The Role of RodA-Conserved Cysteine Residues in the Aspergillus fumigatus Conidial Surface Organization
Abstract
1. Introduction
2. Materials and Methods
3. Results
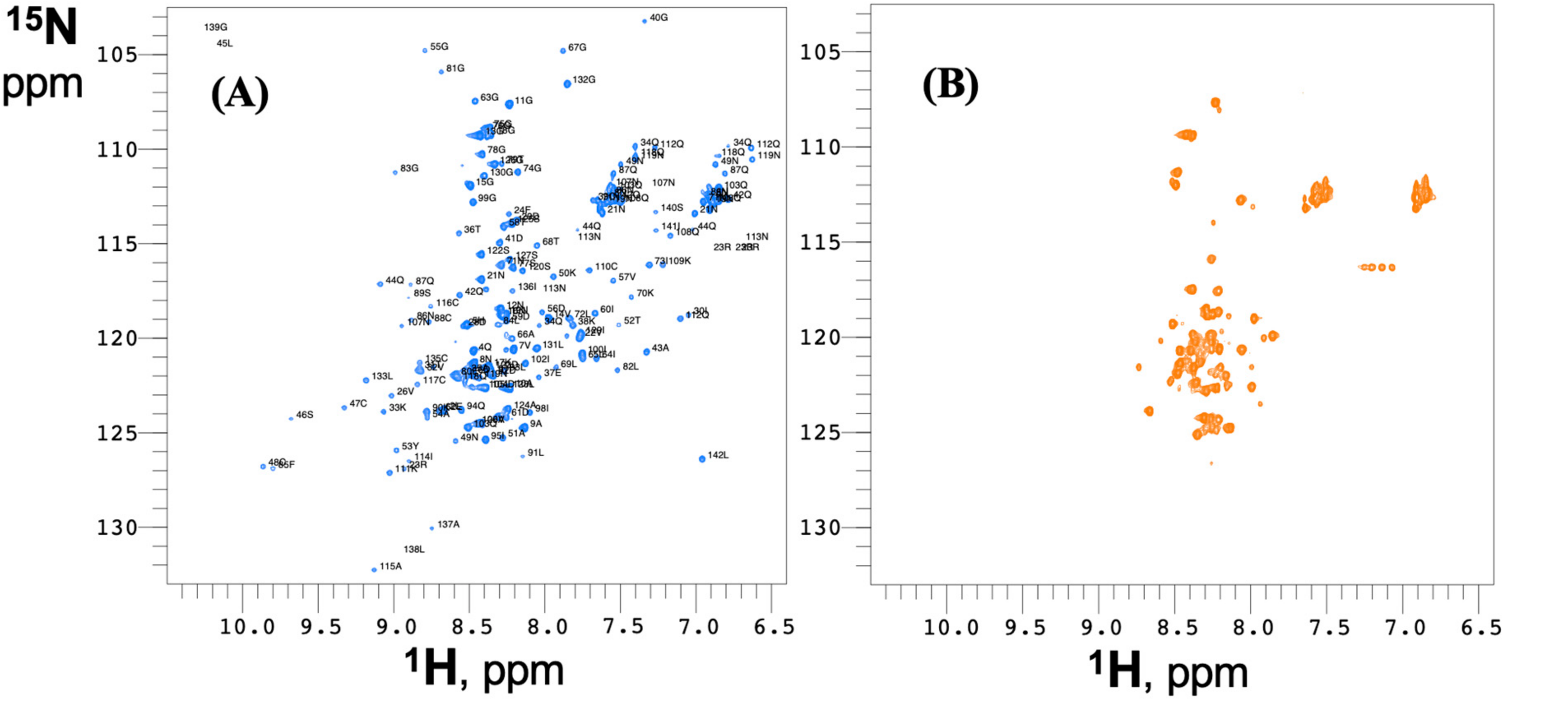
3.1. Disulfide Bonds are Necessary for the Structure of the Spontaneously Self-Assembling RodAp
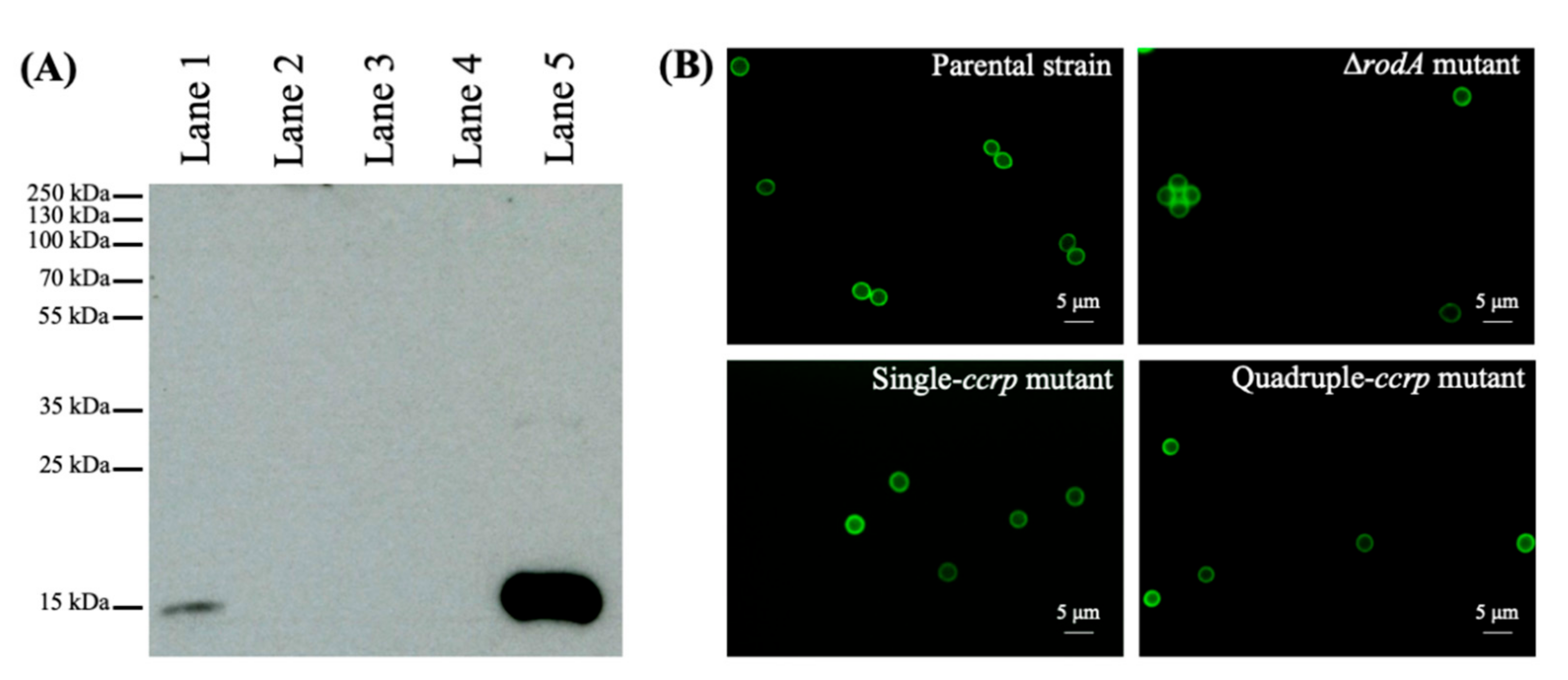
3.2. Point-Mutations of the Conserved Cysteine Residues of RODA Resulted in the Lack of the Rodlet Layer on the Conidial Surface
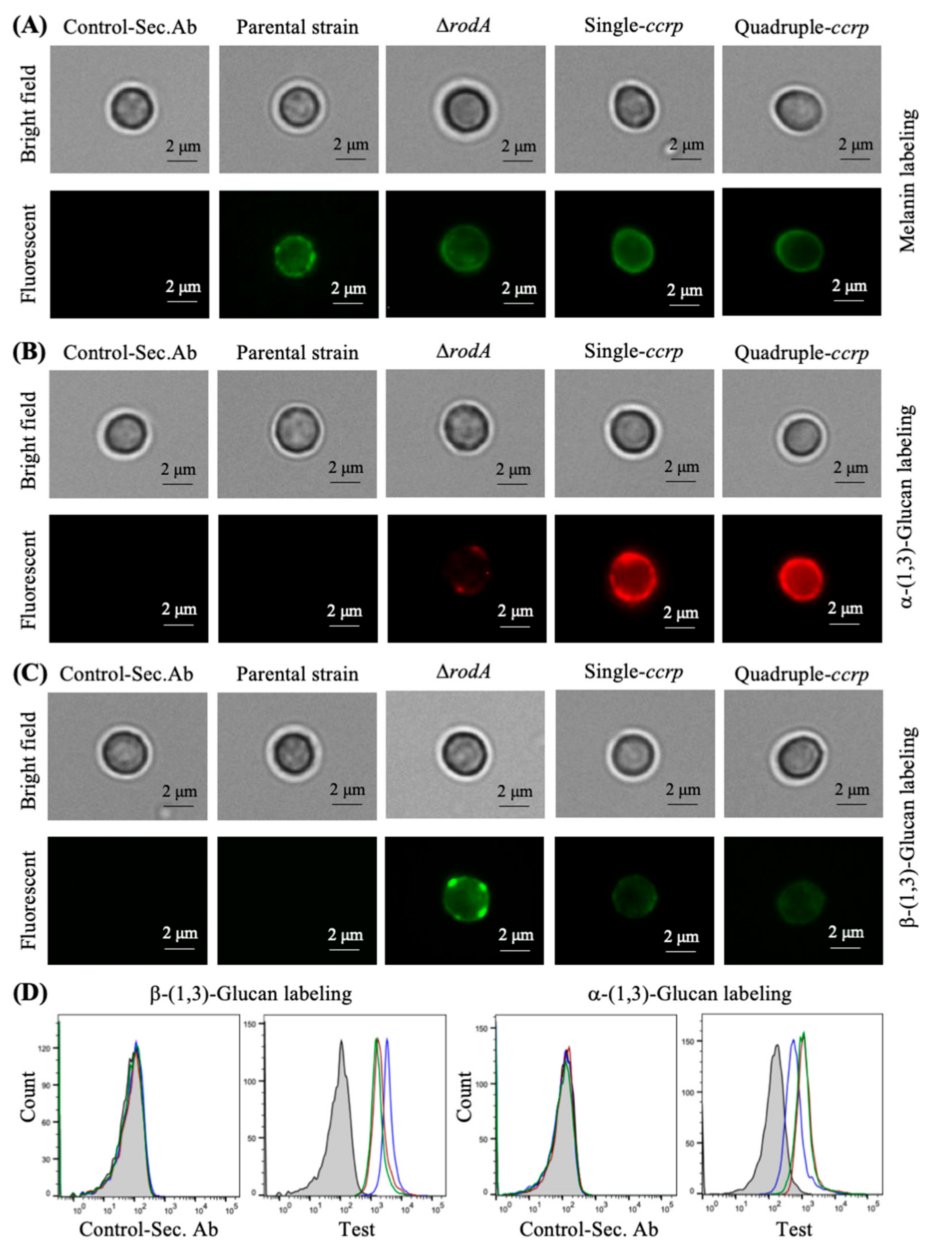
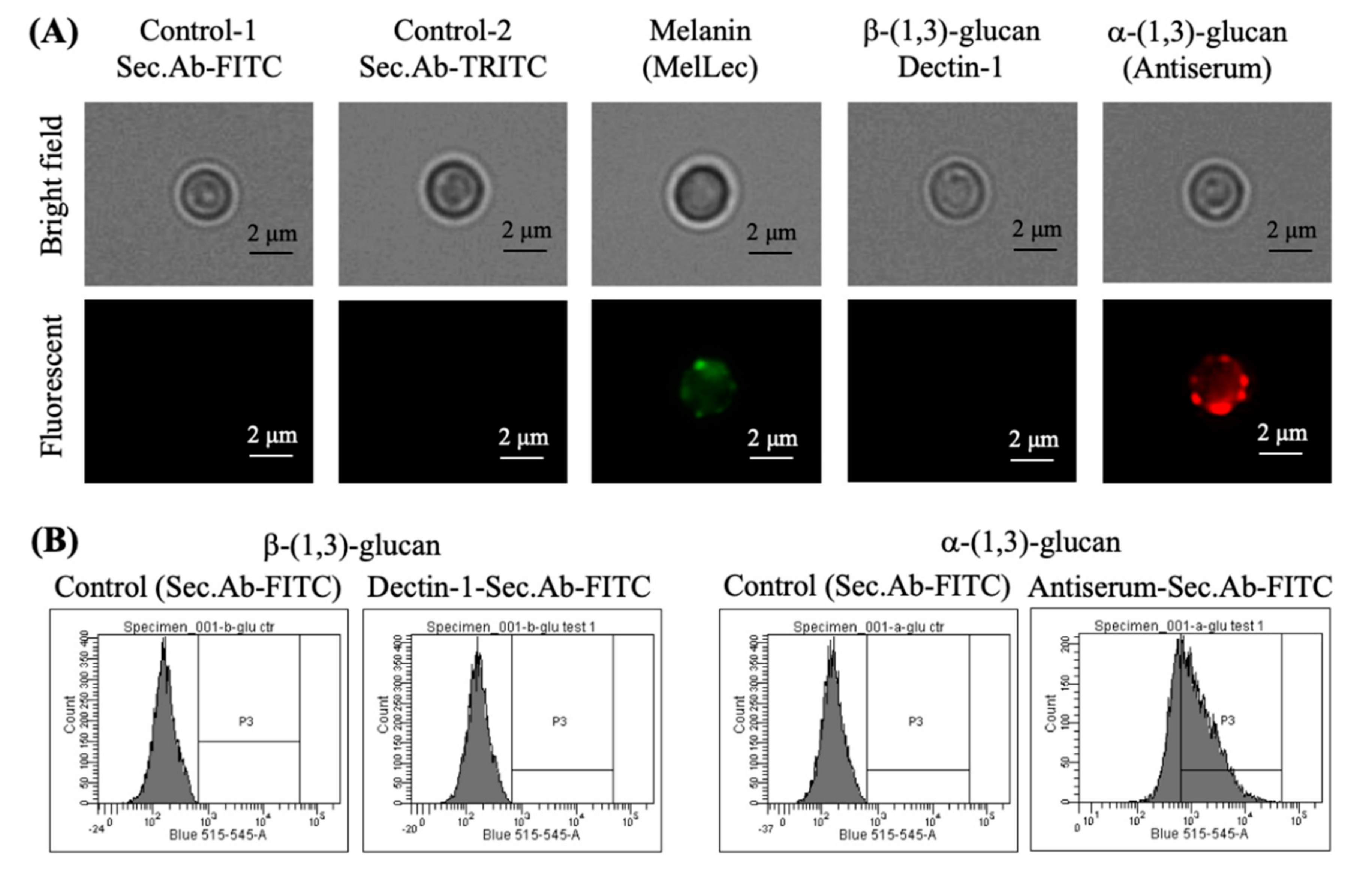
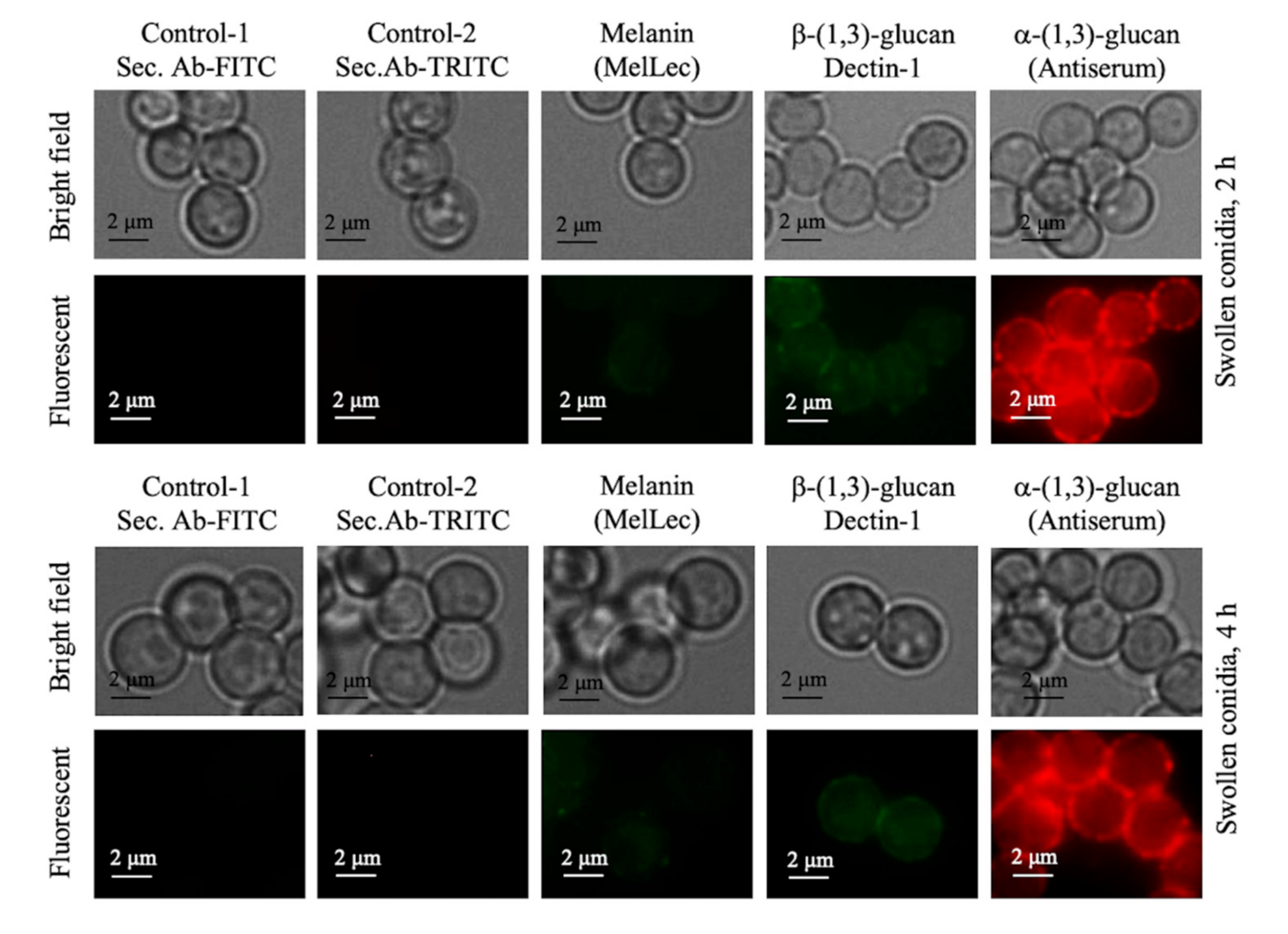
3.3. Absence of Rodlets Exposes α-(1,3)-Glucan on the Conidial Surface
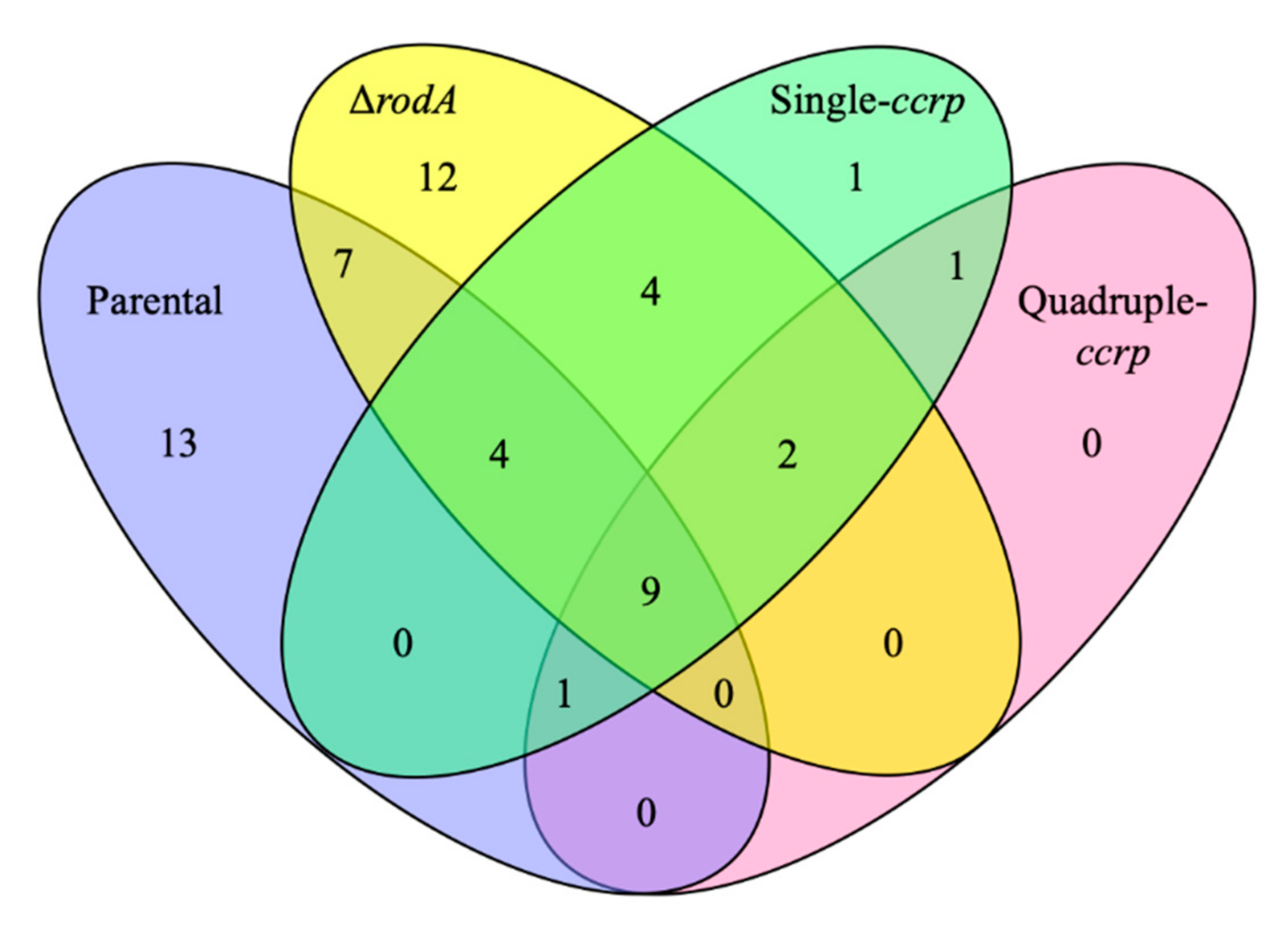
3.4. The Proteins Exposed on the Ccrp-Mutant Conidial Surface are More Similar to Those of the Parental Strain
3.5. Ccrp-Mutant Conidia Are Less Stimulatory than ∆rodA Conidia
3.6. Chemical Removal of the Rodlet Layer Results in the Exposure of α-(1,3)-Glucan on the Conidial Surface
3.7. Cell Wall Components Exposed on the Ccrp-Mutant and Swollen Conidia Are Similar
4. Discussions
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Appendix A
| Accession | Protein | Parental | ΔrodA | Single-ccrp Mutant | Quadruple-ccrp Mutant |
|---|---|---|---|---|---|
| Afu2g05635 | Afu2g05635 | 115 ± 28 | 71 ± 15 | 24 ± 21 | 0 |
| Afu5g09580 | RodA | 160 ± 112 | 0 | 36 ± 13 | 31 ± 7 |
| Afu6g14470 | Afu6g14470 | 55 ± 31 | 44 ± 10 | 15 ± 13 | 8 ± 2 |
| Afu4g09280 | Afu4g09280 | 38 ± 37 | 17 ± 12 | 7 ± 3 | 7 ± 3 |
| Afu4g09310 | Afu4g09310 | 40 ± 28 | 34 ± 26 | 34 ± 10 | 15 ± 3 |
| Afu4g09600 | Afu4g09600 | 132 ± 126 | 7 ± 7 | 0 | 0 |
| Afu1g13670 | CcpA | 33 ± 43 | 8 ± 9 | 0 | 0 |
| Afu6g03210 | ConJ | 10 ± 4 | 39 ± 26 | 20 ± 24 | 5 ± 2 |
| Afu2g09030 | DppV | 88 ± 95 | 104 ± 84 | 38 ± 16 | 23 ± 14 |
| Afu8g01980 | Afu8g01980 | 47 ± 57 | 21 ± 19 | 2 ± 2 | 0 |
