Molecular Epidemiology of Candida Auris Outbreak in a Major Secondary-Care Hospital in Kuwait
Abstract
1. Introduction
2. Materials and Methods
2.1. Outbreak Setting, Case Definitions and Sample Collection
2.2. Laboratory Methods
2.3. Infection Control Measures and Outbreak Management
2.4. Statistical Analysis
3. Results
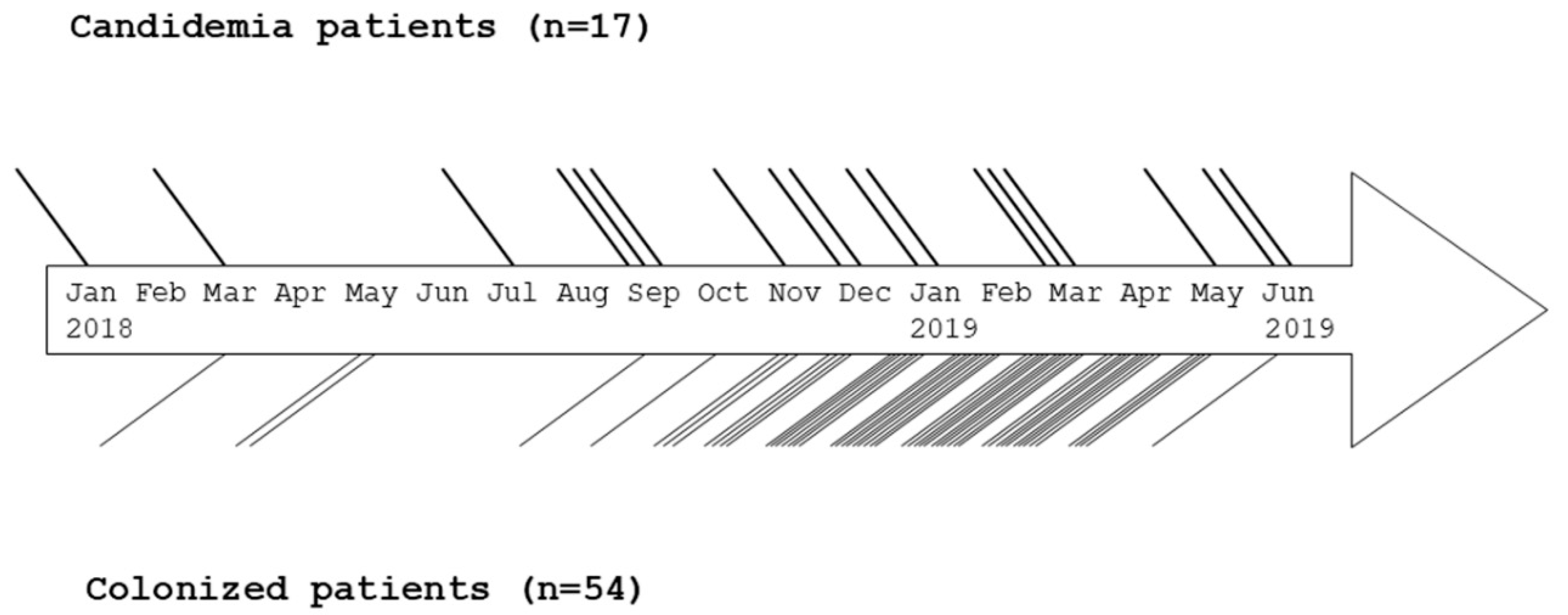
3.1. Patient Characteristics, Outbreak Description, Treatment and Outcome
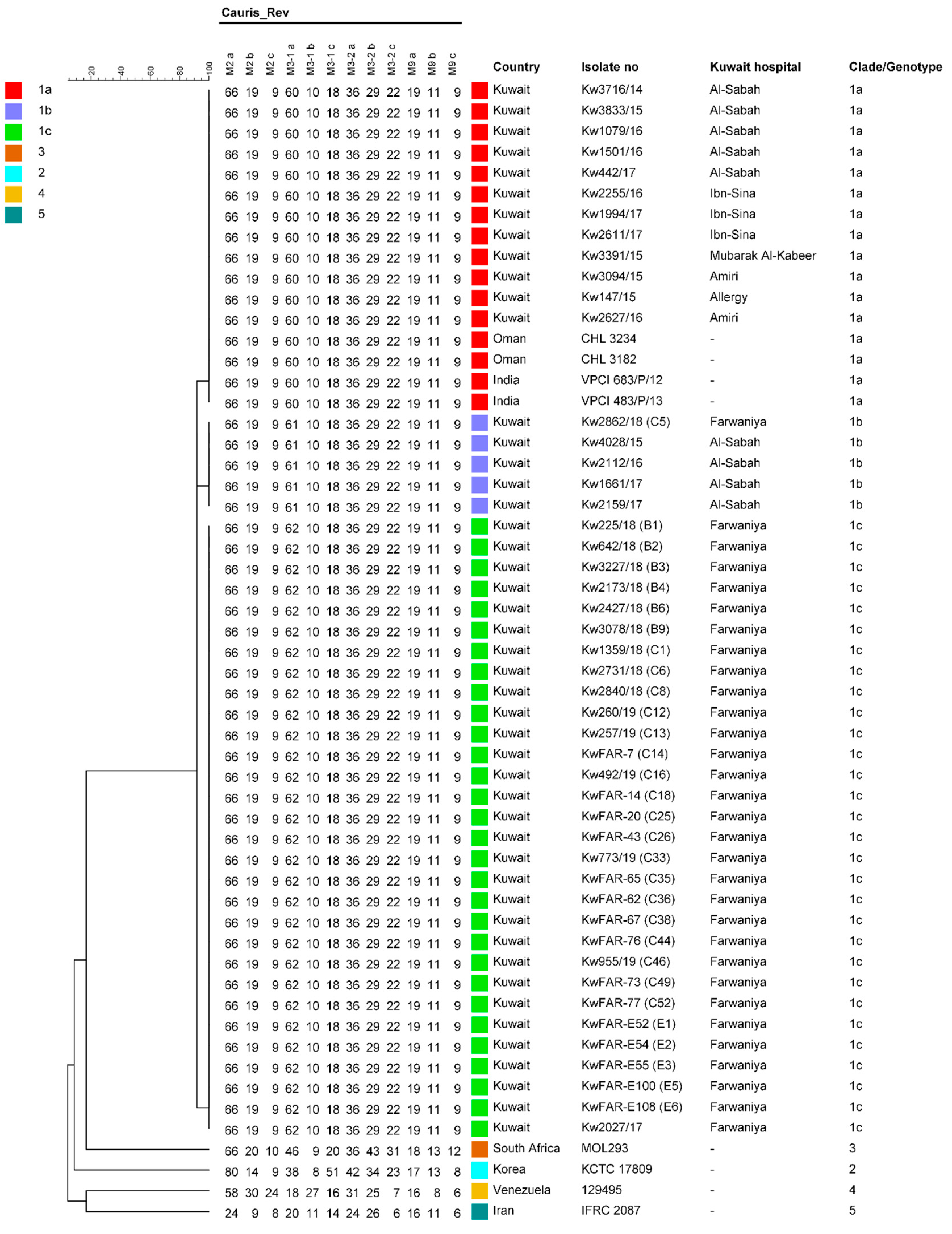
3.2. Culture Identification and Genotyping
3.3. Antifungal Susceptibility Testing and Mutation Analyses of ERG11 and FKS1 Genes
3.4. Infection Control Measures and Outbreak Management
4. Discussion
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Chowdhary, A.; Sharma, C.; Meis, J.F. Candida auris: A rapidly emerging cause of hospital-acquired multidrug-resistant fungal infections globally. PLoS Pathog. 2017, 13, e1006290. [Google Scholar] [CrossRef] [PubMed]
- Khan, Z.; Ahmad, S.; Benwan, K.; Purohit, P.; Al-Obaid, I.; Bafna, R.; Emara, M.; Mokaddas, E.; Abdullah, A.A.; Al-Obaid, K.; et al. Invasive Candida auris infections in Kuwait hospitals: Epidemiology, antifungal treatment and outcome. Infection 2018, 46, 641–650. [Google Scholar] [CrossRef] [PubMed]
- Eyre, D.W.; Sheppard, A.E.; Madder, H.; Moir, I.; Moroney, R.; Quan, T.P.; Griffiths, D.; George, S.; Butcher, L.; Morgan, M.; et al. A Candida auris outbreak and its control in an intensive care setting. N. Engl. J. Med. 2018, 379, 1322–1331. [Google Scholar] [CrossRef] [PubMed]
- Cortegiani, A.; Misseri, G.; Giarratano, A.; Bassetti, M.; Eyre, D. The global challenge of Candida auris in the intensive care unit. Crit. Care 2019, 23, 150. [Google Scholar] [CrossRef] [PubMed]
- Welsh, R.M.; Bentz, M.L.; Shams, A.; Houston, H.; Lyons, A.; Rose, L.J.; Litvintseva, A.P. Survival, persistence, and isolation of the emerging multidrug-resistant pathogenic yeast Candida auris on a plastic health care surface. J. Clin. Microbiol. 2017, 55, 2996–3005. [Google Scholar] [CrossRef]
- Ruiz-Gaitán, A.; Martínez, H.; Moret, A.M.; Calabuig, E.; Tasias, M.; Alastruey-Izquierdo, A.; Zaragoza, Ó.; Mollar, J.; Frasquet, J.; Salavert-Lletí, M.; et al. Detection and treatment of Candida auris in an outbreak situation: Risk factors for developing colonization and candidemia by this new species in critically ill patients. Expert Rev. Anti Infect. Ther. 2019, 17, 295–305. [Google Scholar] [CrossRef]
- Kumar, J.; Eilertson, B.; Cadnum, J.L.; Whitlow, C.S.; Jencson, A.L.; Safdar, N.; Krein, S.L.; Tanner, W.D.; Mayer, J.; Samore, M.H.; et al. Environmental contamination with Candida species in multiple hospitals including a tertiary care hospital with a Candida auris outbreak. Pathog. Immun. 2019, 4, 260–270. [Google Scholar] [CrossRef]
