In Vitro or In Vivo Models, the Next Frontier for Unraveling Interactions between Malassezia spp. and Hosts. How Much Do We Know?
Abstract
1. Introduction
| Disease | Clinical Findings | Species Involved | Most Commonly Affected Population | References |
|---|---|---|---|---|
| Pityriasis versicolor (PV) | Macules on the trunk and arms; the skin lesions are hypopigmented and hyperpigmented | Malassezia globosa, Malassezia sympodialis, and Malassezia furfur | Young adults and rarely children and older adults | [3,22,23,24,25,26,27] |
| Dandruff/seborrheic dermatitis (D/SD) | Flaking and erythema in sebum-rich areas like the scalp, nostrils, chest, and eyebrows | M. globosa, Malassezia restricta, M. furfur, and Malassezia obtusa | Elders, infants, children in puberty and HIV patients | [3,25,27,28,29,30,31,32,33] |
| Atopic dermatitis (AD) | Chronic inflammatory illness with pruritic eczematous lesions. Malassezia has been proposed to act as an exacerbator | M sympodialis, M. globosa, M. furfur, M. restricta, Malassezia japonica, Malassezia yamatoensis, and M. slooffiae | Adults with genetic and environmental predisposing factors | [3,27,34,35,36,37,38] |
| Folliculitis | Small dome-shaped papules localized around follicular areas, mainly in the back, chest, and shoulders. The papules can evolve into pustules | M. globosa, M. restricta, M. sympodialis, M. furfur, and M. pachydermatis | Teenagers and young adult males | [3,25,26,31,32] |
| Psoriasis | Chronic skin disease, characterized by hyperproliferation and hyperkeratinization of the epidermis. Malassezia may augment inflammation and the severity of the disorder | M. globosa, M. furfur, M. sympodialis, M. restricta, and M. slooffiae | Patients with psoriasis, mainly on the scalps of young adults | [3,27,37,38,39,40,41,42] |
| Crohn´s disease | Inflammatory bowel disease characterized by altered immune response to intestinal microbiota. Malassezia yeasts in the gut may increase the severity of the disease | M. restricta | Crohn´s disease patients carrying the CARD9S12N risk allele | [4,43] |
| Parkinson’s disease | Neurodegenerative disease. Seborrheic dermatitis has been strongly associated with this disease | M. globosa, M. restricta, M. furfur, and M. obtusa | Elders. Risk increases after a seborrheic dermatitis diagnosis | [5] |
| Pancreatic ductal adenocarcinoma | Carcinoma due to fungal dysbiosis | M. globosa | Individuals with oncogenic Kras that induces inflammation, resulting in fungal dysbiosis | [6] |
| Invasive infections | Fungemia, endocarditis, bronchopneumonia, respiratory distress, splenic lesions, etc. | M. furfur, M. pachydermatis, M. sympodialis, and M. restricta | Low-weight neonates and immunocompromised patients | [3,7,8,26,44,45,46,47,48,49,50,51] |
2. Infection Models as a Way to Understand Host–Microbe Interactions
2.1. In Vitro Models of Host-Microbe Interaction
2.2. In Vivo Models of Host–Pathogen Interactions
2.2.1. Mammalian Models of Host–Pathogen Interactions
2.2.2. In Vivo Alternative Models of Host–Microbe Interactions
| Infection Model | Cost | Inoculation | Advantages | Disavantages | References |
|---|---|---|---|---|---|
| Keratinocyte culture | High | -Co-culture | -Controlled conditions -Just one type of cell | -It does not represent the complex interactions with the host | [52,54,74,75,76,77,78,79,80,81,83] |
| Murine model | High | -Oral gavage -Inoculation through the tail vein -Inhalation and intranasal administration -Direct inoculation -Ocular -Intracranial -Intraperitoneal | -Well-defined inoculation routes -Immune response is similar to a human’s, with innate and adaptative immune response -Mimics human infection and disease -Annotated genome -Available mutants | -Ethical issues -Bigger space for storage -Longer generation time -Trained personnel to handle the models -Immune suppression required | [57,60,117,118] |
| Amoeboid model (Acanthamoeba castellani) | Low | -Co-incubation | -Controlled conditions -Inoculum quantification -Available mutants -Incubation at 37 °C -Phagocytosis assays -Short life cycle -Annotated genome | -Undesired mutation and loss of phagocytic abilities in long-cultured strains | [53,54,102,119,120] |
| Zebrafish larvae (Danio rerio) | Low | -Microinjection into the caudal vein, notochord, duct of Cuvier, hindbrain ventricle, eye, peritoneal cavity, or muscle - exposure by immersion | -Short generation time -Annotated genome sequence -Available mutants -Transparency -High-throughput screening -Innate immune response similar to that of humans | -In larval stage, there is no adaptative immune response -Ethical issues in some countries -Difficult to handle | [100,118,121,122,123,124,125,126] |
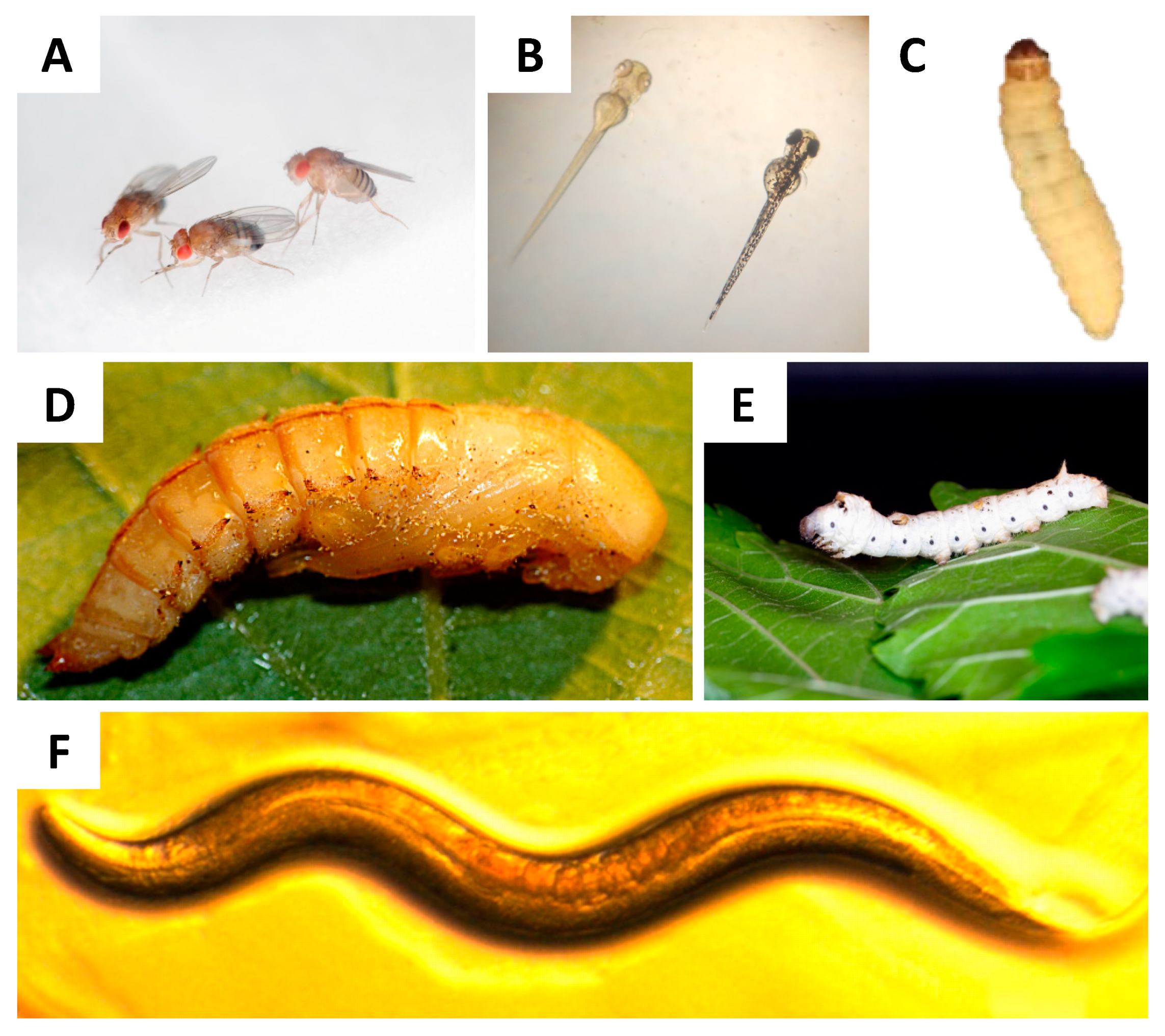
| Caenorhabditis elegans | Low | -Exposure of larvae by immersion (feeding and contact with the cuticle) | -Short generation time -Small size -Easy to grow -Annotated genome sequence -Available mutants -Innate immune response similar to that of humans -Results correlated with results from mammals | -There is no adaptative immune response -Difficult to inoculate and quantify the inoculum | [87,127] |
| Silkworm (Bombyx mori) | Low | -Microinjection into the haemocoel -Oral (puncture) | -Inoculum quantification -High inoculum volume -Results correlated with results from mammals -Innate immune response similar to that of humans | -No adaptative immune response | [60,118,128] |