| Afu2g00680 | Afu2g00680 | 35 ± 40 | 0 | 0 | 0 |
| Afu1g14450 | ExgO | 82 ± 67 | 35 ± 12 | 19 ± 20 | 2 ± 4 |
| Afu2g12630 | Aspf13 | 12 ± 14 | 0 | 0 | 0 |
| Afu2g17530 | Abr2 | 45 ± 49 | 14 ± 17 | 0 | 0 |
| Afu4g01030 | Afu4g01030 | 7 ± 8 | 0 | 0 | 0 |
| Afu7g01200 | Afu7g01200 | 19 ± 28 | 0 | 0 | 0 |
| Afu1g14560 | MsdS | 31 ± 43 | 57 ± 29 | 22 ± 20 | 10 ± 4 |
| Afu4g12450 | Afu4g12450 | 25 ± 36 | 7 ± 6 | 0 | 0 |
| Afu1g07440 | Hsp70 | 29 ± 6 | 33 ± 14 | 14 ± 8 | 5 ± 3 |
| Afu8g05410 | Afu8g05410 | 8 ± 11 | 0 | 0 | 0 |
| Afu3g15090 | Afu3g15090 | 21 ± 24 | 0 | 0 | 0 |
| Afu1g04670 | Afu1g04670 | 29 ± 26 | 0 | 0 | 0 |
| Afu5g11570 | Afu5g11570 | 17 ± 10 | 0 | 0 | 0 |
| Afu2g10670 | Afu2g10670 | 7 ± 8 | 3 ± 2 | 0 | 0 |
| Afu6g12070 | FmqD | 15 ± 21 | 11 ± 13 | 4 ± 4 | 0 |
| Afu2g03590 | Afu2g03590 | 2 ± 2 | 0 | 0 | 0 |
| Afu1g13195 | Afu1g13195 | 2 ± 2 | 3 ± 1 | 2 ± 2 | 0 |
| Afu6g07910 | Afu6g07910 | 10 ± 15 | 0 | 0 | 0 |
| Afu1g10790 | Afu1g10790 | 16 ± 23 | 5 ± 3 | 0 | 0 |
| Afu3g02270 | Cat1 | 13 ± 14 | 30 ± 10 | 9 ± 7 | 8 ± 4 |
| Afu1g10150 | Afu1g10150 | 5 ± 4 | 0 | 0 | 0 |
| Afu5g13920 | Wos2 | 3 ± 3 | 0 | 0 | 0 |
| Afu5g08020 | Afu5g08020 | 3 ± 3 | 0 | 0 | 0 |
| Afu4g06910 | Afu4g06910 | 2 ± 2 | 2 ± 3 | 0 | 0 |
| Afu3g06880 | SnxA | 0 | 19 ± 10 | 9 ± 3 | 8 ± 5 |
| Afu5g09530 | Afu5g09530 | 0 | 0 | 4 ± 4 | 2 ± 2 |
| Afu4g09320 | DppIV | 0 | 23 ± 21 | 4 ± 2 | 3 ± 3 |
| Afu5g14210 | Grg1 | 0 | 12 ± 8 | 6 ± 7 | 0 |
| Afu4g00860 | DprA | 0 | 19 ± 11 | 10 ± 13 | 0 |
| Afu5g12210 | SfaD | 0 | 0 | 1 ± 2 | 0 |
| Afu2g03980 | Afu2g03980 | 0 | 2 ± 2 | 2 ± 2 | 0 |
| Afu6g09980 | Ncr1 | 0 | 7 ± 6 | 3 ± 3 | 0 |
| Afu1g17370 | Scf1 | 0 | 8 ± 1 | 0 | 0 |
| Afu2g01050 | Afu2g01050 | 0 | 3 ± 2 | 0 | 0 |
| Afu5g03540 | Afu5g03540 | 0 | 13 ± 10 | 0 | 0 |
| Afu5g10570 | Afu5g10570 | 0 | 4 ± 5 | 0 | 0 |
| Afu1g05770 | Exg12 | 0 | 24 ± 21 | 0 | 0 |
| Afu8g05020 | NagA | 0 | 14 ± 10 | 0 | 0 |