- Kenters, N.; Kiernan, M.; Chowdhary, A.; Denning, D.W.; Pemán, J.; Saris, K.; Schelenz, S.; Tartari, E.; Widmer, A.; Meis, J.F.; et al. Control of Candida auris in healthcare institutions: Outcome of an International Society for Antimicrobial Chemotherapy expert meeting. Int. J. Antimicrob. Agents 2019, 54, 400–406. [Google Scholar] [CrossRef]
- Lee, W.G.; Shin, J.H.; Uh, Y.; Kang, M.G.; Kim, S.H.; Park, K.H.; Jang, H.-C. First three reported cases of nosocomial fungemia caused by Candida auris. J. Clin. Microbiol. 2011, 49, 3139–3142. [Google Scholar] [CrossRef]
- Saris, K.; Meis, J.F.; Voss, A. Candida auris. Curr. Opin. Infect. Dis. 2018, 31, 334–340. [Google Scholar] [CrossRef]
- Cortegiani, A.; Misseri, G.; Fasciana, T.; Giammanco, A.; Giarratano, A.; Chowdhary, A. Epidemiology, clinical characteristics, resistance, and treatment of infections by Candida auris. J. Intensive Care 2018, 6, 69. [Google Scholar] [CrossRef] [PubMed]
- Chowdhary, A.; Anil Kumar, V.; Sharma, C.; Prakash, A.; Agarwal, K.; Babu, R.; Dinesh, K.R.; Karim, S.; Singh, S.K.; Hagen, F.; et al. Multidrug-resistant endemic clonal strain of Candida auris, India. Eur. J. Clin. Microbiol. Infect. Dis. 2014, 33, 919–926. [Google Scholar] [CrossRef] [PubMed]
- Govender, N.P.; Magobo, R.E.; Mpembe, R.; Mhlanga, M.; Matlapeng, P.; Corcoran, C.; Govind, C.; Lowman, W.; Senekal, M.; Thomas, J. Candida auris in South Africa, 2012–2016. Emerg. Infect. Dis. 2018, 24, 2036–2040. [Google Scholar] [CrossRef] [PubMed]
- Ahmad, S.; Khan, Z.; Al-Sweih, N.; Alfouzan, W.; Joseph, L. Candida auris in various hospitals across Kuwait and their susceptibility and molecular basis of resistance to antifungal drugs. Mycoses 2020, 63, 104–112. [Google Scholar] [CrossRef] [PubMed]
- Van Schalkwyk, E.; Mpembe, R.S.; Thomas, J.; Shuping, L.; Ismail, H.; Lowman, W.; Karstaedt, A.S.; Chibabhai, V.; Wadula, J.; Avenant, T.; et al. Epidemiologic shift in candidemia driven by Candida auris, South Africa, 2016–2017. Emerg. Infect. Dis. 2019, 25, 1698–1707. [Google Scholar] [CrossRef] [PubMed]
- Mathur, P.; Hasan, F.; Singh, P.K.; Malhotra, R.; Walia, K.; Chowdhary, A. Five-year profile of candidaemia at an Indian trauma centre: High rates of Candida auris blood stream infections. Mycoses 2018, 61, 674–680. [Google Scholar] [CrossRef]
- Forsberg, K.; Woodworth, K.; Walters, M.; Berkow, E.L.; Jackson, B.; Chiller, T.; Vallabahaneni, S. Candida auris: The recent emergence of a multidrug-resistant fungal pathogen. Med. Mycol. 2019, 57, 1–12. [Google Scholar] [CrossRef]
- Sharma, C.; Kumar, N.; Pandey, R.; Meis, J.F.; Chowdhary, A. Whole genome sequencing of emerging multidrug-resistant Candida auris isolates in India demonstrates low genetic variation. New Microbes New Infect. 2016, 13, 77–82. [Google Scholar] [CrossRef]