| Drosophila melanogaster | Low | -Puncture in the dorsal side of the thorax | -Annotated genome sequence -Available mutants -Innate immune response similar to that of humans | -No adaptative immune response -Difficult to inoculate and quantify the inoculum | [63,106,129] |
| Galleria mellonella | Low | Microinjection directly to the haemocoel -Topical -Oral | -Inoculum quantification -Wide range of temperatures -Innate immune response similar to that of humans -Available immune response transcriptome -Results correlated with results from mammals | -No adaptative immune response | [67,109,118,119,130,131,132,133] |
3. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Juntachai, W.; Oura, T.; Murayama, S.Y.; Kajiwara, S. The lipolytic enzymes activities of Malassezia species. Med. Mycol. 2009, 47, 477–484. [Google Scholar] [CrossRef] [PubMed]
- Wu, G.; Zhao, H.; Li, C.; Rajapakse, M.P.; Wong, W.C.; Xu, J.; Saunders, C.W.; Reeder, N.L.; Reilman, R.A.; Scheynius, A.; et al. Genus-Wide Comparative Genomics of Malassezia Delineates Its Phylogeny, Physiology, and Niche Adaptation on Human Skin. PLoS Genet. 2015, 11, 1–26. [Google Scholar] [CrossRef] [PubMed]
- Theelen, B.; Cafarchia, C.; Gaitanis, G.; Bassukas, I.D.; Boekhout, T.; Dawson, T.L. Malassezia ecology, pathophysiology, and treatment. Med. Mycol. 2018, 56, S10–S25. [Google Scholar] [CrossRef] [PubMed]
- Limon, J.J.; Tang, J.; Li, D.; Wolf, A.J.; Michelsen, K.S.; Funari, V.; Gargus, M.; Nguyen, C.; Sharma, P.; Maymi, V.I.; et al. Malassezia Is Associated with Crohn’s Disease and Exacerbates Colitis in Mouse Models. Cell Host Microbe 2019, 25, 377–388. [Google Scholar] [CrossRef]
- Laurence, M.; Benito-León, J.; Calon, F. Malassezia and Parkinson’s disease. Front. Neurol. 2019, 10, 758. [Google Scholar] [CrossRef] [PubMed]
- Aykut, B.; Pushalkar, S.; Chen, R.; Li, Q.; Abengozar, R.; Kim, J.I.; Shadaloey, S.A.; Wu, D.; Preiss, P.; Verma, N.; et al. The fungal mycobiome promotes pancreatic oncogenesis via activation of MBL. Nature 2019, 574, 264–267. [Google Scholar] [CrossRef]
- Roman, J.; Bagla, P.; Ren, P.; Blanton, L.S.; Berman, M.A. Malassezia pachydermatis fungemia in an adult with multibacillary leprosy. Med. Mycol. Case Rep. 2016, 12, 1–3. [Google Scholar] [CrossRef]
- Ochman, E.; Podsiadło, B.; Połowniak-Pracka, H.; Hagmajer, E.; Sowiński, P. Malassezia furfur sepsis in a cancer patient. Nowotw. J. Oncol. 2004, 54, 130–134. [Google Scholar]
- Nagata, R.; Nagano, H.; Ogishima, D.; Nakamura, Y.; Hiruma, M.; Sugita, T. Transmission of the major skin microbiota, Malassezia, from mother to neonate. Pediatr. Int. 2012, 54, 350–355. [Google Scholar] [CrossRef]
- DeAngelis, Y.M.; Gemmer, C.M.; Kaczvinsky, J.R.; Kenneally, D.C.; Schwartz, J.R.; Dawson, T.L. Three etiologic facets of dandruff and seborrheic dermatitis: Malassezia fungi, sebaceous lipids, and individual sensitivity. J. Investig. Dermatol. Symp. Proc. 2005, 10, 295–297. [Google Scholar] [CrossRef]
- Borelli, D.; Jacobs, P.H.; Nall, L. Tinea versicolor: Epidemiologic, clinical, and therapeutic aspects. J. Am. Acad. Dermatol. 1991, 25, 300–305. [Google Scholar] [CrossRef]
- Leeming, J.P.; Notman, F.H. Improved Methods for Isolation and Enumeration of Malassezia furfur from Human Skin. J. Clin. Microbiol. 1987, 25, 2017–2019. [Google Scholar] [CrossRef] [PubMed]
- Guého, E.; Midgley, G.; Guillot, J. The genus Malassezia with description of four new species. Antonie Leeuwenhoek Int. J. Gen. Mol. Microbiol. 1996, 69, 337–355. [Google Scholar] [CrossRef] [PubMed]
- Ashbee, H.R.; Evans, E.G.V. Immunology of diseases associated with Malassezia species. Clin. Microbiol. Rev. 2002, 15, 21–57. [Google Scholar] [CrossRef] [PubMed]
- Sparber, F.; De Gregorio, C.; Steckholzer, S.; Ferreira, F.M.; Dolowschiak, T.; Ruchti, F.; Kirchner, F.R.; Mertens, S.; Prinz, I.; Joller, N.; et al. The Skin Commensal Yeast Malassezia Triggers a Type 17 Response that Coordinates Anti-fungal Immunity and Exacerbates Skin Inflammation. Cell Host Microbe 2019, 25, 389–403. [Google Scholar] [CrossRef] [PubMed]
- Sparber, F.; Ruchti, F.; Leibund Gut-Landmann, S. Host Immunity to Malassezia in Health and Disease. Front. Cell. Infect. Microbiol. 2020, 10, 198. [Google Scholar] [CrossRef]
- Vlachos, C.; Schulte, B.M.; Magiatis, P.; Adema, G.J.; Gaitanis, G. Malassezia-derived indoles activate the aryl hydrocarbon receptor and inhibit Toll-like receptor-induced maturation in monocyte-derived dendritic cells. Br. J. Dermatol. 2012, 167, 496–505. [Google Scholar] [CrossRef]
- Kim, S.-H.; Ko, H.-C.; Kim, M.-B.; Kwon, K.-S.; Oh, C.-K. The Effect of Detergents on the Morphology and Immunomodulatory Activity of Malassezia furfur. Ann. Dermatol. 2009, 21, 130–135. [Google Scholar] [CrossRef]
- Kesavan, S.; Holland, K.T.; Ingham, E. The effects of lipid extraction on the immunomodulatory activity of Malassezia species in vitro. Med. Mycol. 2000, 38, 239–247. [Google Scholar] [CrossRef]
- Youngchim, S.; Nosanchuk, J.D.; Pornsuwan, S.; Kajiwara, S.; Vanittanakom, N. The Role of L-DOPA on Melanization and Mycelial Production in Malassezia furfur. PLoS ONE 2013, 8, e63764. [Google Scholar] [CrossRef]
- Sparber, F.; LeibundGut-Landmann, S. Host responses to Malassezia spp. in the mammalian skin. Front. Immunol. 2017, 8, 1614. [Google Scholar] [CrossRef] [PubMed]