| Afu6g13300 | Afu6g13300 | 0 | 3 ± 3 | 0 | 0 |
| Afu8g07060 | RodC | 0 | 2 ± 2 | 0 | 0 |
| Afu2g00967 | Afu2g00967 | 0 | 2 ± 2 | 0 | 0 |
| Afu3g07160 | Afu3g07160 | 0 | 4 ± 3 | 0 | 0 |
| Afu8g05320 | Afu8g05320 | 0 | 5 ± 3 | 0 | 0 |
| Afu1g06470 | Afu1g06470 | 0 | 6 ± 6 | 0 | 0 |
References
- Aimanianda, V.; Bayry, J.; Bozza, S.; Kniemeyer, O.; Perruccio, K.; Elluru, S.R.; Clavaud, C.; Paris, S.; Brakhage, A.A.; Kaveri, S.V.; et al. Surface hydrophobin prevents immune recognition of airborne fungal spores. Nature 2009, 460, 1117–1121. [Google Scholar] [CrossRef] [PubMed]
- Bertuzzi, M.; Hayes, G.E.; Icheoku, U.J.; van Rhijn, N.; Denning, D.W.; Osherov, N.; Bignell, E.M. Anti-aspergillus activities of the respiratory epithelium in health and disease. J. Fungi 2018, 4, 8. [Google Scholar] [CrossRef] [PubMed]
- van de Veerdonk, F.L.; Gresnigt, M.S.; Romani, L.; Netea, M.G.; Latge, J.P. Aspergillus fumigatus morphology and dynamic host interactions. Nat. Rev. Microbiol. 2017, 15, 661–674. [Google Scholar] [CrossRef]
- Hohl, T.M.; Van Epps, H.L.; Rivera, A.; Morgan, L.A.; Chen, P.L.; Feldmesser, M.; Pamer, E.G. Aspergillus fumigatus triggers inflammatory responses by stage-specific beta-glucan display. PLoS Pathog. 2005, 1, e30. [Google Scholar] [CrossRef] [PubMed]
- Slesiona, S.; Gressler, M.; Mihlan, M.; Zaehle, C.; Schaller, M.; Barz, D.; Hube, B.; Jacobsen, I.D.; Brock, M. Persistence versus escape: Aspergillus terreus and Aspergillus fumigatus employ different strategies during interactions with macrophages. PLoS ONE 2012, 7, e31223. [Google Scholar] [CrossRef]
- Bayry, J.; Aimanianda, V.; Guijarro, J.I.; Sunde, M.; Latge, J.P. Hydrophobins—Unique fungal proteins. PLoS Pathog. 2012, 8, e1002700. [Google Scholar] [CrossRef]