- Lockhart, S.R.; Etienne, K.A.; Vallabhaneni, S.; Farooqi, J.; Chowdhary, A.; Govender, N.P.; Colombo, A.; Calvo, B.; Cuomo, C.A.; Desjardins, C.A.; et al. Simultaneous emergence of multidrug-resistant Candida auris on 3 continents confirmed by whole-genome sequencing and epidemiological analyses. Clin. Infect. Dis. 2017, 64, 134–140. [Google Scholar] [CrossRef]
- Chow, N.A.; de Groot, T.; Badali, H.; Abastabar, M.; Chiller, T.M.; Meis, J.F. Potential fifth clade of Candida auris, Iran, 2018. Emerg. Infect. Dis. 2019, 25, 1780–1781. [Google Scholar] [CrossRef]
- Al Maani, A.; Paul, H.; Al-Rashdi, A.; Wahaibi, A.A.; Al-Jardani, A.; Al Abri, A.M.A.; AlBalushi, M.A.H.; Al-Abri, S.; Al Reesi, M.; Al Maqbali, A.; et al. Ongoing challenges with healthcare-associated Candida auris outbreaks in Oman. J. Fungi (Basel) 2019, 5, 101. [Google Scholar] [CrossRef] [PubMed]
- Alfouzan, W.; Dhar, R.; Albarrag, A.; Al-Abdely, H. The emerging pathogen Candida auris: A focus on the Middle-Eastern countries. J. Infect. Public Health 2019, 12, 451–459. [Google Scholar] [CrossRef] [PubMed]
- Chowdhary, A.; Prakash, A.; Sharma, C.; Kordalewska, M.; Kumar, A.; Sarma, S.; Tarai, B.; Singh, A.; Upadhyaya, G.; Upadhyay, S.; et al. A multicentre study of antifungal susceptibility patterns among 350 Candida auris isolates (2009–17) in India: Role of ERG11 and FKS1 genes in azole and echinocandin resistance. J. Antimicrob. Chemother. 2018, 73, 891–899. [Google Scholar] [CrossRef] [PubMed]
- Kwon, Y.J.; Shin, J.H.; Byun, S.A.; Choi, M.J.; Won, E.J.; Lee, D.; Lee, S.Y.; Chun, S.; Lee, J.H.; Choi, H.J.; et al. Candida auris clinical isolates from South Korea: Identification, antifungal susceptibility, and genotyping. J. Clin. Microbiol. 2019, 57, e01624-18. [Google Scholar] [CrossRef]
- Chowdhary, A.; Sharma, C.; Duggal, S.; Agarwal, K.; Prakash, A.; Singh, P.K.; Jain, S.; Kathuria, S.; Randhawa, H.S.; Hagen, F.; et al. New clonal strain of Candida auris, Delhi, India. Emerg. Infect. Dis. 2013, 19, 1670–1673. [Google Scholar] [CrossRef]
- Adams, E.; Quinn, M.; Tsay, S.; Poirot, E.; Chaturvedi, S.; Southwick, K.; Greenko, J.; Fernandez, R.; Kallen, A.; Vallabhaneni, S.; et al. Candida auris in healthcare facilities, New York, USA, 2013–2017. Emerg. Infect. Dis. 2018, 24, 1816–1824. [Google Scholar] [CrossRef]
- Ku, T.S.N.; Walraven, C.J.; Lee, S.A. Candida auris: Disinfectants and implications for infection control. Front. Microbiol. 2018, 9, 726. [Google Scholar] [CrossRef]
- Zhu, Y.; O’Brien, B.; Leach, L.; Clarke, A.; Bates, M.; Adams, E.; Ostrowsky, B.; Quinn, M.; Dufort, E.; Southwick, K.; et al. Laboratory analysis of an outbreak of Candida auris in New York from 2016 to 2018: Impact and lessons learned. J. Clin. Microbiol. 2020, 58, e01503-19. [Google Scholar] [CrossRef]
