Loss of Kex2 Affects the Candida albicans Cell Wall and Interaction with Innate Immune Cells
Abstract
1. Introduction
2. Materials and Methods
2.1. Strains and Culture Conditions
2.2. Generation of a Ura + kex2Δ Null Mutant and the Re-Integrant Control Strain
2.3. Analysis of Cell Wall Composition
2.4. Alcian Blue Binding Assays
2.5. Fluorochrome Staining
2.6. Cell Wall Porosity Assay
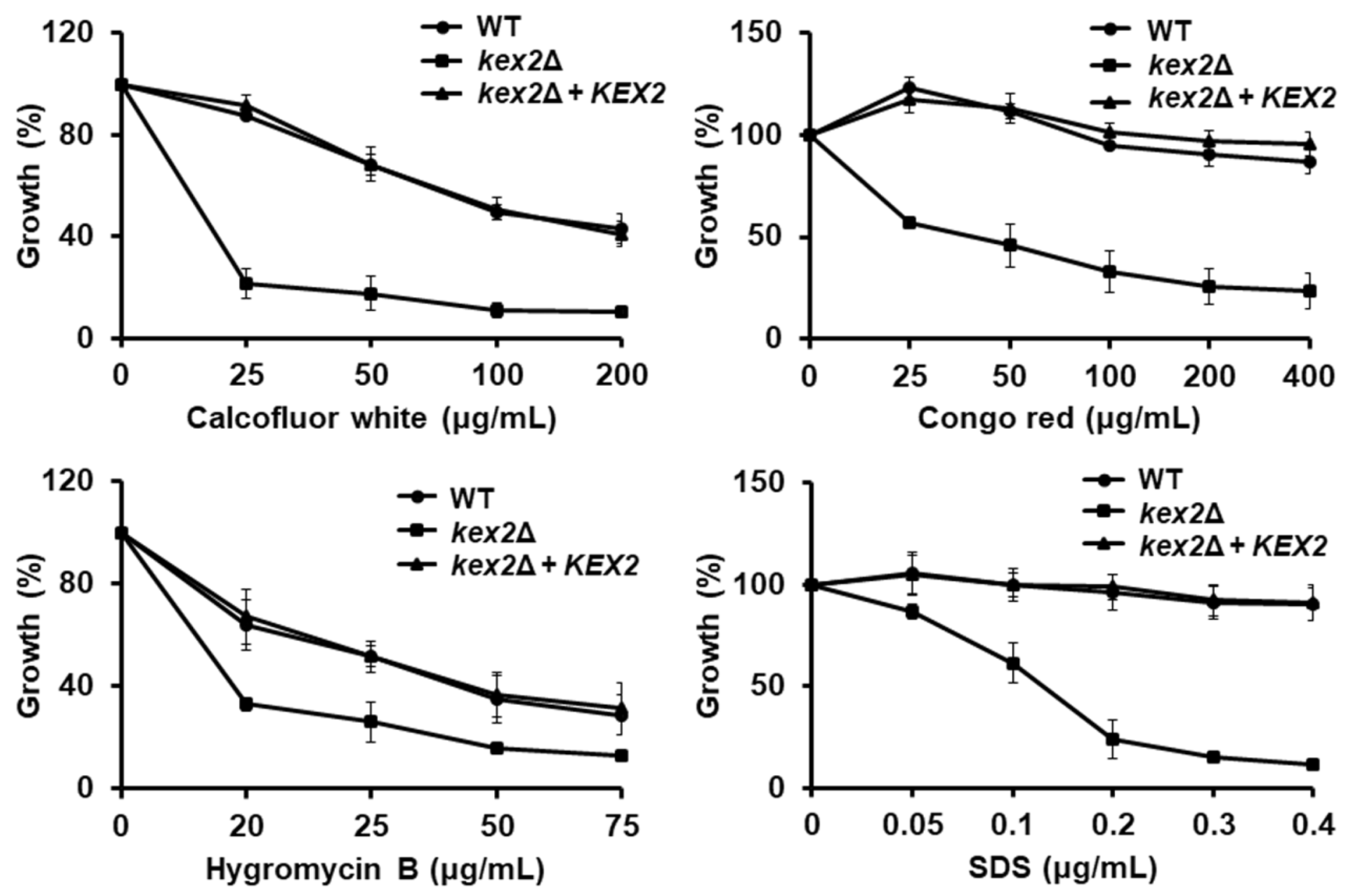
2.7. Susceptibility to Cell Wall Perturbing Agents
2.8. Electrophoretic Mobility Shift Assays of Hex1
2.9. Antifungal Drug Susceptibility Assays
2.10. Protoplast Generation and Differential Centrifugation
2.11. Enzyme Activity Assays
2.12. Ethics Statement
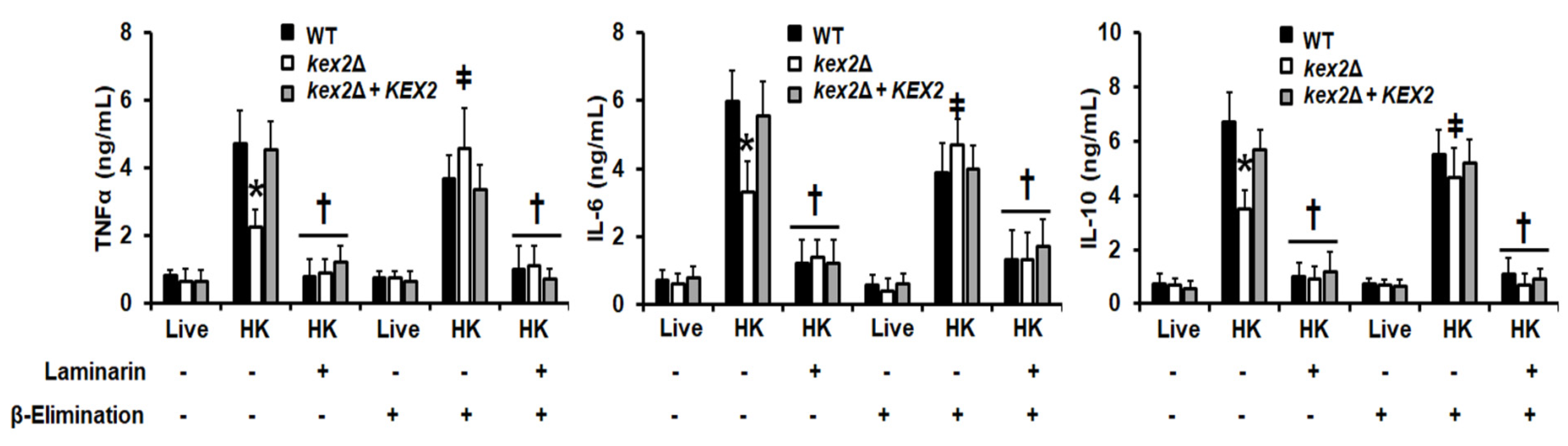
2.13. Stimulation of Cytokine Production by Human PBMCs
2.14. Differentiation of Human PBMCs to Macrophages
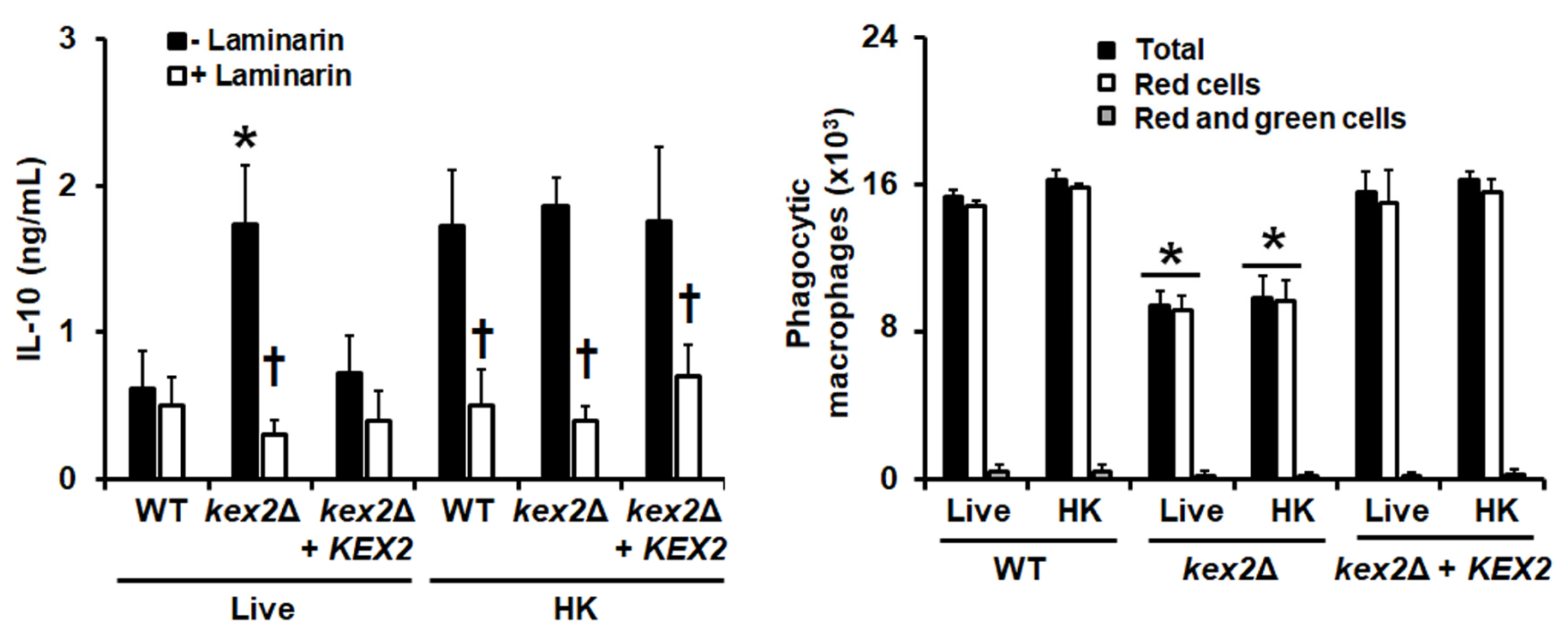
2.15. Analysis of C. albicans Phagocytosis by Human PBMC-Derived Macrophages
2.16. G. mellonella Survival Assays
2.17. Cytotoxicity Assays in Hemocytes of G. mellonella
2.18. Statistical Analysis
3. Results
3.1. Generation of kex2Δ Null Mutant Strains with URA3 Integrated into the Neutral Locus RPS1
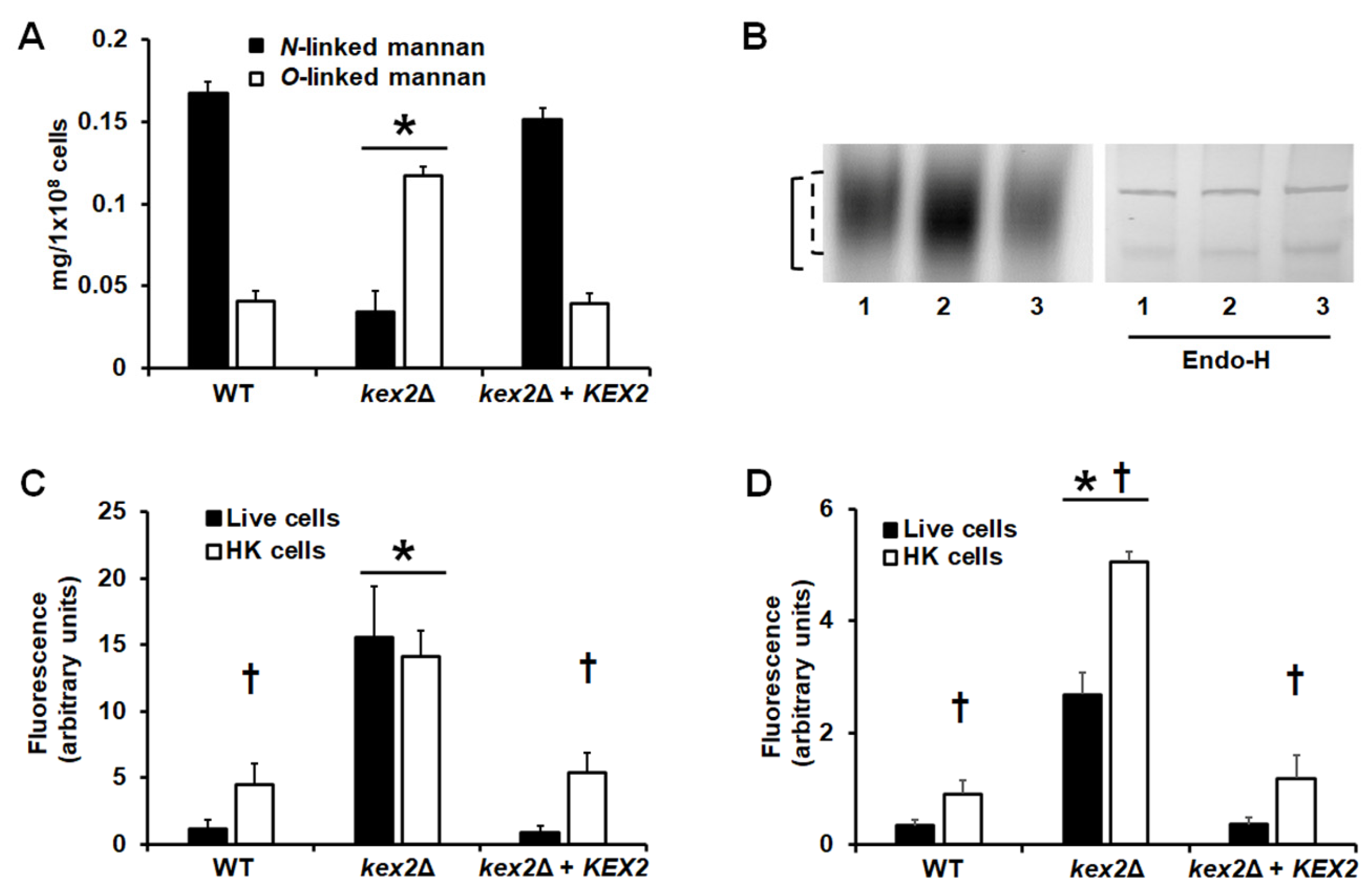
3.2. Loss of Kex2 Affects the C. albicans Cell Wall and Protein Glycosylation Pathways
3.3. Loss of Kex2 Affects the Localization and the Activity of Mannosyltransferases
3.4. The C. albicans kex2Δ Null Mutant Shows Abnormal Interactions with Human PMBCs and Monocyte-Derived Macrophages
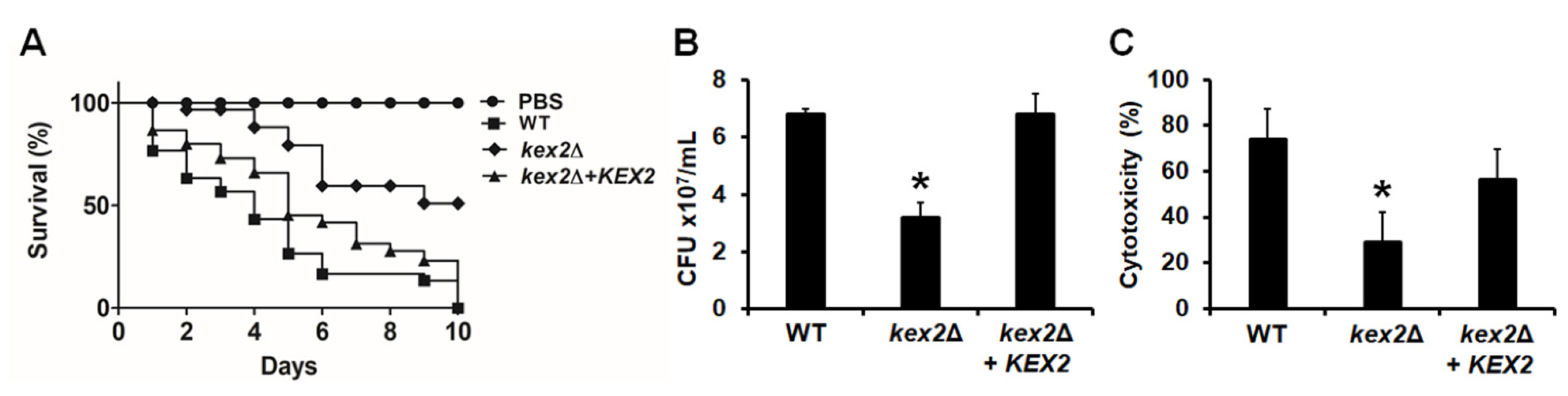
3.5. The C. albicans kex2Δ Null Mutant Shows Defects in the Ability to Kill G. mellonella Larvae
4. Discussion