- Pille, A.; Kwan, A.H.; Cheung, I.; Hampsey, M.; Aimanianda, V.; Delepierre, M.; Latge, J.P.; Sunde, M.; Guijarro, J.I. (1)H, (13)C and (15)N resonance assignments of the RodA hydrophobin from the opportunistic pathogen Aspergillus fumigatus. Biomol. NMR Assign. 2015, 9, 113–118. [Google Scholar] [CrossRef]
- Jensen, B.G.; Andersen, M.R.; Pedersen, M.H.; Frisvad, J.C.; Sondergaard, I. Hydrophobins from Aspergillus species cannot be clearly divided into two classes. BMC Res. Notes 2010, 3, 344. [Google Scholar] [CrossRef]
- Valsecchi, I.; Lai, J.I.; Stephen-Victor, E.; Pille, A.; Beaussart, A.; Lo, V.; Pham, C.L.L.; Aimanianda, V.; Kwan, A.H.; Duchateau, M.; et al. Assembly and disassembly of aspergillus fumigatus conidial rodlets. Cell Surf. 2019, 5, 100023. [Google Scholar] [CrossRef]
- da Silva Ferreira, M.E.; Kress, M.R.; Savoldi, M.; Goldman, M.H.; Hartl, A.; Heinekamp, T.; Brakhage, A.A.; Goldman, G.H. The akuB(KU80) mutant deficient for nonhomologous end joining is a powerful tool for analyzing pathogenicity in Aspergillus fumigatus. Eukaryot Cell 2006, 5, 207–211. [Google Scholar] [CrossRef]
- Thau, N.; Monod, M.; Crestani, B.; Rolland, C.; Tronchin, G.; Latge, J.P.; Paris, S. Rodletless mutants of Aspergillus fumigatus. Infect. Immun. 1994, 62, 4380–4388. [Google Scholar] [CrossRef]
- Rosas, A.L.; Nosanchuk, J.D.; Gomez, B.L.; Edens, W.A.; Henson, J.M.; Casadevall, A. Isolation and serological analyses of fungal melanins. J. Immunol. Methods 2000, 244, 69–80. [Google Scholar] [CrossRef]
- Stappers, M.H.T.; Clark, A.E.; Aimanianda, V.; Bidula, S.; Reid, D.M.; Asamaphan, P.; Hardison, S.E.; Dambuza, I.M.; Valsecchi, I.; Kerscher, B.; et al. Recognition of DHN-melanin by a C-type lectin receptor is required for immunity to Aspergillus. Nature 2018, 555, 382–386. [Google Scholar] [CrossRef] [PubMed]
- Komarova, B.S.; Orekhova, M.V.; Tsvetkov, Y.E.; Beau, R.; Aimanianda, V.; Latge, J.P.; Nifantiev, N.E. Synthesis of a pentasaccharide and neoglycoconjugates related to fungal alpha-(1–>3)-glucan and their use in the generation of antibodies to trace Aspergillus fumigatus cell wall. Chemistry 2015, 21, 1029–1035. [Google Scholar] [CrossRef] [PubMed]