- Sekyere, J.O. Candida auris: A systematic review and meta-analysis of current updates on an emerging multidrug-resistant pathogen. Microbiol. Open 2018, 7, e578. [Google Scholar] [CrossRef]
- Ruiz-Gaitán, A.; Moret, A.M.; Tasias-Pitarch, M.; Aleixandre-López, A.I.; Martínez-Morel, H.; Calabuig, E.; Salavert-Lletí, M.; Ramírez, P.; López-Hontangas, J.L.; Hagen, F.; et al. An outbreak due to Candida auris with prolonged colonisation and candidaemia in a tertiary care European hospital. Mycoses 2018, 61, 498–505. [Google Scholar] [CrossRef]
- Khan, Z.; Ahmad, S. Candida auris: An emerging multidrug-resistant pathogen of global significance. Clin. Med. Res. Pract. 2017, 7, 240–248. [Google Scholar] [CrossRef]
- Schelenz, S.; Hagen, F.; Rhodes, J.L.; Abdolrasouli, A.; Chowdhary, A.; Hall, A.; Ryan, L.; Shackleton, J.; Trimlett, R.; Meis, J.F.; et al. First hospital outbreak of the globally emerging Candida auris in a European hospital. Antimicrob. Resist. Infect. Control 2016, 5, 35. [Google Scholar] [CrossRef] [PubMed]
- Rhodes, J.; Abdolrasouli, A.; Farrer, R.A.; Cuomo, C.A.; Aanensen, D.M.; Armstrong-James, D.; Fischer, M.C.; Schelenz, S. Genomic epidemiology of the UK outbreak of the emerging human fungal pathogen Candida auris. Emerg. Microbes Infect. 2018, 7, 43. [Google Scholar] [CrossRef] [PubMed]
- Tsay, S.; Welsh, R.M.; Adams, E.H.; Chow, N.A.; Gade, L.; Berkow, E.L.; Poirot, E.; Lutterloh, E.; Quinn, M.; Chaturvedi, S.; et al. Notes from the field: Ongoing transmission of Candida auris in health care facilities—United States, June 2016-May 2017. Morb. Mortal. Wkly. Rep. 2017, 66, 514–515. [Google Scholar] [CrossRef] [PubMed]
- Theodoropoulos, N.M.; Bolstorff, B.; Bozorgzadeh, A.; Brandeburg, C.; Cumming, M.; Daly, J.S.; Ellison, R.T., 3rd; Forsberg, K.; Gade, L.; Gibson, L.; et al. Candida auris outbreak involving liver transplant recipients in a surgical intensive care unit. Am. J. Transplant. 2020. [Google Scholar] [CrossRef] [PubMed]
- Mulet Bayona, J.V.; Tormo Palop, N.; Salvador García, C.; Herrero Rodríguez, P.; Abril López de Medrano, V.; Ferrer Gómez, C.; Gimeno Cardona, C. Characteristics and management of candidaemia episodes in an established Candida auris outbreak. Antibiotics 2020, 9, 558. [Google Scholar] [CrossRef] [PubMed]
- Armstrong, P.A.; Rivera, S.M.; Escandon, P.; Caceres, D.H.; Chow, N.; Stuckey, M.J.; Díaz, J.; Gomez, A.; Vélez, N.; Espinosa-Bode, A.; et al. Hospital-associated multicenter outbreak of emerging fungus Candida auris, Colombia, 2016. Emerg. Infect. Dis. 2019, 25, 1339–1346. [Google Scholar] [CrossRef] [PubMed]