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Brown, G.D.; Denning, D.W.; Gow, N.A.; Levitz, S.M.; Netea, M.G.; White, T.C. Hidden killers: Human fungal infections. Sci. Transl. Med. 2012, 4, 165rv113. [Google Scholar]
- Moyes, D.L.; Runglall, M.; Murciano, C.; Shen, C.; Nayar, D.; Thavaraj, S.; Kohli, A.; Islam, A.; Mora-Montes, H.; Challacombe, S.J.; et al. A biphasic innate immune MAPK response discriminates between the yeast and hyphal forms of Candida albicans in epithelial cells. Cell Host Microbe 2010, 8, 225–235. [Google Scholar] [CrossRef]
- Brown, G.D. Innate antifungal immunity: The key role of phagocytes. Annu. Rev. Immunol. 2011, 29, 1–21. [Google Scholar] [CrossRef]
- Díaz-Jiménez, D.F.; Pérez-García, L.A.; Martínez-Álvarez, J.A.; Mora-Montes, H.M. Role of the fungal cell wall in pathogenesis and antifungal resistance. Curr. Fungal Infect. Rep. 2012, 6, 275–282. [Google Scholar]
- Mora-Montes, H.M.; Ponce-Noyola, P.; Villagómez-Castro, J.C.; Gow, N.A.R.; Flores-Carreón, A.; López-Romero, E. Protein glycosylation in Candida. Future Microbiol. 2009, 4, 1167–1183. [Google Scholar] [CrossRef]
- Gow, N.A.R.; Latge, J.P.; Munro, C.A. The Fungal Cell Wall: Structure, Biosynthesis, and Function. Microbiol. Spectr. 2017, 5. [Google Scholar] [CrossRef]
- Martinez-Alvarez, J.A.; Perez-Garcia, L.A.; Flores-Carreon, A.; Mora-Montes, H.M. The immune response against Candida spp. and Sporothrix schenckii. Rev. Iberoam. Micol. 2014, 31, 62–66. [Google Scholar] [CrossRef]
- Hernandez-Chavez, M.J.; Perez-Garcia, L.A.; Nino-Vega, G.A.; Mora-Montes, H.M. Fungal strategies to evade the host immune recognition. J. Fungi 2017, 3, 51. [Google Scholar] [CrossRef]
- Klis, F.M.; de Groot, P.; Hellingwerf, K. Molecular organization of the cell wall of Candida albicans. Med. Mycol. 2001, 39 (Suppl. 1), 1–8. [Google Scholar]