- Voltersen, V.; Blango, M.G.; Herrmann, S.; Schmidt, F.; Heinekamp, T.; Strassburger, M.; Kruger, T.; Bacher, P.; Lother, J.; Weiss, E.; et al. Proteome analysis reveals the conidial surface protein ccpa essential for virulence of the pathogenic fungus aspergillus fumigatus. mBio 2018, 9. [Google Scholar] [CrossRef] [PubMed]
- Blango, M.G.; Pschibul, A.; Rivieccio, F.; Kruger, T.; Rafiq, M.; Jia, L.J.; Zheng, T.; Goldmann, M.; Voltersen, V.; Li, J.; et al. Dynamic surface proteomes of allergenic fungal conidia. J. Proteome Res. 2020, 19, 2092–2104. [Google Scholar] [CrossRef]
- Brakhage, A.A.; Van den Brulle, J. Use of reporter genes to identify recessive trans-acting mutations specifically involved in the regulation of Aspergillus nidulans penicillin biosynthesis genes. J. Bacteriol. 1995, 177, 2781–2788. [Google Scholar] [CrossRef]
- Maerker, C.; Rohde, M.; Brakhage, A.A.; Brock, M. Methylcitrate synthase from Aspergillus fumigatus. Propionyl-CoA affects polyketide synthesis, growth and morphology of conidia. FEBS J. 2005, 272, 3615–3630. [Google Scholar] [CrossRef]
- Perez-Riverol, Y.; Csordas, A.; Bai, J.; Bernal-Llinares, M.; Hewapathirana, S.; Kundu, D.J.; Inuganti, A.; Griss, J.; Mayer, G.; Eisenacher, M.; et al. The PRIDE database and related tools and resources in 2019: Improving support for quantification data. Nucleic Acids Res. 2019, 47, D442–D450. [Google Scholar] [CrossRef]
- Holla, S.; Sharma, M.; Vani, J.; Kaveri, S.V.; Balaji, K.N.; Bayry, J. GM-CSF along with IL-4 but not alone is indispensable for the differentiation of human dendritic cells from monocytes. J. Allergy Clin. Immunol. 2014, 133, 1500–1502.e1. [Google Scholar] [CrossRef]
