L.; Trippel, C.; Mendoza-Mendoza, A.; Ullmann, S.; Moretti, M.; Carsten, A.; Kahnt, J.; Reissmann, S.; Zechmann, B.; et al. The Ustilago maydis repetitive effector Rsp3 blocks the antifungal activity of mannose-binding maize proteins. Nat. Commun. 2018, 9, 1711. [Google Scholar] [CrossRef] [PubMed]
- Benevenuto, J.; Longatto, D.P.; Reis, G.V.; Mielnichuk, N.; Palhares, A.C.; Carvalho, G.; Saito, S.; Quecine, M.C.; Sanguino, A.; Vieira, M.L.C.; et al. Molecular variability and genetic relationship among Brazilian strains of the sugarcane smut fungus. FEMS Microbiol. Lett. 2016, 363, fnw277. [Google Scholar] [CrossRef] [PubMed]
- Wei, C.-F.; Kvitko, B.H.; Shimizu, R.; Crabill, E.; Alfano, J.R.; Lin, N.-C.; Martin, G.B.; Huang, H.-C.; Collmer, A. A Pseudomonas syringae pv. tomato DC3000 mutant lacking the type III effector HopQ1-1 is able to cause disease in the model plant Nicotiana benthamiana. Plant J. 2007, 51, 32–46. [Google Scholar] [CrossRef] [PubMed]
- Su, Y.; Xu, L.; Wang, S.; Wang, Z.; Yang, Y.; Chen, Y.; Que, Y. Identification, phylogeny, and transcript of chitinase family genes in sugarcane. Sci. Rep. 2015, 5, 10708. [Google Scholar] [CrossRef] [PubMed]
- Passarinho, P.A.; de Vries, S.C. Arabidopsis chitinases: A genomic survey. Arab. B 2002, 1, e0023. [Google Scholar] [CrossRef]
- Bateman, A.; Coin, L.; Durbin, R.; Finn, R.D.; Hollich, V.; Griffiths-Jones, S.; Khanna, A.; Marshall, M.; Moxon, S.; Sonnhammer, E.L.; et al. The Pfam protein families database. Nucleic Acids Res. 2004, 32, D141. [Google Scholar] [CrossRef]
- Almagro Armenteros, J.J.; Tsirigos, K.D.; Sønderby, C.K.; Petersen, T.N.; Winther, O.; Brunak, S.; von Heijne, G.; Nielsen, H. SignalP 5.0 improves signal peptide predictions using deep neural networks. Nat. Biotechnol. 2019, 37, 420–423. [Google Scholar] [CrossRef]
- Farkas, I.; Dombrádi, V.; Miskei, M.; Szabados, L.; Koncz, C. Arabidopsis PPP family of serine/threonine phosphatases. Trends Plant Sci. 2007, 12, 169–176. [Google Scholar] [CrossRef]
- Sharpee, W.C.; Dean, R.A. Form and function of fungal and oomycete effectors. Fungal Biol. Rev. 2016, 30, 62–73. [Google Scholar] [CrossRef]
- Vicente, C.; Alarcón, B.; Santiago, R.; Armas, R.D.; Sánchez-Elordi, E.; Legaz, M.E. An elicitor isolated from Sporisoriun scitamineum multiplies xylem bridges between vascular neighboring bundles in sugarcane leaves. Res. Rev. Biosci. 2017, 12, 114. [Google Scholar]
- Santiago, R.; de Armas, R.; Blanch, M.; Vicente, C.; Legaz, M.-E. In vitro effects of caffeic acid upon growth of the fungi Sporisorium scitamineum. J. Plant Interact 2010, 5, 233–240. [Google Scholar] [CrossRef]
- Barnabas, L.; Ashwin, N.M.R.; Kaverinathan, K.; Trentin, A.R.; Pivato, M.; Sundar, A.R.; Malathi, P.; Viswanathan, R.; Carletti, P.; Arrigoni, G.; et al. In vitro secretomic analysis identifies putative pathogenicity-related proteins of Sporisorium scitamineum—The sugarcane smut fungus. Fungal Biol. 2017, 121, 199–211. [Google Scholar] [CrossRef]