- Lyakhovitsky, A.; Shemer, A.; Amichai, B.; Boaz Amichai, C. Molecular analysis of Malassezia species isolated from Israeli patients with pityriasis versicolor. Int. J. Dermatol. 2013, 52, 231–233. [Google Scholar] [CrossRef]
- Borelli, D. Pitiriasis versicolor por Malassezia ovalis. Mycopathologia 1985, 89, 147–153. [Google Scholar] [CrossRef] [PubMed]
- Gupta, A.K.; Kohliyz, Y.; Faergemann, J.; Summerbell, R.C. Epidemiology of Malassezia yeasts associated with pityriasis versicolor in Ontario, Canada. Med. Mycol. 2001, 39, 199–206. [Google Scholar] [CrossRef] [PubMed]
- Magiatis, P.; Pappas, P.; Gaitanis, G.; Mexia, N.; Melliou, E.; Galanou, M.; Vlachos, C.; Stathopoulou, K.; Skaltsounis, A.L.; Marselos, M.; et al. Malassezia yeasts produce a collection of exceptionally potent activators of the ah (dioxin) receptor detected in diseased human skin. J. Investig. Dermatol. 2013, 133, 2023–2030. [Google Scholar] [CrossRef] [PubMed]
- Morrison, V.A.; Weisdorf, D.J. The spectrum of Malassezia infections in the bone marrow transplant population. Bone Marrow Transplant. 2000, 26, 645–648. [Google Scholar] [CrossRef]
- Gupta, A.K.; Kohliy, Y.; Summerbell, R.C.; Faergemann, J. Quantitative culture of Malassezia species from different body sites of individuals with or without dermatoses. Med. Mycol. 2001, 39, 243–251. [Google Scholar] [CrossRef]
- Hiruma, M.; Cho, O.; Hiruma, M.; Kurakado, S.; Sugita, T.; Ikeda, S. Genotype Analyses of Human Commensal Scalp Fungi, Malassezia globosa, and Malassezia restricta on the Scalps of Patients with Dandruff and Healthy Subjects. Mycopathologia 2014, 177, 263–269. [Google Scholar] [CrossRef]
- Ro, B.I.; Dawson, T.L. The Role of Sebaceous Gland Activity and Scalp Microfloral Metabolism in the Etiology of Seborrheic Dermatitis and Dandruff. J. Investig. Dermatol. Symp. Proc. 2005, 10, 194–197. [Google Scholar] [CrossRef]
- Nakabayashi, A.; Sei, Y.; Guillot, J. Identification of Malassezia species isolated from patients with seborrhoeic dermatitis, atopic dermatitis, pityriasis versicolor and normal subjects. Med. Mycol. 2000, 38, 337–341. [Google Scholar] [CrossRef]
- Ashbee, H.R. Update on the genus Malassezia. Med. Mycol. 2007, 45, 287–303. [Google Scholar] [CrossRef] [PubMed]
- Dokos, C.; Pana, Z.D.; Tragiannidis, A. Malassezia species: A rare cause of invasive fungal infections in immunocompromised patients. Curr. Fungal Infect. Rep. 2011, 5, 18–22. [Google Scholar] [CrossRef]
- Kamamoto, C.S.L.; Nishikaku, A.S.; Gompertz, O.F.; Melo, A.S.; Hassun, K.M.; Bagatin, E. Cutaneous fungal microbiome: Malassezia yeasts in seborrheic dermatitis scalp in a randomized, comparative and therapeutic trial. Dermatoendocrinology 2018, 9, e1361573. [Google Scholar] [CrossRef] [PubMed]
- Johansson, H.J.; Vallhov, H.; Holm, T.; Gehrmann, U.; Andersson, A.; Johansson, C.; Blom, H.; Carroni, M.; Lehtiö, J.; Scheynius, A. Extracellular nanovesicles released from the commensal yeast Malassezia sympodialis are enriched in allergens and interact with cells in human skin. Sci. Rep. 2018, 8, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Jain, C.; Das, S.; Ramachandran, V.G.; Saha, R.; Bhattacharya, S.N.; Dar, S. Malassezia yeast and cytokine gene polymorphism in atopic dermatitis. J. Clin. Diagn. Res. 2017, 11, DC01–DC05. [Google Scholar] [CrossRef]
- Glatz, M.; Bosshard, P.; Hoetzenecker, W.; Schmid-Grendelmeier, P. The Role of Malassezia spp. in Atopic Dermatitis. J. Clin. Med. 2015, 4, 1217–1228. [Google Scholar] [CrossRef]
- Prohic, A.; Jovovic Sadikovic, T.; Krupalija-Fazlic, M.; Kuskunovic-Vlahovljak, S. Malassezia species in healthy skin and in dermatological conditions. Int. J. Dermatol. 2016, 55, 494–504. [Google Scholar] [CrossRef]
- Kanda, N.; Tani, K.; Enomoto, U.; Nakai, K.; Watanabe, S. The skin fungus-induced Th1- and Th2-related cytokine, chemokine and prostaglandin E2 production in peripheral blood mononuclear cells from patients with atopic dermatitis and psoriasis vulgaris. Clin. Exp. Allergy 2002, 32, 1243–1250. [Google Scholar] [CrossRef]
- Jagielski, T.; Rup, E.; Ziółkowska, A.; Roeske, K.; Macura, A.B.; Bielecki, J. Distribution of Malassezia species on the skin of patients with atopic dermatitis, psoriasis, and healthy volunteers assessed by conventional and molecular identification methods. BMC Dermatol. 2014, 14, 3. [Google Scholar] [CrossRef]
- Rudramurthy, S.M.; Honnavar, P.; Chakrabarti, A.; Dogra, S.; Singh, P.; Handa, S. Association of Malassezia species with psoriatic lesions. Mycoses 2014, 57, 483–488. [Google Scholar] [CrossRef]
- Hurabielle, C.; Link, V.M.; Bouladoux, N.; Han, S.-J.; Dean Merrill, E.; Lightfoot, Y.L.; Seto, N.; Bleck, C.K.E.; Smelkinson, M.; Harrison, O.J.; et al. Immunity to commensal skin fungi promotes psoriasiform skin inflammation. Proc. Natl. Acad. Sci. USA 2020, 117, 16465–16474. [Google Scholar] [CrossRef] [PubMed]
- Lober, C.W.; Belew, P.W.; Rosenberg, W.; Bale, G. Patch Tests With Killed Sonicated Microflora in Patients With Psoriasis from the Departments of Medicine. Arch. Dermatol. 1982, 118, 322–325. [Google Scholar] [CrossRef] [PubMed]
- Kellermayer, R.; Mir, S.A.V.; Nagy-Szakal, D.; Cox, S.B.; Dowd, S.E.; Kaplan, J.L.; Sun, Y.; Reddy, S.; Bronsky, J.; Winter, H.S. Microbiota separation and C-reactive protein elevation in treatment-naïve pediatric granulomatous crohn disease. J. Pediatr. Gastroenterol. Nutr. 2012, 55, 243–250. [Google Scholar] [CrossRef] [PubMed]