- Stephen-Victor, E.; Karnam, A.; Fontaine, T.; Beauvais, A.; Das, M.; Hegde, P.; Prakhar, P.; Holla, S.; Balaji, K.N.; Kaveri, S.V.; et al. Aspergillus fumigatus cell wall alpha-(1,3)-glucan stimulates regulatory T-Cell polarization by Inducing PD-L1 expression on human dendritic cells. J. Infect. Dis. 2017, 216, 1281–1294. [Google Scholar] [CrossRef] [PubMed]
- Steele, C.; Rapaka, R.R.; Metz, A.; Pop, S.M.; Williams, D.L.; Gordon, S.; Kolls, J.K.; Brown, G.D. The beta-glucan receptor dectin-1 recognizes specific morphologies of Aspergillus fumigatus. PLoS Pathog. 2005, 1, e42. [Google Scholar] [CrossRef] [PubMed]
- Pihet, M.; Vandeputte, P.; Tronchin, G.; Renier, G.; Saulnier, P.; Georgeault, S.; Mallet, R.; Chabasse, D.; Symoens, F.; Bouchara, J.P. Melanin is an essential component for the integrity of the cell wall of Aspergillus fumigatus conidia. BMC Microbiol. 2009, 9, 177. [Google Scholar] [CrossRef] [PubMed]
- Bozza, S.; Clavaud, C.; Giovannini, G.; Fontaine, T.; Beauvais, A.; Sarfati, J.; D’Angelo, C.; Perruccio, K.; Bonifazi, P.; Zagarella, S.; et al. Immune sensing of Aspergillus fumigatus proteins, glycolipids, and polysaccharides and the impact on Th immunity and vaccination. J. Immunol. 2009, 183, 2407–2414. [Google Scholar] [CrossRef] [PubMed]
- O’Brien, X.M.; Heflin, K.E.; Lavigne, L.M.; Yu, K.; Kim, M.; Salomon, A.R.; Reichner, J.S. Lectin site ligation of CR3 induces conformational changes and signaling. J. Biol. Chem. 2012, 287, 3337–3348. [Google Scholar] [CrossRef]
- Fontaine, T.; Beauvais, A.; Loussert, C.; Thevenard, B.; Fulgsang, C.C.; Ohno, N.; Clavaud, C.; Prevost, M.C.; Latge, J.P. Cell wall alpha1-3glucans induce the aggregation of germinating conidia of Aspergillus fumigatus. Fungal Genet. Biol. 2010, 47, 707–712. [Google Scholar] [CrossRef]