- Barnabas, L.; Ashwin, N.M.R.; Ramesh Sundar, A.; Malathi, P.; Viswanathan, R. Putative orthologs of Ustilago maydis effectors screened from the genome of sugarcane smut fungus—Sporisorium scitamineum. Australas Plant Pathol. 2017, 46, 147–156. [Google Scholar] [CrossRef]
- Que, Y.; Xu, L.; Wu, Q.; Liu, Y.; Ling, H.; Liu, Y.; Zhang, Y.; Guo, J.; Su, Y.; Chen, J.; et al. Genome sequencing of Sporisorium scitamineum provides insights into the pathogenic mechanisms of sugarcane smut. BMC Genom. 2014, 15, 996. [Google Scholar] [CrossRef] [PubMed]
- Xia, W.; Yu, X.; Ye, Z. Smut fungal strategies for the successful infection. Microb. Pathog. 2020, 142, 104039. [Google Scholar] [CrossRef] [PubMed]
- Toruño, T.Y.; Stergiopoulos, I.; Coaker, G. Plant-pathogen effectors: Cellular probes interfering with plant defenses in spatial and temporal manners. Annu. Rev. Phytopathol. 2016, 54, 419–441. [Google Scholar] [CrossRef]
- Brefort, T.; Doehlemann, G.; Mendoza-Mendoza, A.; Reissmann, S.; Djamei, A.; Kahmann, R. Ustilago maydis as a Pathogen. Annu. Rev. Phytopathol. 2009, 47, 423–445. [Google Scholar] [CrossRef]
- Doehlemann, G.; Wahl, R.; Horst, R.J.; Voll, L.M.; Usadel, B.; Poree, F.; Stitt, M.; Pons-Kühnemann, J.; Sonnewald, U.; Kahmann, R.; et al. Reprogramming a maize plant: Transcriptional and metabolic changes induced by the fungal biotroph Ustilago maydis. Plant J. 2008, 56, 181–195. [Google Scholar] [CrossRef]
- Kämper, J.; Kahmann, R.; Bölker, M.; Ma, L.J.; Brefort, T.; Saville, B.J.; Banuett, F.; Kronstad, J.W.; Gold, S.E.; Müller, O.; et al. Insights from the genome of the biotrophic fungal plant pathogen Ustilago maydis. Nature 2006, 444, 97–101. [Google Scholar] [CrossRef]
- Doehlemann, G.; Van Der Linde, K.; Aßmann, D.; Schwammbach, D.; Hof, A.; Mohanty, A.; Jackson, D.; Kahmann, R. Pep1, a secreted effector protein of Ustilago maydis, is required for successful invasion of plant cells. PLoS Pathog. 2009, 5, e1000290. [Google Scholar] [CrossRef]
- Barrett, L.G.; Thrall, P.H.; Dodds, P.N.; van der Merwe, M.; Linde, C.C.; Lawrence, G.J.; Burdon, J.J. Diversity and evolution of effector loci in natural populations of the plant pathogen Melampsora lini. Mol. Biol. Evol. 2009, 26, 2499–2513. [Google Scholar] [CrossRef] [PubMed]
- Schweizer, G.; Münch, K.; Mannhaupt, G.; Schirawski, J.; Kahmann, R.; Dutheil, J.Y. Positively selected effector genes and their contribution to virulence in the smut fungus Sporisorium reilianum. Genome Biol. Evol. 2018, 10, 629–645. [Google Scholar] [CrossRef] [PubMed]
- Lin, M.; Ficke, A.; Cockram, J.; Lillemo, M. Genetic structure of the norwegian Parastagonospora nodorum population. Front Microbiol. 2020, 11, 1280. [Google Scholar] [CrossRef] [PubMed]
- Dutheil, J.Y.; Mannhaupt, G.; Schweizer, G.; Sieber, C.M.K.; Münsterkötter, M.; Güldener, U.; Schirawski, J.; Kahmann, R. A tale of genome compartmentalization: The evolution of virulence clusters in smut fungi. Genome Biol. Evol. 2016, 8, 681–704. [Google Scholar] [CrossRef] [PubMed]