- Iatta, R.; Battista, M.; Miragliotta, G.; Boekhout, T.; Otranto, D.; Cafarchia, C. Blood culture procedures and diagnosis of Malassezia furfur bloodstream infections: Strength and weakness. Med. Mycol. 2018, 56, 828–833. [Google Scholar] [CrossRef] [PubMed]
- De St Maurice, A.; Frangoul, H.; Coogan, A.; Williams, J.V. Prolonged fever and splenic lesions caused by Malassezia restricta in an immunocompromised patient. Pediatr. Transplant. 2014, 18, E283–E286. [Google Scholar] [CrossRef]
- Lee, J.; Cho, Y.G.; Kim, D.S.; Choi, S.I.; Lee, H.S. First case of catheter-related Malassezia pachydermatis fungemia in an adult. Ann. Lab. Med. 2018, 39, 99–101. [Google Scholar] [CrossRef]
- Al-Sweih, N.; Ahmad, S.; Joseph, L.; Khan, S.; Khan, Z. Malassezia pachydermatis fungemia in a preterm neonate resistant to fluconazole and flucytosine. Med. Mycol. Case Rep. 2014, 5, 9–11. [Google Scholar] [CrossRef] [PubMed]
- Iatta, R.; Cafarchia, C.; Cuna, T.; Montagna, O.; Laforgia, N.; Gentile, O.; Rizzo, A.; Boekhout, T.; Otranto, D.; Montagna, M.T. Bloodstream infections by Malassezia and Candida species in critical care patients. Med. Mycol. 2014, 52, 264–269. [Google Scholar] [CrossRef] [PubMed]
- Chen, I.T.; Chen, C.C.; Huang, H.C.; Kuo, K.C. Malassezia furfur Emergence and Candidemia Trends in a Neonatal Intensive Care Unit during 10 Years: The Experience of Fluconazole Prophylaxis in a Single Hospital. Adv. Neonatal Care 2020, 20, E3–E8. [Google Scholar] [CrossRef]
- Aguirre, C.; Euliarte, C.; Finquelievich, J.; de los Ángeles Sosa, M.; Giusiano, G. Fungemia and interstitial lung compromise caused by Malassezia sympodialis in a pediatric patient. Rev. Iberoam. Micol. 2015, 32, 118–121. [Google Scholar] [CrossRef]
- Patron, R.L. A 34-Year-Old Man With Cough, Lung Nodules, Fever, and Eosinophilia. Clin. Infect. Dis. 2016, 63, 1525–1526. [Google Scholar] [CrossRef] [PubMed]
- Swearengen, J.R. Choosing the right animal model for infectious disease research. Anim. Model. Exp. Med. 2018, 1, 100–108. [Google Scholar] [CrossRef] [PubMed]
- Mylonakis, E.; Casadevall, A.; Ausubel, F.M. Exploiting amoeboid and non-vertebrate animal model systems to study the virulence of human pathogenic fungi. PLoS Pathog. 2007, 3, e101. [Google Scholar] [CrossRef] [PubMed]
- Capilla, J.; Clemons, K.V.; Stevens, D.A. Animal models: An important tool in mycology. Med. Mycol. 2007, 45, 657–684. [Google Scholar] [CrossRef] [PubMed]
- White, R.L. What In Vitro Models of Infection Can and Cannot Do. Pharmacotherapy 2001, 21, 292S–301S. [Google Scholar] [CrossRef]
- Dalziel, J.E.; Dunstan, K.E.; Finch, S.C. Combined effects of fungal alkaloids on intestinal motility in an in vitro rat model. J. Anim. Sci 2013, 91, 5177–5182. [Google Scholar] [CrossRef]
- Van Dijck, P.; Sjollema, J.; Cammue, B.P.A.; Lagrou, K.; Berman, J.; d’Enfert, C.; Andes, D.R.; Arendrup, M.C.; Brakhage, A.A.; Calderone, R.; et al. Methodologies for in vitro and in vivo evaluation of efficacy of antifungal and antibiofilm agents and surface coatings against fungal biofilms. Microb. Cell 2018, 5, 300–326. [Google Scholar] [CrossRef]
- Holland, D.B.; Bojar, R.A.; Jeremy, A.H.T.; Ingham, E.; Holland, K.T. Microbial colonization of an in vitro model of a tissue engineered human skin equivalent—A novel approach. FEMS Microbiol. Lett. 2008, 279, 110–115. [Google Scholar] [CrossRef]
- Pedrosa, A.F.; Lisboa, C.; Branco, J.; Pellevoisin, C.; Miranda, I.M.; Rodrigues, A.G. Malassezia interaction with a reconstructed human epidermis: Keratinocyte immune response. Mycoses 2019, 62, 932–936. [Google Scholar] [CrossRef]
- Matsumoto, Y.; Sekimizu, K. Silkworm as an experimental animal for research on fungal infections. Microbiol. Immunol. 2019, 63, 41–50. [Google Scholar] [CrossRef]
- Arvanitis, M.; Glavis-Bloom, J.; Mylonakis, E. Invertebrate models of fungal infection. Biochim. Biophys. Acta Mol. Basis Dis. 2013, 1832, 1378–1383. [Google Scholar] [CrossRef] [PubMed]
- Lemaitre, B.; Nicolas, E.; Michaut, L.; Reichhart, J.-M.; Hoffmann, J.A. The dorsoventral regulatory gene cassette spätzle/Toll/cactus controls the potent antifungal response in Drosophila adults. Cell 1996, 86, 973–983. [Google Scholar] [CrossRef]
- Merkel, S.; Heidrich, D.; Danilevicz, C.K.; Scroferneker, M.L.; Zanette, R.A. Drosophila melanogaster as a model for the study of Malassezia pachydermatis infections. Vet. Microbiol. 2018, 224, 31–33. [Google Scholar] [CrossRef]
- Tenor, J.L.; Aballay, A. A conserved Toll-like receptor is required for Caenorhabditis elegans innate immunity. EMBO Rep. 2008, 9, 103–109. [Google Scholar] [CrossRef]
- Trevijano-Contador, N.; Zaragoza, O. Immune response of Galleria mellonella against human fungal pathogens. J. Fungi 2019, 5, 3. [Google Scholar] [CrossRef] [PubMed]
- Wojda, I. Immunity of the greater wax moth Galleria mellonella. Insect Sci. 2017, 24, 342–357. [Google Scholar] [CrossRef] [PubMed]
- Pereira, T.; de Barros, P.; Fugisaki, L.; Rossoni, R.; Ribeiro, F.; de Menezes, R.; Junqueira, J.; Scorzoni, L. Recent advances in the use of Galleria mellonella model to study immune responses against human pathogens. J. Fungi 2018, 4, 128. [Google Scholar] [CrossRef]
- Boguś, M.I.; Ligęza-Żuber, M.; Polańska, M.A.; Mosiewicz, M.; Włóka, E.; Sobocińska, M. Fungal infection causes changes in the number, morphology and spreading ability of Galleria mellonella haemocytes. Physiol. Entomol. 2018, 43, 214–226. [Google Scholar] [CrossRef]