- Beauvais, A.; Bozza, S.; Kniemeyer, O.; Formosa, C.; Balloy, V.; Henry, C.; Roberson, R.W.; Dague, E.; Chignard, M.; Brakhage, A.A.; et al. Deletion of the alpha-(1,3)-glucan synthase genes induces a restructuring of the conidial cell wall responsible for the avirulence of Aspergillus fumigatus. PLoS Pathog. 2013, 9, e1003716. [Google Scholar] [CrossRef]
- Dichtl, K.; Samantaray, S.; Aimanianda, V.; Zhu, Z.; Prevost, M.C.; Latge, J.P.; Ebel, F.; Wagener, J. Aspergillus fumigatus devoid of cell wall beta-1,3-glucan is viable, massively sheds galactomannan and is killed by septum formation inhibitors. Mol. Microbiol. 2015, 95, 458–471. [Google Scholar] [CrossRef]
- Henry, C.; Fontaine, T.; Heddergott, C.; Robinet, P.; Aimanianda, V.; Beau, R.; Beauvais, A.; Mouyna, I.; Prevost, M.C.; Fekkar, A.; et al. Biosynthesis of cell wall mannan in the conidium and the mycelium of Aspergillus fumigatus. Cell Microbiol. 2016, 18, 1881–1891. [Google Scholar] [CrossRef]
- Herrero, A.B.; Magnelli, P.; Mansour, M.K.; Levitz, S.M.; Bussey, H.; Abeijon, C. KRE5 gene null mutant strains of Candida albicans are avirulent and have altered cell wall composition and hypha formation properties. Eukaryot. Cell 2004, 3, 1423–1432. [Google Scholar] [CrossRef]
- Gilbert, N.M.; Donlin, M.J.; Gerik, K.J.; Specht, C.A.; Djordjevic, J.T.; Wilson, C.F.; Sorrell, T.C.; Lodge, J.K. KRE genes are required for beta-1,6-glucan synthesis, maintenance of capsule architecture and cell wall protein anchoring in Cryptococcus neoformans. Mol. Microbiol. 2010, 76, 517–534. [Google Scholar] [CrossRef] [PubMed]
- Dennehy, K.M.; Willment, J.A.; Williams, D.L.; Brown, G.D. Reciprocal regulation of IL-23 and IL-12 following co-activation of Dectin-1 and TLR signaling pathways. Eur. J. Immunol. 2009, 39, 1379–1386. [Google Scholar] [CrossRef] [PubMed]
- Kang, X.; Kirui, A.; Muszynski, A.; Widanage, M.C.D.; Chen, A.; Azadi, P.; Wang, P.; Mentink-Vigier, F.; Wang, T. Molecular architecture of fungal cell walls revealed by solid-state NMR. Nat. Commun. 2018, 9, 2747. [Google Scholar] [CrossRef]