- Bhuiyan, S.A.; Croft, B.J.; Stringer, J.K.; Deomano, E.C. Pathogenic variation in spore populations of Sporisorium scitamineum, causal agent of sugarcane smut in Australia. Plant Dis. 2015, 99, 93–99. [Google Scholar] [CrossRef] [PubMed]
- Djamei, A.; Kahmann, R. Ustilago maydis: Dissecting the molecular interface between pathogen and plant. PLoS Pathog. 2012, 8, 11–14. [Google Scholar] [CrossRef]
- Valent, B.; Khang, C.H. Recent advances in rice blast effector research. Curr. Opin. Plant Biol. 2010, 13, 434–441. [Google Scholar] [CrossRef]
- Guzmán-Guzmán, P.; Alemán-Duarte, M.I.; Delaye, L.; Herrera-Estrella, A.; Olmedo-Monfil, V. Identification of effector-like proteins in Trichoderma spp. and role of a hydrophobin in the plant-fungus interaction and mycoparasitism. BMC Genet 2017, 18, 16. [Google Scholar]
- Kleemann, J.; Rincon-Rivera, L.J.; Takahara, H.; Neumann, U.; Ver Loren van Themaat, E.; van der Does, H.C.; Hacquard, S.; Stüber, K.; Will, I.; Schmalenbach, W.; et al. Sequential delivery of host-induced virulence effectors by appressoria and intracellular hyphae of the phytopathogen Colletotrichum higginsianum. PLoS Pathog. 2012, 8, e1002643. [Google Scholar] [CrossRef]
- Lanver, D.; Berndt, P.; Tollot, M.; Naik, V.; Vranes, M.; Warmann, T.; Münch, K.; Rössel, N.; Kahmann, R. Plant surface cues prime Ustilago maydis for biotrophic development. PLoS Pathog. 2014, 10, e1004272. [Google Scholar] [CrossRef]
- O’Connell, R.J.; Thon, M.R.; Hacquard, S.; Amyotte, S.G.; Kleemann, J.; Torres, M.F.; Damm, U.; Buiate, E.A.; Epstein, L.; Alkan, N.; et al. Lifestyle transitions in plant pathogenic Colletotrichum fungi deciphered by genome and transcriptome analyses. Nat. Genet 2012, 44, 1060–1065. [Google Scholar] [CrossRef] [PubMed]
- Tan, K.-C.; Oliver, R.P. Regulation of proteinaceous effector expression in phytopathogenic fungi. PLoS Pathog. 2017, 13, e1006241. [Google Scholar] [CrossRef] [PubMed]
- Tollot, M.; Assmann, D.; Becker, C.; Altmüller, J.; Dutheil, J.Y.; Wegner, C.E.; Kahmann, R. The WOPR protein Ros1 is a master regulator of sporogenesis and late effector gene expression in the maize pathogen Ustilago maydis. PLoS Pathog. 2016, 12, e1005697. [Google Scholar] [CrossRef] [PubMed]
- Ashwin, N.M.R.; Barnabas, L.; Sundar, A.R.; Malathi, P.; Viswanathan, R.; Masi, A.; Agrawal, G.K.; Rakwal, R. CfPDIP1, a novel secreted protein of Colletotrichum falcatum, elicits defense responses in sugarcane and triggers hypersensitive response in tobacco. Appl. Microbiol. Biotechnol. 2018, 102, 6001–6021. [Google Scholar] [CrossRef] [PubMed]
- Gan, P.; Rafiqui, M.; Hardham, A.; Dodds, P.N. Effectors of biotrophic fungal plant pathogens. Funct. Plant Biol. 2010, 37, 913–918. [Google Scholar] [CrossRef]