- Adam, R.D.; Revathi, G.; Okinda, N.; Fontaine, M.; Shah, J.; Kagotho, E.; Castanheira, M.; Pfaller, M.A.; Maina, D. Analysis of Candida auris fungemia at a single facility in Kenya. Int. J. Infect. Dis. 2019, 85, 182–187. [Google Scholar] [CrossRef]
- Barantsevich, N.E.; Vetokhina, A.V.; Ayushinova, N.I.; Orlova, O.E.; Barantsevich, E.P. Candida auris bloodstream infections in Russia. Antibiotics 2020, 9, 557. [Google Scholar] [CrossRef]
- Alshamrani, M.M.; El-Saed, A.; Mohammed, A.; Alghoribi, M.F.; Al Johani, S.M.; Cabanalan, H.; Balkhy, H.H. Management of Candida auris outbreak in a tertiary-care setting in Saudi Arabia. Infect. Control Hosp. Epidemiol. 2020, 3, 1–7. [Google Scholar] [CrossRef]
- Mohsin, J.; Weerakoon, S.; Ahmed, S.; Puts, Y.; Al Balushi, Z.; Meis, J.F.; Al-Hatmi, A.M.S. A cluster of Candida auris blood stream infections in a tertiary care hospital in Oman from 2016 to 2019. Antibiotics 2020, 9, 638. [Google Scholar] [CrossRef] [PubMed]
- Khan, Z.; Ahmad, S.; Al-Sweih, N.; Mokaddas, E.; Al-Banwan, K.; Alfouzan, W.; Al-Obaid, I.; Al-Obaid, K.; Asadzadeh, M.; Jeragh, A.; et al. Changing trends in epidemiology and antifungal susceptibility patterns of six bloodstream Candida species isolates over a 12-year period in Kuwait. PLoS ONE 2019, 14, e0216250. [Google Scholar] [CrossRef] [PubMed]
- Jamal, W.Y.; Ahmad, S.; Khan, Z.U.; Rotimi, V.O. Comparative evaluation of two matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOF MS) systems for the identification of clinically significant yeasts. Int. J. Infect. Dis. 2014, 26, 167–170. [Google Scholar] [CrossRef] [PubMed]
- Asadzadeh, M.; Ahmad, S.; Hagen, F.; Meis, J.F.; Al-Sweih, N.; Khan, Z. Simple, low-cost detection of Candida parapsilosis complex isolates and molecular fingerprinting of Candida orthopsilosis strains in Kuwait by ITS region sequencing and amplified fragment length polymorphism analysis. PLoS ONE 2015, 10, e0142880. [Google Scholar] [CrossRef] [PubMed]
- Khan, Z.U.; Ahmad, S.; Al-Sweih, N.; Joseph, L.; Alfouzan, F.; Asadzadeh, M. Increasing prevalence, molecular characterization and antifungal drug susceptibility of serial Candida auris isolates in Kuwait. PLoS ONE 2018, 13, e0195743. [Google Scholar] [CrossRef] [PubMed]
- Ahmad, S.; Khan, Z.; Asadzadeh, M.; Theyyathel, A.; Chandy, R. Performance comparison of phenotypic and molecular methods for detection and differentiation of Candida albicans and Candida dubliniensis. BMC Infect. Dis. 2012, 12, 230. [Google Scholar] [CrossRef]
- European Committee on Antimicrobial Susceptibility Testing (EUCAST). Breakpoint Tables for Interpretation of MICs for Antifungal Agents. Version 10.0. 2020. Available online: http://www.eucast.org/astoffungi/clinicalbreakpointsforantifungals/ (accessed on 14 October 2020).