- Estrada-Mata, E.; Navarro-Arias, M.J.; Perez-Garcia, L.A.; Mellado-Mojica, E.; Lopez, M.G.; Csonka, K.; Gacser, A.; Mora-Montes, H.M. Members of the Candida parapsilosis complex and Candida albicans are differentially recognized by human peripheral blood mononuclear cells. Front. Microbiol. 2015, 6, 1527. [Google Scholar]
- Navarro-Arias, M.J.; Hernandez-Chavez, M.J.; Garcia-Carnero, L.C.; Amezcua-Hernandez, D.G.; Lozoya-Perez, N.E.; Estrada-Mata, E.; Martinez-Duncker, I.; Franco, B.; Mora-Montes, H.M. Differential recognition of Candida tropicalis, Candida guilliermondii, Candida krusei, and Candida auris by human innate immune cells. Infect. Drug Resist. 2019, 12, 783–794. [Google Scholar] [CrossRef]
- Martinez-Duncker, I.; Diaz-Jimenez, D.F.; Mora-Montes, H.M. Comparative analysis of protein glycosylation pathways in humans and the fungal pathogen Candida albicans. Int. J. Microbiol. 2014, 2014, 267497. [Google Scholar] [CrossRef]
- Netea, M.G.; Gow, N.A.; Munro, C.A.; Bates, S.; Collins, C.; Ferwerda, G.; Hobson, R.P.; Bertram, G.; Hughes, H.B.; Jansen, T.; et al. Immune sensing of Candida albicans requires cooperative recognition of mannans and glucans by lectin and Toll-like receptors. J. Clin. Investig. 2006, 116, 1642–1650. [Google Scholar] [CrossRef]
- Cambi, A.; Netea, M.G.; Mora-Montes, H.M.; Gow, N.A.; Hato, S.V.; Lowman, D.W.; Kullberg, B.J.; Torensma, R.; Williams, D.L.; Figdor, C.G. Dendritic cell interaction with Candida albicans critically depends on N-linked mannan. J. Biol. Chem. 2008, 283, 20590–20599. [Google Scholar] [CrossRef]