- Urbański, A.; Adamski, Z.; Rosiński, G. Developmental changes in haemocyte morphology in response to Staphylococcus aureus and latex beads in the beetle Tenebrio molitor L. Micron 2018, 104, 8–20. [Google Scholar] [CrossRef]
- Firat, Y.H.; Simanski, M.; Rademacher, F.; Schröder, L.; Brasch, J.; Harder, J. Infection of keratinocytes with Trichophytum rubrum induces epidermal growth factor-dependent RNase 7 and human beta-defensin-3 expression. PLoS ONE 2014, 9, e93941. [Google Scholar] [CrossRef]
- Wollina, U.; Kü, W.; Bulling, L.; Fü Nfstü Ck, C.; Knö, B.; Vennewald, I.; Hipler, U.-C.; Wollina, U. Candida albicans-induced inflammatory response in human keratinocytes. Mycoses 2004, 47, 193–199. [Google Scholar] [CrossRef]
- Lopez, C.M.; Wallich, R.; Riesbeck, K.; Skerka, C.; Zipfel, P.F. Candida albicans uses the surface protein Gpm1 to attach to human endothelial cells and to keratinocytes via the adhesive protein vitronectin. PLoS ONE 2014, 9, e90796. [Google Scholar] [CrossRef] [PubMed]
- Ryu, S.; Choi, S.Y.; Acharya, S.; Chun, Y.J.; Gurley, C.; Park, Y.; Armstrong, C.A.; Song, P.I.; Kim, B.J. Antimicrobial and anti-inflammatory effects of cecropin A(1-8)-magainin2(1-12) hybrid peptide analog P5 against Malassezia furfur infection in human keratinocytes. J. Investig. Dermatol. 2011, 131, 1677–1683. [Google Scholar] [CrossRef]
- Watanabe, S.; Kano, R.; Sato, H.; Nakamura, Y.; Hasegawa, A. The effects of Malassezia yeasts on cytokine production by human keratinocytes. J. Investig. Dermatol. 2001, 116, 769–773. [Google Scholar] [CrossRef] [PubMed]
- Donnarumma, G.; Perfetto, B.; Paoletti, I.; Oliviero, G.; Clavaud, C.; Del Bufalo, A.; Guéniche, A.; Jourdain, R.; Tufano, M.A.; Breton, L. Analysis of the response of human keratinocytes to Malassezia globosa and restricta strains. Arch. Dermatol. Res. 2014, 306, 763–768. [Google Scholar] [CrossRef]
- Ishibashi, Y.; Sugita, T.; Nishikawa, A. Cytokine secretion profile of human keratinocytes exposed to Malassezia yeasts. FEMS Immunol. Med. Microbiol. 2006, 48, 400–409. [Google Scholar] [CrossRef]
- Akaza, N.; Akamatsu, H.; Takeoka, S.; Mizutani, H.; Nakata, S.; Matsunaga, K. Increased hydrophobicity in Malassezia species correlates with increased proinflammatory cytokine expression in human keratinocytes. Med. Mycol. 2012, 87, 802–810. [Google Scholar] [CrossRef] [PubMed]
- Baroni, A.; Perfetto, B.; Paoletti, I.; Ruocco, E.; Canozo, N.; Orlando, M.; Buommino, E. Malassezia furfur invasiveness in a keratinocyte cell line (HaCat): Effects on cytoskeleton and on adhesion molecule and cytokine expression. Arch. Dermatol. Res. 2001, 293, 414–419. [Google Scholar] [CrossRef]
- Buommino, E.; De Filippis, A.; Parisi, A.; Nizza, S.; Martano, M.; Iovane, G.; Donnarumma, G.; Tufano, M.A.; De Martino, L. Innate immune response in human keratinocytes infected by a feline isolate of Malassezia pachydermatis. Vet. Microbiol. 2013, 163, 90–96. [Google Scholar] [CrossRef]
- Buommino, E.; Baroni, A.; Papulino, C.; Nocera, F.P.; Coretti, L.; Donnarumma, G.; De Filippis, A.; De Martino, L. Malassezia pachydermatis up-regulates AhR related CYP1A1 gene and epidermal barrier markers in human keratinocytes. Med. Mycol. 2018, 56, 987–993. [Google Scholar] [CrossRef]
- Thomas, D.S.; Ingham, E.; Bojar, R.A.; Holland, K.T. In vitro modulation of human keratinocyte pro- and anti-inflammatory cytokine production by the capsule of Malassezia species. FEMS Immunol. Med. Microbiol. 2008, 54, 203–214. [Google Scholar] [CrossRef] [PubMed]
- Vallhov, H.; Johansson, C.; Veerman, R.E.; Scheynius, A. Extracellular Vesicles Released from the Skin Commensal Yeast Malassezia sympodialis Activate Human Primary Keratinocytes. Front. Cell. Infect. Microbiol. 2020, 10, 6. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.J.; Han, Y.; Sun, Y.Z.; Jiang, H.H.; Liu, M.; Qi, R.Q.; Gao, X.H. Extracellular vesicles derived from Malassezia furfur stimulate IL-6 production in keratinocytes as demonstrated in in vitro and in vivo models. J. Dermatol. Sci. 2019, 93, 168–175. [Google Scholar] [CrossRef] [PubMed]
- McCarthy, M.W.; Denning, D.W.; Walsh, T.J. Future research priorities in fungal resistance. J. Infect. Dis. 2017, 216, S484–S492. [Google Scholar] [CrossRef] [PubMed]
- Moore, M.; Louis, S.; Pullar, A.; Hardwicke, R. LXXXIII.-Malassezia furfur, the cause of tinea versicolor cultivation of the organism and experimental production of the disease. Arch. Dermatol. Syphilol. 1940, 41, 253–260. [Google Scholar] [CrossRef]
- Zhang, F.; An, Y.; Li, Z.; Zhao, C. A novel model of invasive fungal rhinosinusitis in rats. Am. J. Rhinol. Allergy 2013, 27, 361–366. [Google Scholar] [CrossRef]
- Kurtz, M.B.; Bernard, E.M.; Edwards, F.F.; Marrinan, J.A.; Dropinski, J.; Douglas, C.M.; Armstrong, A.D. Aerosol and Parenteral Pneumocandins Are Effective in a Rat Model of Pulmonary Aspergillosis. Antimicrob. Agents Chemother. 1995, 39, 1784–1789. [Google Scholar] [CrossRef]
- Zhang, H.; Wang, L.; Li, Z.; Liu, S.; Xie, Y.; He, S.; Deng, X.; Yang, B.; Liu, H.; Chen, G.; et al. A novel murine model of Fusarium solani keratitis utilizing fluorescent labeled fungi. Exp. Eye Res. 2013, 110, 107–112. [Google Scholar] [CrossRef]
- Van Cutsem, J.; Van Gerven, F.; Fransen, J.; Schrooten, P.; Janssen, P.A.J. The in vitro antifungal activity of ketoconazole, zinc pyrithione, and selenium sulfide against Pityrosporum and their efficacy as a shampoo in the treatment of experimental pityrosporosis in guinea pigs. J. Am. Acad. Dermatol. 1990, 22, 993–998. [Google Scholar] [CrossRef]