- Kathuria, S.; Singh, P.K.; Sharma, C.; Prakash, A.; Masih, A.; Kumar, A.; Meis, J.F.; Chowdhary, A. Multidrug-resistant Candida auris misidentified as Candida haemulonii: Characterization by matrix-assisted laser desorption ionization-time of flight mass spectrometry and DNA sequencing and its antifungal susceptibility profile variability by Vitek 2, CLSI broth microdilution, and Etest method. J. Clin. Microbiol. 2015, 53, 1823–1830. [Google Scholar]
- Arendrup, M.C.; Prakash, A.; Meletiadis, J.; Sharma, C.; Chowdhary, A. Comparison of EUCAST and CLSI reference microdilution MICs of eight antifungal compounds for Candida auris and associated tentative epidemiological cutoff values. Antimicrob. Agents Chemother. 2017, 61, e00485-17. [Google Scholar] [CrossRef]
- Centers for Disease Control and Prevention (CDC). Recommendations for Identification of Candida auris. Available online: https://www.cdc.gov/fungal/diseases/candidiasis/recommendations.html (accessed on 12 October 2020).
- De Groot, T.; Puts, Y.; Barrio, I.; Chowdhary, A.; Meis, J.F. Development of Candida auris short tandem repeat typing and its application to a global collection of isolates. mBio 2020, 11, e02971-19. [Google Scholar] [CrossRef]
- Asadzadeh, M.; Ahmad, S.; Al-Sweih, N.; Hagen, F.; Meis, J.F.; Khan, Z. High-resolution fingerprinting of Candida parapsilosis isolates suggests persistence and transmission of infections among neonatal intensive care unit patients in Kuwait. Sci. Rep. 2019, 9, 1340. [Google Scholar] [CrossRef]
- Centers for Disease Control and Prevention: Infection Prevention and Control for Candida auris. Available online: https://www.cdc.gov/fungal/candida-auris/c-auris-infection-control.html (accessed on 12 October 2020).
- Infection Control Directorate, Ministry of Health, Kuwait. Candida auris in Health Care Setting—Infection Control Policy. 2018. Available online: http://www.icdkwt.com/pdf/policiesandguidelines/InfectionPreventionandControl/c_auris_policy.pdf (accessed on 12 October 2020).
- Emara, M.; Ahmad, S.; Khan, Z.; Joseph, L.; Al-Obaid, I.; Purohit, P.; Bafna, R. Candida auris candidemia in Kuwait, 2014. Emerg. Infect. Dis. 2015, 21, 1091–1092. [Google Scholar] [CrossRef] [PubMed]
- Dannaoui, E.; Espinal-Ingroff, A. Antifungal susceptibility testing by concentration gradient strip Etest method for fungal isolates: A review. J. Fungi 2019, 5, 108. [Google Scholar] [CrossRef] [PubMed]
- Pfaller, M.A. Antifungal drug resistance: Mechanisms, epidemiology, and consequences for treatment. Am. J. Med. 2012, 125, S3–S13. [Google Scholar] [CrossRef] [PubMed]
- Asadzadeh, M.; Ahmad, S.; Al-Sweih, N.; Khan, Z. Epidemiology and molecular basis of resistance to fluconazole among clinical Candida parapsilosis isolates in Kuwait. Microb. Drug Resist. 2017, 23, 966–972. [Google Scholar] [CrossRef]
- Khan, Z.; Ahmad, S.; Al-Sweih, N.; Khan, S.; Joseph, L. Candida lusitaniae in Kuwait: Prevalence, antifungal susceptibility and role in neonatal fungemia. PLoS ONE 2019, 14, e0213532. [Google Scholar] [CrossRef]
- Healey, K.R.; Kordalewska, M.; Jiménez Ortigosa, C.; Singh, A.; Berrío, I.; Chowdhary, A.; Perlin, D.S. Limited ERG11 mutations identified in isolates of Candida auris directly contribute to reduced azole susceptibility. Antimicrob. Agents Chemother. 2018, 62, e01427-18. [Google Scholar] [CrossRef]