- Saijo, S.; Ikeda, S.; Yamabe, K.; Kakuta, S.; Ishigame, H.; Akitsu, A.; Fujikado, N.; Kusaka, T.; Kubo, S.; Chung, S.-h.; et al. Dectin-2 recognition of alpha-mannans and induction of Th17 cell differentiation is essential for host defense against Candida albicans. Immunity 2010, 32, 681–691. [Google Scholar] [CrossRef]
- Zhu, L.-L.; Zhao, X.-Q.; Jiang, C.; You, Y.; Chen, X.-P.; Jiang, Y.-Y.; Jia, X.-M.; Lin, X. C-type lectin receptors Dectin-3 and Dectin-2 form a heterodimeric pattern-recognition receptor for host defense against fungal infection. Immunity 2013, 39, 324–334. [Google Scholar] [CrossRef]
- Haider, M.; Dambuza, I.M.; Asamaphan, P.; Stappers, M.; Reid, D.; Yamasaki, S.; Brown, G.D.; Gow, N.A.R.; Erwig, L.P. The pattern recognition receptors dectin-2, mincle, and FcRγ impact the dynamics of phagocytosis of Candida, Saccharomyces, Malassezia, and Mucor species. PLoS ONE 2019, 14, e0220867. [Google Scholar] [CrossRef]
- Netea, M.G.; Brown, G.D.; Kullberg, B.J.; Gow, N.A. An integrated model of the recognition of Candida albicans by the innate immune system. Nat. Rev. Microbiol. 2008, 6, 67–78. [Google Scholar] [CrossRef]
- Delic, M.; Valli, M.; Graf, A.B.; Pfeffer, M.; Mattanovich, D.; Gasser, B. The secretory pathway: Exploring yeast diversity. FEMS Microbiol. Rev. 2013, 37, 872–914. [Google Scholar] [CrossRef]
- Fuller, R.S.; Brake, A.J.; Thorner, J. Intracellular targeting and structural conservation of a prohormone-processing endoprotease. Science 1989, 246, 482–486. [Google Scholar] [CrossRef]
- Bryant, N.J.; Stevens, T.H. Two separate signals act independently to localize a yeast late Golgi membrane protein through a combination of retrieval and retention. J. Cell Biol. 1997, 136, 287–297. [Google Scholar] [CrossRef]