- Koga, H.; Munechika, Y.; Matsumoto, H.; Nanjoh, Y.; Harada, K.; Makimura, K.; Tsuboi, R. Guinea pig seborrheic dermatitis model of Malassezia restricta and the utility of luliconazole. Med. Mycol. 2020, 58, 820–826. [Google Scholar] [CrossRef]
- Uchida, Y.; Mizutani, M.; Kubo, T.; Nakade, T.; Otomo, K. Otitis External Induced with Malassezia pachydermatis in Dogs and the Efficacy of Pimaricin. J. Vet. Med. Sci. 1992, 54, 611–614. [Google Scholar] [CrossRef][Green Version]
- Rosenberg, E.W.; Belew, P.; Bale, G. Effect of topical applications of heavy suspensions of killed malassezia ovalis on rabbit skin. Mycopathologia 1980, 72, 147–154. [Google Scholar] [CrossRef] [PubMed]
- Faergemann, J. Experimental tinea versicolor in rabbits and humans with Pityrosporum orbiculare. J. Investig. Dermatol. 1979, 72, 326–329. [Google Scholar] [CrossRef] [PubMed]
- Faergemann, J.; Fredriksson, T. Experimental Infections in Rabbits and Humans with Pityrosporum orbiculare and P. ovale. J. Investig. Dermatol. 1981, 77, 314–318. [Google Scholar] [CrossRef] [PubMed]
- Groisser, D.; Bottone, E.J.; Lebwohl, M. Association of Pityrosporum orbiculare (Malassezia furfur) with seborrheic dermatitis in patients with acquired immunodeficiency syndrome (AIDS). J. Am. Acad. Dermatol. 1989, 20, 770–773. [Google Scholar] [CrossRef]
- Conant, M.A. The AIDS epidemic. J. Am. Acad. Dermatol. 1994, 31, S47–S50. [Google Scholar] [CrossRef]
- Oble, D.A.; Collett, E.; Hsieh, M.; Ambjrn, M.; Law, J.; Dutz, J.; Teh, H.-S. A Novel T Cell Receptor Transgenic Animal Model of Seborrheic Dermatitis-Like Skin Disease. J. Investig. Dermatol. 2005, 124, 151–159. [Google Scholar] [CrossRef]
- Yamasaki, S.; Matsumoto, M.; Takeuchi, O.; Matsuzawa, T.; Ishikawa, E.; Sakuma, M.; Tateno, H.; Uno, J.; Hirabayashi, J.; Mikami, Y.; et al. C-type lectin Mincle is an activating receptor for pathogenic fungus, Malassezia. Proc. Natl. Acad. Sci. USA 2009, 106, 1897–1902. [Google Scholar] [CrossRef]
- Schlemmer, K.B.; Jesus, F.P.K.; Loreto, É.S.; Tondolo, J.S.M.; Ledur, P.C.; Dallabrida, A.; da Silva, T.M.; Kommers, G.D.; Alves, S.H.; Santurio, J.M. An experimental murine model of otitis and dermatitis caused by Malassezia pachydermatis. Mycoses 2018, 61, 954–958. [Google Scholar] [CrossRef]
- Rosowski, E.E.; Knox, B.P.; Archambault, L.S.; Huttenlocher, A.; Keller, N.P.; Wheeler, R.T.; Davis, J.M. The zebrafish as a model host for invasive fungal infections. J. Fungi 2018, 4, 136. [Google Scholar] [CrossRef]
- Doke, S.K.; Dhawale, S.C. Alternatives to animal testing: A review. Saudi Pharm. J. 2015, 23, 223–229. [Google Scholar] [CrossRef] [PubMed]
- Novohradská, S.; Ferling, I.; Hillmann, F. Exploring virulence determinants of filamentous fungal pathogens through interactions with soil amoebae. Front. Cell. Infect. Microbiol. 2017, 7, 497. [Google Scholar] [CrossRef] [PubMed]
- Johnson, C.H.; Ayyadevara, S.; McEwen, J.E.; Shmookler Reis, R.J. Histoplasma capsulatum and Caenorhabditis elegans: A simple nematode model for an innate immune response to fungal infection. Med. Mycol. 2009, 47, 808–813. [Google Scholar] [CrossRef] [PubMed]
- Mylonakis, E.; Ausubel, F.M.; Perfect, J.R.; Heitman, J.; Calderwood, S.B. Killing of Caenorhabditis elegans by Cryptococcus neoformans as a model of yeast pathogenesis. Proc. Natl. Acad. Sci. USA 2002, 99, 15675–15680. [Google Scholar] [CrossRef]
- Scorzoni, L.; de Lucas, M.P.; de Lacorte Singulani, J.; de Oliveira, H.C.; Assato, P.A.; Fusco-Almeida, A.M.; Mendes-Giannini, M.J.S. Evaluation of Caenorhabditis elegans as a host model for Paracoccidioides brasiliensis and Paracoccidioides lutzii. Pathog. Dis. 2018, 76, fty004. [Google Scholar] [CrossRef]
- Alarco, A.-M.; Marcil, A.; Chen, J.; Suter, B.; Thomas, D.; Whiteway, M. Immune-Deficient Drosophila melanogaster: A Model for the Innate Immune Response to Human Fungal Pathogens. J. Immunol. 2004, 172, 5622–5628. [Google Scholar] [CrossRef]
- Glittenberg, M.T.; Silas, S.; MacCallum, D.M.; Gow, N.A.R.; Ligoxygakis, P. Wild-type Drosophila melanogaster as an alternative model system for investigating the pathogenicity of Candida albicans. DMM Dis. Model. Mech. 2011, 4, 504–514. [Google Scholar] [CrossRef]
- De Souza, P.C.; Caloni, C.C.; Wilson, D.; Almeida, R.S. An invertebrate host to study fungal infections, mycotoxins and antifungal drugs: Tenebrio molitor. J. Fungi 2018, 4, 125. [Google Scholar] [CrossRef]
- Singkum, P.; Suwanmanee, S.; Pumeesat, P.; Luplertlop, N. A powerful in vivo alternative model in scientific research: Galleria mellonella. Acta Microbiol. Immunol. Hung. 2019, 66, 31–55. [Google Scholar] [CrossRef]
- Jemel, S.; Guillot, J.; Kallel, K.; Botterel, F.; Dannaoui, E. Galleria mellonella for the evaluation of antifungal efficacy against medically important fungi, a narrative review. Microorganisms 2020, 8, 390. [Google Scholar] [CrossRef]
- Amorim-Vaz, S.; Delarze, E.; Ischer, F.; Sanglard, D.; Coste, A.T. Examining the virulence of Candida albicans transcription factor mutants using Galleria mellonella and mouse infection models. Front. Microbiol. 2015, 6, 367. [Google Scholar] [CrossRef] [PubMed]