- Kean, R.; Sherry, L.; Townsend, E.; McKloud, E.; Short, B.; Akinbobola, A.; Mackay, W.G.; Williams, C.; Jones, B.L.; Ramage, G. Surface disinfection challenges for Candida auris: An in-vitro study. J. Hosp. Infect. 2018, 98, 433–436. [Google Scholar] [CrossRef]
- Abdolrasouli, A.; Armstrong-James, D.; Ryan, L.; Schelenz, S. In vitro efficacy of disinfectants utilised for skin decolonisation and environmental decontamination during a hospital outbreak with Candida auris. Mycoses 2017, 60, 758–763. [Google Scholar] [CrossRef]
- Singh, R.; Kaur, M.; Chakrabarti, A.; Shankarnarayan, S.A.; Rudramurthy, S.M. Biofilm formation by Candida auris isolated from colonising sites and candidemia cases. Mycoses 2019, 62, 706–709. [Google Scholar] [CrossRef]
- Kordalewska, M.; Lee, A.; Park, S.; Berrio, I.; Chowdhary, A.; Zhao, Y.; Perlin, D.S. Understanding echinocandin resistance in the emerging pathogen Candida auris. Antimicrob. Agents Chemother. 2018, 62, e00238-18. [Google Scholar] [CrossRef]
- Carolus, H.; Pierson, S.; Muñoz, J.F.; Subotić, A.; Cruz, R.B.; Cuomo, C.A.; Van Dijck, P. Genome-wide analysis of experimentally evolved Candida auris reveals multiple novel mechanisms of multidrug-resistance. bioRxiv 2020. [Google Scholar] [CrossRef]
- Al-Baqsami, Z.; Ahmad, S.; Khan, Z. Antifungal drug susceptibility, molecular basis of resistance to echinocandins and molecular epidemiology of fluconazole resistance among clinical Candida glabrata isolates in Kuwait. Sci. Rep. 2020, 10, 6238. [Google Scholar] [CrossRef] [PubMed]
- Biagi, M.J.; Wiederhold, N.P.; Gibas, C.; Wickes, B.; Lozano, V.; Bleasdale, S.C.; Danziger, L. Development of high-level echinocandin resistance in a patient with recurrent Candida auris candidemia secondary to chronic candiduria. Open Forum Infect. Dis. 2019, 6, ofz262. [Google Scholar] [CrossRef] [PubMed]
- Khan, Z.; Ahmad, S.; Mokaddas, E.; Meis, J.F.; Joseph, L.; Abdullah, A.; Vayalil, S. Development of echinocandin resistance in Candida tropicalis following short-term exposure to caspofungin for empiric therapy. Antimicrob. Agents Chemother. 2018, 62, e01926-17. [Google Scholar] [CrossRef] [PubMed]
- Ahmad, S.; Joseph, L.; Parker, J.E.; Asadzadeh, M.; Kelly, S.L.; Meis, J.F.; Khan, Z. ERG6 and ERG2 are major targets conferring reduced susceptibility to amphotericin B in clinical Candida glabrata isolates in Kuwait. Antimicrob. Agents Chemother. 2019, 63, e01900-18. [Google Scholar] [CrossRef]
- Khan, Z.; Ahmad, S.; Al-Sweih, N.; Mokaddas, E.; Al-Banwan, K.; Alfouzan, W.; Al-Obaid, I.; Al-Obaid, K.; Varghese, S. Increasing Trends of Reduced Susceptibility to Antifungal Drugs Among Clinical Candida glabrata Isolates in Kuwait. Microb. Drug Resist. 2020, 26, 982–990. [Google Scholar] [CrossRef] [PubMed]
| Variables | Candidemia Patients | Colonized Patients | p Value |
|---|---|---|---|
| n = 17 (%) | n = 54 (%) | ||
| Gender | |||
| Male | 10 (58.8) | 37 (68.5) | 0.461 |
| Female | 7 (41.2) | 17 (31.5) | |
| Age (years) groups | |||
| <50 | 3 (17.6) | 16 (29.6) | 0.381 |
| 50–65 | 3 (17.6) | 15 (27.8) | |
| 66–80 | 8 (47.1) | 19 (35.2) | |
| >80 | 3 (17.6) | 4 (7.4) | |
| Comorbidities | |||
| Hypertension | 10 (58.8) | 30 (55.6) | 0.813 |
| Diabetes mellitus | 9 (52.9) | 30 (55.6) | 0.85 |
| Dyslipidemia | 4 (23.5) | 8 (14.3) | 0.403 |
| Cardiovascular diseases | 7 (41.2) | 21 (38.9) | 0.866 |