- Chai, L.Y.; Kullberg, B.J.; Vonk, A.G.; Warris, A.; Cambi, A.; Latge, J.P.; Joosten, L.A.; van der Meer, J.W.; Netea, M.G. Modulation of Toll-like receptor 2 (TLR2) and TLR4 responses by Aspergillus fumigatus. Infect. Immun. 2009, 77, 2184–2192. [Google Scholar] [CrossRef] [PubMed]

© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Valsecchi, I.; Stephen-Victor, E.; Wong, S.S.W.; Karnam, A.; Sunde, M.; Guijarro, J.I.; Rodríguez de Francisco, B.; Krüger, T.; Kniemeyer, O.; Brown, G.D.; et al. The Role of RodA-Conserved Cysteine Residues in the Aspergillus fumigatus Conidial Surface Organization. J. Fungi 2020, 6, 151. https://doi.org/10.3390/jof6030151
Valsecchi I, Stephen-Victor E, Wong SSW, Karnam A, Sunde M, Guijarro JI, Rodríguez de Francisco B, Krüger T, Kniemeyer O, Brown GD, et al. The Role of RodA-Conserved Cysteine Residues in the Aspergillus fumigatus Conidial Surface Organization. Journal of Fungi. 2020; 6(3):151. https://doi.org/10.3390/jof6030151
Chicago/Turabian StyleValsecchi, Isabel, Emmanuel Stephen-Victor, Sarah Sze Wah Wong, Anupama Karnam, Margaret Sunde, J. Iñaki Guijarro, Borja Rodríguez de Francisco, Thomas Krüger, Olaf Kniemeyer, Gordon D. Brown, and et al. 2020. "The Role of RodA-Conserved Cysteine Residues in the Aspergillus fumigatus Conidial Surface Organization" Journal of Fungi 6, no. 3: 151. https://doi.org/10.3390/jof6030151
APA StyleValsecchi, I., Stephen-Victor, E., Wong, S. S. W., Karnam, A., Sunde, M., Guijarro, J. I., Rodríguez de Francisco, B., Krüger, T., Kniemeyer, O., Brown, G. D., Willment, J. A., Latgé, J.-P., Brakhage, A. A., Bayry, J., & Aimanianda, V. (2020). The Role of RodA-Conserved Cysteine Residues in the Aspergillus fumigatus Conidial Surface Organization. Journal of Fungi, 6(3), 151. https://doi.org/10.3390/jof6030151