- Cotter, G.; Doyle, S.; Kavanagh, K. Development of an insect model for the in vivo pathogenicity testing of yeasts. FEMS Immunol. Med. Microbiol. 2000, 27, 163–169. [Google Scholar] [CrossRef] [PubMed]
- Sheehan, G.; Tully, L.; Kavanagh, K.A. Candida albicans increases the pathogenicity of Staphylococcus aureus during polymicrobial infection of Galleria mellonella larvae. Microbiology 2020, 166, 375–385. [Google Scholar] [CrossRef]
- Sheehan, G.; Kavanagh, K. Analysis of the early cellular and humoral responses of Galleria mellonella larvae to infection by Candida albicans. Virulence 2018, 9, 163–172. [Google Scholar] [CrossRef] [PubMed]
- Brennan, M.; Thomas, D.Y.; Whiteway, M.; Kavanagh, K. Correlation between virulence of Candida albicans mutants in mice and Galleria mellonella larvae. FEMS Immunol. Med. Microbiol. 2002, 34, 153–157. [Google Scholar] [CrossRef] [PubMed]
- Slater, J.L.; Gregson, L.; Denning, D.W.; Warn, P.A. Pathogenicity of Aspergillus fumigatus mutants assessed in Galleria mellonella matches that in mice. Med. Mycol. 2011, 49, S107–S113. [Google Scholar] [CrossRef]
- Hohl, T.M. Overview of vertebrate animal models of fungal infection. J. Immunol. Methods 2014, 410, 100–112. [Google Scholar] [CrossRef]
- Malavia, D.; Gow, N.A.R.; Usher, J. Advances in molecular tools and in vivo models for the study of human fungal pathogenesis. Microorganisms 2020, 8, 803. [Google Scholar] [CrossRef]
- Singulani, J.L.; Scorzoni, L.; de Oliveira, H.C.; Marcos, C.M.; Assato, P.A.; Fusco-Almeida, A.M.; Mendes-Giannini, M.J.S. Applications of invertebrate animal models to dimorphic fungal infections. J. Fungi 2018, 4, 118. [Google Scholar] [CrossRef]
- Guimaraes, A.J.; Gomes, K.X.; Cortines, J.R.; Peralta, J.M.; Peralta, R.H.S. Acanthamoeba spp. as a universal host for pathogenic microorganisms: One bridge from environment to host virulence. Microbiol. Res. 2016, 193, 30–38. [Google Scholar] [CrossRef]
- Rakus, K.; Adamek, M.; Mojżesz, M.; Podlasz, P.; Chmielewska-Krzesińska, M.; Naumowicz, K.; Kasica-Jarosz, N.; Kłak, K.; Rakers, S.; Way, K.; et al. Evaluation of zebrafish (Danio rerio) as an animal model for the viral infections of fish. J. Fish Dis. 2019, 42, 923–934. [Google Scholar] [CrossRef] [PubMed]
- Avendaño-Herrera, R.; Benavides, I.; Espina, J.A.; Soto-Comte, D.; Poblete-Morales, M.; Valdés, J.A.; Feijóo, C.G.; Reyes, A.E. Zebrafish (Danio rerio) as an animal model for bath infection by Flavobacterium psychrophilum. J. Fish Dis. 2020, 43, 561–570. [Google Scholar] [CrossRef] [PubMed]
- Saraceni, P.R.; Romero, A.; Figueras, A.; Novoa, B. Establishment of infection models in zebrafish larvae (Danio rerio) to study the pathogenesis of Aeromonas hydrophila. Front. Microbiol. 2016, 7, 1219. [Google Scholar] [CrossRef] [PubMed]
- Chao, C.C.; Hsu, P.C.; Jen, C.F.; Chen, I.H.; Wang, C.H.; Chan, H.C.; Tsai, P.W.; Tung, K.C.; Wang, C.H.; Lan, C.Y.; et al. Zebrafish as a model host for Candida albicans infection. Infect. Immun. 2010, 78, 2512–2521. [Google Scholar] [CrossRef]
- Davis, J.M.; Huang, M.; Botts, M.R.; Hull, C.M.; Huttenlocher, A. A zebrafish model of cryptococcal infection reveals roles for macrophages, endothelial cells, and neutrophils in the establishment and control of sustained fungemia. Infect. Immun. 2016, 84, 3047–3062. [Google Scholar] [CrossRef]
- Sabiiti, W.; May, R.C.; Pursall, E.R. Experimental models of cryptococcosis. Int. J. Microbiol. 2012, 2012, 626745. [Google Scholar] [CrossRef]
- Powell, J.R.; Ausubel, F.M. Models of Caenorhabditis elegans Infection by Bacterial and Fungal Pathogens. In Innate Immunity. Methods in Molecular BiologyTM; Ewbank, J., Vivier, E., Eds.; Humana Press: Totowa, NJ, USA, 2008; pp. 403–427. [Google Scholar] [CrossRef]
- Matsumoto, Y.; Sumiya, E.; Sugita, T.; Sekimizu, K. An invertebrate hyperglycemic model for the identification of anti-diabetic drugs. PLoS ONE 2011, 6, e18292. [Google Scholar] [CrossRef]
- Brunke, S.; Quintin, J.; Kasper, L.; Jacobsen, I.D.; Richter, M.E.; Hiller, E.; Schwarzmüller, T.; D’Enfert, C.; Kuchler, K.; Rupp, S.; et al. Of mice, flies—And men? Comparing fungal infection models for large-scale screening efforts. DMM Dis. Model. Mech. 2015, 8, 473–486. [Google Scholar] [CrossRef]
- Fallon, J.; Kelly, J.; Kavanagh, K. Galleria mellonella as a model for fungal pathogenicity testing. In Host-Fungus Interactions: Methods and Protocols; Brand, A., MacCallum, D., Eds.; Humana Press: Totowa, NJ, USA, 2012; Volume 845, pp. 469–485. ISBN 9781617795381. [Google Scholar] [CrossRef]
- Fuchs, B.B.; O’Brien, E.; El Khoury, J.B.; Mylonakis, E. Methods for using Galleria mellonella as a model host to study fungal pathogenesis. Virulence 2010, 1, 475–482. [Google Scholar] [CrossRef]
- Scorzoni, L.; De Paula E Silva, A.C.A.; De Oliveira, H.C.; Marcos, C.M.; De Lacorte Singulani, J.; Fusco-Almeida, A.M.; Mendes-Giannini, M.J.S. Can passage in Galleria mellonella activate virulence factors of Paracoccidioides brasiliensis as in the murine model? Med. Mycol. 2018, 56, 374–377. [Google Scholar] [CrossRef]
- Benaducci, T.; Sardi, J.d.C.O.; Lourencetti, N.M.S.; Scorzoni, L.; Gullo, F.P.; Rossi, S.A.; Derissi, J.B.; de Azevedo Prata, M.C.; Fusco-Almeida, A.M.; Mendes-Giannini, M.J.S. Virulence of Cryptococcus sp. biofilms in vitro and in vivo using Galleria mellonella as an alternative model. Front. Microbiol. 2016, 7, 290. [Google Scholar] [CrossRef] [PubMed]