- Alfano, J. Roadmap for future research on plant pathogen effectors. Trends Microbiol. 2009, 10, 805–813. [Google Scholar] [CrossRef] [PubMed]
- Win, J.; Krasileva, K.V.; Kamoun, S.; Shirasu, K.; Staskawicz, B.J.; Banfield, M.J. Pearls sequence divergent RXLR effectors share a structural fold conserved across plant pathogenic oomycete species. PLoS Pathog. 2012, 8, e1002400. [Google Scholar] [CrossRef] [PubMed]
- Dodds, P.N. The Melampsora lini AvrL567 avirulence genes are expressed in haustoria and their products are recognized inside plant cells. Plant Cell 2004, 16, 755–768. [Google Scholar] [CrossRef]
- Kuppireddy, V.; Uversky, V.; Toh, S.; Tsai, M.C.; Beckerson, W.C.; Cahill, C.; Carman, B.; Perlin, M.H. Identification and initial characterization of the effectors of an anther smut fungus and potential host target proteins. Int. J. Mol. Sci. 2017, 18, 2489. [Google Scholar] [CrossRef]
- Qi, M.; Grayczyk, J.P.; Seitz, J.M.; Lee, Y.; Link, T.I.; Choi, D.; Pedley, K.F.; Voegele, R.T.; Baum, T.J.; Whitham, S.A. Suppression or activation of immune responses by predicted secreted proteins of the soybean rust pathogen Phakopsora pachyrhizi. Mol. Plant Microbe Interact 2018, 31, 163–174. [Google Scholar] [CrossRef]
- Verma, A.; Lee, C.; Morriss, S.; Odu, F.; Kenning, C.; Rizzo, N.; Spollen, W.G.; Lin, M.; McRae, A.G.; Givan, S.A.; et al. The novel cyst nematode effector protein 30D08 targets host nuclear functions to alter gene expression in feeding sites. New Phytol. 2018, 219, 697–713. [Google Scholar] [CrossRef] [PubMed]
- Chaudhari, P.; Ahmed, B.; Joly, D.L.; Germain, H. Effector biology during biotrophic invasion of plant cells. Virulence 2014, 5, 703–709. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Peng, H.; Cui, J.; Huang, W.; Kong, L.; Clarke, J.L.; Jian, H.; Wang, G.L.; Peng, D. Molecular characterization of a novel effector expansin-like protein from Heterodera avenae that induces cell death in Nicotiana benthamiana. Sci. Rep. 2016, 6, 35677. [Google Scholar] [CrossRef] [PubMed]
- Petre, B.; Lorrain, C.; Saunders, D.G.O.; Win, J.; Sklenar, J.; Duplessis, S.; Kamoun, S. Rust fungal effectors mimic host transit peptides to translocate into chloroplasts. Cell Microbiol. 2016, 18, 453–465. [Google Scholar] [CrossRef]
- Tonkin, C.J.; Foth, B.J.; Ralph, S.A.; Struck, N.; Cowman, A.F.; McFadden, G.I. Evolution of malaria parasite plastid targeting sequences. Proc. Natl. Acad. Sci. USA 2008, 105, 4781–4785. [Google Scholar] [CrossRef]
- Mukhtar, M.S.; McCormack, M.E.; Argueso, C.T.; Pajerowska-Mukhtar, K.M. Pathogen tactics to manipulate plant cell death. Curr. Biol. 2016, 26, R608–R619. [Google Scholar] [CrossRef]
- Block, A.; Guo, M.; Li, G.; Elowsky, C.; Clemente, T.E.; Alfano, J.R. The Pseudomonas syringae type III effector HopG1 targets mitochondria, alters plant development and suppresses plant innate immunity. Cell Microbiol. 2010, 12, 318–330. [Google Scholar] [CrossRef]