| Others a | 9 (52.9) | 24 (44.4) | 0.54 |
| Length of hospital stay b (days) | |||
| <15 | 1 (5.9) | 16 (29.3) | 0.068 |
| 15–35 | 5 (29.4) | 17 (32.5) | |
| 36–55 | 3 (17.6) | 11 (20.4) | |
| >55 | 8 (47.1) | 26 (48.2) | |
| Treatment with antifungals | |||
| None | 0 (0.0) | 17 (31.4) | 0.002 |
| One antifungal drug | 14 (82.4) | 19 (35.2) | |
| Two antifungal drugs | 3 (17.6) | 18 (33.3) | |
| Mortality | |||
| Yes | 10 (58.8) | 27 (50) | 0.44 |
| No | 7 (41.2) | 27 (50) |
| Antifungal | Number of Isolates with Minimum Inhibitory Concentration (MIC) Value (µg/mL) of | Geometric | ||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Drug | 0.002 | 0.004 | 0.008 | 0.016 | 0.031 | 0.062 | 0.125 | 0.25 | 0.5 | 1 | 2 | 4 | 8 | 16 | 32 | 64 | 128 | Mean (µg/mL) |
| Fluconazole | 1 * | 1 | 12 | 2 | 61.29 | |||||||||||||
| Voriconazole | 1 | 15 | 2.95 | |||||||||||||||
| Itraconazole | 1 | 1 | 14 | 2.59 | ||||||||||||||
| Posaconazole | 1 | 2 | 1 | 4 | 7 * | 1 | 0.39 | |||||||||||
| Anidulafungin | 10 | 6 | 0.02 | |||||||||||||||
| Micafungin | 12 | 4 | 0.02 | |||||||||||||||
| Caspofungin | 1 | 1 | 2 * | 3 | 5 | 4 | 2.18 | |||||||||||
| Amphotericin B | 8 | 8 * | 0.71 | |||||||||||||||
| Antifungal | Number of Isolates with Minimum Inhibitory Concentration (MIC) Value (µg/mL) of | Geometric | ||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Drug | 0.002 | 0.004 | 0.008 | 0.016 | 0.031 | 0.06 | 0.125 | 0.25 | 0.5 | 1 | 2 | 4 | 8 | 16 | 32 | 64 | >128 | Mean (µg/mL) |
| Fluconazole | 3 * | 3 * | 4 | 31 | 5 | 51.83 | ||||||||||||
| Voriconazole | 1 | 8 | 9 | 5 | 6 | 4 | 1 | 12 | 0.27 | |||||||||
| Itraconazole | 12 | 10 | 1 | 1 | 3 | 1 * | 1 | 17 | 0.36 | |||||||||
| Posaconazole | 27 | 4 | 6 | 1 | 1 | 2 | 4 * | 1 | 0.02 | |||||||||
| Anidulafungin | 26 | 13 | 3 | 1 | 1 * | 1 | 1 | 0.03 | ||||||||||
| Micafungin | 37 | 6 | 1 * | 2 | 0.02 | |||||||||||||
| Caspofungin | 12 | 10 | 4 * | 2 | 9 | 9 | 1.22 | |||||||||||
| Amphotericin B | 28 | 18 * | 0.66 | |||||||||||||||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Alfouzan, W.; Ahmad, S.; Dhar, R.; Asadzadeh, M.; Almerdasi, N.; Abdo, N.M.; Joseph, L.; de Groot, T.; Alali, W.Q.; Khan, Z.; et al. Molecular Epidemiology of Candida Auris Outbreak in a Major Secondary-Care Hospital in Kuwait. J. Fungi 2020, 6, 307. https://doi.org/10.3390/jof6040307
Alfouzan W, Ahmad S, Dhar R, Asadzadeh M, Almerdasi N, Abdo NM, Joseph L, de Groot T, Alali WQ, Khan Z, et al. Molecular Epidemiology of Candida Auris Outbreak in a Major Secondary-Care Hospital in Kuwait. Journal of Fungi. 2020; 6(4):307. https://doi.org/10.3390/jof6040307
Chicago/Turabian StyleAlfouzan, Wadha, Suhail Ahmad, Rita Dhar, Mohammad Asadzadeh, Noura Almerdasi, Naglaa M. Abdo, Leena Joseph, Theun de Groot, Walid Q. Alali, Ziauddin Khan, and et al. 2020. "Molecular Epidemiology of Candida Auris Outbreak in a Major Secondary-Care Hospital in Kuwait" Journal of Fungi 6, no. 4: 307. https://doi.org/10.3390/jof6040307
APA StyleAlfouzan, W., Ahmad, S., Dhar, R., Asadzadeh, M., Almerdasi, N., Abdo, N. M., Joseph, L., de Groot, T., Alali, W. Q., Khan, Z., Meis, J. F., & Al-Rashidi, M. R. (2020). Molecular Epidemiology of Candida Auris Outbreak in a Major Secondary-Care Hospital in Kuwait. Journal of Fungi, 6(4), 307. https://doi.org/10.3390/jof6040307