- Brilhante, R.S.N.; da Rocha, M.G.; de Melo Guedes, G.M.; de Oliveira, J.S.; dos Santos Araújo, G.; España, J.D.A.; Sales, J.A.; de Aguiar, L.; de Araújo Neto Paiva, M.; de Aguiar Cordeiro, R.; et al. Malassezia pachydermatis from animals: Planktonic and biofilm antifungal susceptibility and its virulence arsenal. Vet. Microbiol. 2018, 220, 47–52. [Google Scholar] [CrossRef] [PubMed]
- Silva Rabelo, A.P.; Valério, A.; Viana, R.O.; Ricoy, A.C.D.S.; Johann, S.; Alves, V.D.S. Caenorhabditis Elegans and Tenebrio Molitor—New Tools to Investigate Malassezia Species. Preprints 2018, 2018100001. [Google Scholar] [CrossRef]
- Thomaz, L.; García-Rodas, R.; Guimarães, A.J.; Taborda, C.P.; Zaragoza, O.; Nosanchuk, J.D. Galleria mellonella as a model host to study Paracoccidioides Lutzii and Histoplasma Capsulatum. Virulence 2013, 4, 139–146. [Google Scholar] [CrossRef] [PubMed]
- Coleman, J.J.; Muhammed, M.; Kasperkovitz, P.V.; Vyas, J.M.; Mylonakis, E. Fusarium pathogenesis investigated using Galleria mellonella as a heterologous host. Fungal Biol. 2011, 115, 1279–1289. [Google Scholar] [CrossRef] [PubMed]
- Kloezen, W.; van Helvert-van Poppel, M.; Fahal, A.H.; van de Sande, W.W.J. A Madurella mycetomatis grain model in Galleria mellonella larvae. PLoS Negl. Trop. Dis. 2015, 9, e0003926. [Google Scholar] [CrossRef]
- Scorzoni, L.; de Lucas, M.P.; Mesa-Arango, A.C.; Fusco-Almeida, A.M.; Lozano, E.; Cuenca-Estrella, M.; Mendes-Giannini, M.J.; Zaragoza, O. Antifungal Efficacy during Candida krusei Infection in Non-Conventional Models Correlates with the Yeast In Vitro Susceptibility Profile. PLoS ONE 2013, 8, e60047. [Google Scholar] [CrossRef]
- Sheehan, G.; Kavanagh, K. Proteomic analysis of the responses of Candida albicans during infection of Galleria mellonella larvae. J. Fungi 2019, 5, 7. [Google Scholar] [CrossRef]
- Mowlds, P.; Coates, C.; Renwick, J.; Kavanagh, K. Dose-dependent cellular and humoral responses in Galleria mellonella larvae following β-glucan inoculation. Microbes Infect. 2010, 12, 146–153. [Google Scholar] [CrossRef]
- Fallon, J.P.; Troy, N.; Kavanagh, K. Pre-exposure of Galleria mellonella larvae to different doses of Aspergillus fumigatus conidia causes differential activation of cellular and humoral immune responses. Virulence 2011, 2, 413–421. [Google Scholar] [CrossRef]
- Bergin, D.; Murphy, L.; Keenan, J.; Clynes, M.; Kavanagh, K. Pre-exposure to yeast protects larvae of Galleria mellonella from a subsequent lethal infection by Candida albicans and is mediated by the increased expression of antimicrobial peptides. Microbes Infect. 2006, 8, 2105–2112. [Google Scholar] [CrossRef] [PubMed]
- Mesa-Arango, A.C.; Forastiero, A.; Bernal-Martínez, L.; Cuenca-Estrella, M.; Mellado, E.; Zaragoza, O. The non-mammalian host Galleria mellonella can be used to study the virulence of the fungal pathogen Candida tropicalis and the efficacy of antifungal drugs during infection by this pathogenic yeast. Med. Mycol. 2013, 51, 461–472. [Google Scholar] [CrossRef] [PubMed]
- De Lacorte Singulani, J.; Scorzoni, L.; de Paula e Silva, A.C.A.; Fusco-Almeida, A.M.; Mendes-Giannini, M.J.S. Evaluation of the efficacy of antifungal drugs against Paracoccidioides brasiliensis and Paracoccidioides lutzii in a Galleria mellonella model. Int. J. Antimicrob. Agents 2016, 48, 292–297. [Google Scholar] [CrossRef] [PubMed]
- Astvad, K.M.T.; Meletiadis, J.; Whalley, S.; Arendrup, M.C. Fluconazole pharmacokinetics in Galleria mellonella larvae and performance evaluation of a bioassay compared to liquid chromatography-tandem mass spectrometry for hemolymph specimens. Antimicrob. Agents Chemother. 2017, 61, 1–8. [Google Scholar] [CrossRef]
- Vogel, H.; Altincicek, B.; Glöckner, G.; Vilcinskas, A. A comprehensive transcriptome and immune-gene repertoire of the lepidopteran model host Galleria mellonella. BMC Genom. 2011, 12, 308. [Google Scholar] [CrossRef]
- Mukherjee, K.; Vilcinskas, A. Development and immunity-related microRNAs of the lepidopteran model host Galleria mellonella. BMC Genom. 2014, 15, 1–12. [Google Scholar] [CrossRef]
- Torres, M.; Pinzón, E.N.; Rey, F.M.; Martinez, H.; Parra Giraldo, C.M.; Celis Ramírez, A.M. Galleria mellonella as a Novelty in vivo Model of Host-Pathogen Interaction for Malassezia furfur CBS 1878 and Malassezia pachydermatis CBS 1879. Front. Cell. Infect. Microbiol. 2020, 10, 199. [Google Scholar] [CrossRef]
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Torres, M.; de Cock, H.; Celis Ramírez, A.M. In Vitro or In Vivo Models, the Next Frontier for Unraveling Interactions between Malassezia spp. and Hosts. How Much Do We Know? J. Fungi 2020, 6, 155. https://doi.org/10.3390/jof6030155
Torres M, de Cock H, Celis Ramírez AM. In Vitro or In Vivo Models, the Next Frontier for Unraveling Interactions between Malassezia spp. and Hosts. How Much Do We Know? Journal of Fungi. 2020; 6(3):155. https://doi.org/10.3390/jof6030155
Chicago/Turabian StyleTorres, Maritza, Hans de Cock, and Adriana Marcela Celis Ramírez. 2020. "In Vitro or In Vivo Models, the Next Frontier for Unraveling Interactions between Malassezia spp. and Hosts. How Much Do We Know?" Journal of Fungi 6, no. 3: 155. https://doi.org/10.3390/jof6030155
APA StyleTorres, M., de Cock, H., & Celis Ramírez, A. M. (2020). In Vitro or In Vivo Models, the Next Frontier for Unraveling Interactions between Malassezia spp. and Hosts. How Much Do We Know? Journal of Fungi, 6(3), 155. https://doi.org/10.3390/jof6030155