- Julius, D.; Brake, A.; Blair, L.; Kunisawa, R.; Thorner, J. Isolation of the putative structural gene for the lysine-arginine-cleaving endopeptidase required for processing of yeast prepro-alpha-factor. Cell 1984, 37, 1075–1089. [Google Scholar] [CrossRef]
- Newport, G.; Agabian, N. KEX2 influences Candida albicans proteinase secretion and hyphal formation. J. Biol Chem. 1997, 272, 28954–28961. [Google Scholar] [CrossRef]
- Newport, G.; Kuo, A.; Flattery, A.; Gill, C.; Blake, J.J.; Kurtz, M.B.; Abruzzo, G.K.; Agabian, N. Inactivation of Kex2p diminishes the virulence of Candida albicans. J. Biol. Chem. 2003, 278, 1713–1720. [Google Scholar] [CrossRef] [PubMed]
- Staniszewska, M.; Bondaryk, M.; Kazek, M.; Gliniewicz, A.; Braunsdorf, C.; Schaller, M.; Mora-Montes, H.M.; Ochal, Z. Effect of serine protease KEX2 on Candida albicans virulence under halogenated methyl sulfones. Future Microbiol. 2017, 12, 285–306. [Google Scholar] [CrossRef]
- Staniszewska, M.; Gizinska, M.; Kazek, M.; de Jesus Gonzalez-Hernandez, R.; Ochal, Z.; Mora-Montes, H.M. New antifungal 4-chloro-3-nitrophenyldifluoroiodomethyl sulfone reduces the Candida albicans pathogenicity in the Galleria mellonella model organism. Braz. J. Microbiol. 2019. [Google Scholar] [CrossRef]
- Richardson, J.P.; Mogavero, S.; Moyes, D.L.; Blagojevic, M.; Krüger, T.; Verma, A.H.; Coleman, B.M.; De La Cruz Diaz, J.; Schulz, D.; Ponde, N.O.; et al. Processing of Candida albicans Ece1p is critical for candidalysin maturation and fungal virulence. mBio 2018, 9, e02178-17. [Google Scholar] [CrossRef]