- Neuhaus, J.M.; Fritig, B.; Linthorst, H.J.M.; Meins, F.; Mikkelsen, J.D.; Ryals, J. A revised nomenclature for chitinase genes. Plant Mol. Biol. Rep. 1996, 14, 102–104. [Google Scholar] [CrossRef]
- Kasprzewska, A. Plant chitinases—Regulation and function. Cell Mol. Biol. Lett. 2003, 8, 809–924. [Google Scholar]
- Punja, Z.K.; Zhang, Y.Y. Plant chitinases and their roles in resistance to fungal diseases. J. Nematol. 1993, 25, 540. [Google Scholar]
- Kaffamik, F.A.R.; Jones, A.M.E.; Rathjen, J.P.; Peck, S.C. Effector proteins of the bacterial pathogen Pseudomonas syringae alter the extracellular proteome of the host plant, Arabidopsis thaliana. Mol. Cell Proteom. 2009, 8, 145–156. [Google Scholar] [CrossRef] [PubMed]
- Langin, G.; Gouguet, P.; Üstün, S. Microbial effector proteins—A journey through the proteolytic landscape. Trends Microbiol. 2020, 28, 523–535. [Google Scholar] [CrossRef] [PubMed]
- Máthé, C.; Garda, T.; Freytag, C.; M-Hamvas, M. The role of serine-threonine protein phosphatase pp2a in plant oxidative stress signaling—facts and hypotheses. Int. J. Mol. Sci. 2019, 20, 3028. [Google Scholar] [CrossRef] [PubMed]
- Bheri, M.; Pandey, G.K. PP2A phosphatases take a giant leap in the post-genomics era. Curr. Genom. 2019, 20, 154–171. [Google Scholar] [CrossRef] [PubMed]
- Schweighofer, A.; Kazanaviciute, V.; Scheikl, E.; Teige, M.; Doczi, R.; Hirt, H.; Schwanninger, M.; Kant, M.; Schuurink, R.; Mauch, F.; et al. The PP2C-type phosphatase AP2C1, which negatively regulates MPK4 and MPK6, modulates innate immunity, jasmonic acid, and ethylene levels in Arabidopsis. Plant Cell 2007, 19, 2213–2224. [Google Scholar] [CrossRef]
- Zhu, X.; Wang, Y.; Su, Z.; Lv, L.; Zhang, Z. Silencing of the wheat protein phosphatase 2A catalytic subunit TaPP2AC enhances host resistance to the necrotrophic pathogen Rhizoctonia cerealis. Front. Plant Sci. 2018, 9, 1437. [Google Scholar] [CrossRef]
- He, X.; Anderson, J.C.; del Pozo, O.; Gu, Y.Q.; Tang, X.; Martin, G.B. Silencing of subfamily I of protein phosphatase 2A catalytic subunits results in activation of plant defense responses and localized cell death. Plant J. 2004, 38, 563–577. [Google Scholar] [CrossRef]
- Ahn, H.K.; Yoon, J.T.; Choi, I.; Kim, S.; Lee, H.-S.; Pai, H.-S. Functional characterization of chaperonin containing T-complex polypeptide-1 and its conserved and novel substrates in Arabidopsis. J. Exp. Bot. 2019, 70, 2741–2757. [Google Scholar] [CrossRef]
- Wang, H.; Hao, L.; Shung, C.Y.; Sunter, G.; Bisaroa, D.M. Adenosine kinase is inactivated by Geminivirus AL2 and L2 proteins. Plant Cell 2003, 15, 3020–3032. [Google Scholar] [CrossRef]


| Gene ID | CDS Size (nt) | Protein Size (aa) | Molar Weight (kD) | Effector Reference | Selection Features ** | Genomic Island ** | NLS | Cysteine % | Repeats | SNP | Disorder Domain | In Silico Location |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| g2 | 711 | 236 | 26,290 | - | PE 5 dai; DE 200 dai; UF | x | x | 2.50 | x | x | ✓ | Apoplastic |