- Brand, A.; MacCallum, D.M.; Brown, A.J.P.; Gow, N.A.R.; Odds, F.C. Ectopic expression of URA3 can influence the virulence phenotypes and proteome of Candida albicans but can be overcome by targeted reintegration of URA3 at the RPS10 locus. Eukaryot Cell 2004, 3, 900–909. [Google Scholar] [CrossRef] [PubMed]
- Bates, S.; Hughes, H.B.; Munro, C.A.; Thomas, W.P.H.; MacCallum, D.M.; Bertram, G.; Atrih, A.; Ferguson, M.A.J.; Brown, A.J.P.; Odds, F.C.; et al. Outer chain N-glycans are required for cell wall integrity and virulence of Candida albicans. J. Biol. Chem. 2006, 281, 90–98. [Google Scholar] [CrossRef] [PubMed]
- Murad, A.M.; Lee, P.R.; Broadbent, I.D.; Barelle, C.J.; Brown, A.J. CIp10, an efficient and convenient integrating vector for Candida albicans. Yeast 2000, 16, 325–327. [Google Scholar] [CrossRef]
- Mora-Montes, H.M.; Bates, S.; Netea, M.G.; Diaz-Jimenez, D.F.; Lopez-Romero, E.; Zinker, S.; Ponce-Noyola, P.; Kullberg, B.J.; Brown, A.J.; Odds, F.C.; et al. Endoplasmic reticulum alpha-glycosidases of Candida albicans are required for N glycosylation, cell wall integrity, and normal host-fungus interaction. Eukaryot. Cell 2007, 6, 2184–2193. [Google Scholar] [CrossRef]
- Plaine, A.; Walker, L.; Da Costa, G.; Mora-Montes, H.M.; McKinnon, A.; Gow, N.A.; Gaillardin, C.; Munro, C.A.; Richard, M.L. Functional analysis of Candida albicans GPI-anchored proteins: Roles in cell wall integrity and caspofungin sensitivity. Fungal. Genet. Biol. 2008, 45, 1404–1414. [Google Scholar] [CrossRef]
- Perez-Garcia, L.A.; Csonka, K.; Flores-Carreon, A.; Estrada-Mata, E.; Mellado-Mojica, E.; Nemeth, T.; Lopez-Ramirez, L.A.; Toth, R.; Lopez, M.G.; Vizler, C.; et al. Role of protein glycosylation in Candida parapsilosis cell wall integrity and host interaction. Front. Microbiol. 2016, 7, 306. [Google Scholar] [CrossRef]
- Diaz-Jimenez, D.F.; Mora-Montes, H.M.; Hernandez-Cervantes, A.; Luna-Arias, J.P.; Gow, N.A.; Flores-Carreon, A. Biochemical characterization of recombinant Candida albicans mannosyltransferases Mnt1, Mnt2 and Mnt5 reveals new functions in O- and N-mannan biosynthesis. Biochem. Biophys. Res. Commun. 2012, 419, 77–82. [Google Scholar] [CrossRef]
- Navarro-Arias, M.J.; Defosse, T.A.; Dementhon, K.; Csonka, K.; Mellado-Mojica, E.; Dias Valerio, A.; Gonzalez-Hernandez, R.J.; Courdavault, V.; Clastre, M.; Hernandez, N.V.; et al. Disruption of protein mannosylation affects Candida guilliermondii cell wall, immune sensing, and virulence. Front. Microbiol. 2016, 7, 1951. [Google Scholar] [CrossRef]
- Lozoya-Perez, N.E.; Casas-Flores, S.; de Almeida, J.R.F.; Martinez-Alvarez, J.A.; Lopez-Ramirez, L.A.; Jannuzzi, G.P.; Trujillo-Esquivel, E.; Estrada-Mata, E.; Almeida, S.R.; Franco, B.; et al. Silencing of OCH1 unveils the role of Sporothrix schenckii N-linked glycans during the host-fungus interaction. Infect. Drug Resist. 2019, 12, 67–85. [Google Scholar] [CrossRef]
- Hobson, R.P.; Munro, C.A.; Bates, S.; MacCallum, D.M.; Cutler, J.E.; Heinsbroek, S.E.M.; Brown, G.D.; Odds, F.C.; Gow, N.A.R. Loss of Cell Wall Mannosylphosphate in Candida albicans Does Not Influence Macrophage Recognition. J. Biol. Chem. 2004, 279, 39628–39635. [Google Scholar] [CrossRef]
- Mora-Montes, H.M.; Netea, M.G.; Ferwerda, G.; Lenardon, M.D.; Brown, G.D.; Mistry, A.R.; Kullberg, B.J.; O’Callaghan, C.A.; Sheth, C.C.; Odds, F.C.; et al. Recognition and Blocking of Innate Immunity Cells by Candida albicans Chitin. Infect. Immun. 2011, 79, 1961–1970. [Google Scholar] [CrossRef]