| g1052 | 423 | 140 | 15,539 | [27] | Single.; EE; UF | x | x | 2.86 | x | ✓ | x | Non-apoplastic |
| g1084 | 930 | 309 | 34,473 | - | Single., EE, UF | ✓ | x | 0.00 | ✓ 2 | x | ✓ | Non-apoplastic |
| g1513 | 648 | 215 | 23,140 | [27] | PE 5 dai; DE 5 dai; DE 200 dai; UF | x | x | 1.40 | x | x | ✓ | Apoplastic |
| g2666 | 429 | 142 | 16,443 | - | Single.; EE; UF | ✓ | x | 0.00 | x | x | x | Non-apoplastic |
| g3890 | 408 | 135 | 14,860 | [27] | PE 5 dai; EE; UF | x | x | 3.70 | x | ✓ | x | Non-apoplastic |
| g3970 | 2238 | 745 | 79,540 | [46] | PE 5 dai; DE 5 dai; DE 200 dai; UF | x | x | 1.30 | ✓ | ✓ | ✓ | Non-apoplastic |
| g4255 | 2301 | 766 | 86,484 | - | Single.; EE; UF | ✓ | Monopartite Score 9 1 | 0.78 | x | x | ✓ | Non-apoplastic |
| g4549 | 345 | 114 | 12,406 | [27] | Single.; EE; UF | ✓ | x | 1.75 | x | x | x | Non-apoplastic |
| g4554 | 396 | 131 | 14,068 | [27] | Single.; EE; UF | ✓ | x | 1.53 | x | x | x | Non-apoplastic |
| g5159 | 1293 | 430 | 48,247 | - | Single.; EE; UF | ✓ | x | 0.23 | ✓ 3 | ✓ | ✓ | Non-apoplastic |
| g6610 | 624 | 207 | 21,596 | - | Single.; EE; UF | x | x | 0.48 | x | x | ✓ | Non-apoplastic |
| Candidate Effector | PL1 | PL2 |
|---|---|---|
| g1052 1 | A9T | R158G |
| g3890 | H35R | L112Q |
| g5159 | Y230W | - |
| Protein ID | Subcellular Location | |
|---|---|---|
| N-tag | C-tag | |
| g1513 | Cytoplasmic | Nucleocytoplasmic |
| g2666 | Nucleocytoplasmic | Nucleocytoplasmic |
| g3890 | Nucleocytoplasmic | Nucleocytoplasmic 1, mitochondria |
| g3970 | Aggregates | Cytoplasm |
| g4549 | Nucleocytoplasmic | Nucleocytoplasmic |
| g4554 | Nucleocytoplasmic | Nucleocytoplasmic |
| g5159 | Cell wall | Cell wall |
| g6610 | Aggregates | Cytoplasm |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Teixeira-Silva, N.S.; Schaker, P.D.C.; Rody, H.V.S.; Maia, T.; Garner, C.M.; Gassmann, W.; Monteiro-Vitorello, C.B. Leaping into the Unknown World of Sporisorium scitamineum Candidate Effectors. J. Fungi 2020, 6, 339. https://doi.org/10.3390/jof6040339
Teixeira-Silva NS, Schaker PDC, Rody HVS, Maia T, Garner CM, Gassmann W, Monteiro-Vitorello CB. Leaping into the Unknown World of Sporisorium scitamineum Candidate Effectors. Journal of Fungi. 2020; 6(4):339. https://doi.org/10.3390/jof6040339
Chicago/Turabian StyleTeixeira-Silva, Natália Sousa, Patrícia Dayane Carvalho Schaker, Hugo Vianna Silva Rody, Thiago Maia, Christopher M. Garner, Walter Gassmann, and Claudia Barros Monteiro-Vitorello. 2020. "Leaping into the Unknown World of Sporisorium scitamineum Candidate Effectors" Journal of Fungi 6, no. 4: 339. https://doi.org/10.3390/jof6040339
APA StyleTeixeira-Silva, N. S., Schaker, P. D. C., Rody, H. V. S., Maia, T., Garner, C. M., Gassmann, W., & Monteiro-Vitorello, C. B. (2020). Leaping into the Unknown World of Sporisorium scitamineum Candidate Effectors. Journal of Fungi, 6(4), 339. https://doi.org/10.3390/jof6040339