- Graham, L.M.; Tsoni, S.V.; Willment, J.A.; Williams, D.L.; Taylor, P.R.; Gordon, S.; Dennehy, K.; Brown, G.D. Soluble Dectin-1 as a tool to detect beta-glucans. J. Immunol. Methods 2006, 314, 164–169. [Google Scholar] [CrossRef]
- Marakalala, M.J.; Vautier, S.; Potrykus, J.; Walker, L.A.; Shepardson, K.M.; Hopke, A.; Mora-Montes, H.M.; Kerrigan, A.; Netea, M.G.; Murray, G.I.; et al. Differential adaptation of Candida albicans in vivo modulates immune recognition by dectin-1. PLoS Pathog. 2013, 9, e1003315. [Google Scholar] [CrossRef]
- De Nobel, J.G.; Klis, F.M.; Munnik, T.; Priem, J.; van den Ende, H. An assay of relative cell wall porosity in Saccharomyces cerevisiae, Kluyveromyces lactis and Schizosaccharomyces pombe. Yeast 1990, 6, 483–490. [Google Scholar] [CrossRef]
- Bates, S.; MacCallum, D.M.; Bertram, G.; Munro, C.A.; Hughes, H.B.; Buurman, E.T.; Brown, A.J.P.; Odds, F.C.; Gow, N.A.R. Candida albicans Pmr1p, a secretory pathway P-type Ca2+/Mn2+-ATPase, is required for glycosylation and virulence. J. Biol. Chem. 2005, 280, 23408–23415. [Google Scholar] [CrossRef]
- Hernandez-Cervantes, A.; Mora-Montes, H.M.; Alvarez-Vargas, A.; Jimenez, D.F.; Robledo-Ortiz, C.I.; Flores-Carreon, A. Isolation of Sporothrix schenckii MNT1 and the biochemical and functional characterization of the encoded alpha1,2-mannosyltransferase activity. Microbiology 2012, 158, 2419–2427. [Google Scholar] [CrossRef]
- Garcia-Effron, G.; Park, S.; Perlin, D.S. Improved detection of Candida sp. fks hot spot mutants by using the method of the CLSI M27-A3 document with the addition of bovine serum albumin. Antimicrob. Agents Chemother. 2011, 55, 2245–2255. [Google Scholar] [CrossRef][Green Version]
- Mora-Montes, H.M.; Bader, O.; Lopez-Romero, E.; Zinker, S.; Ponce-Noyola, P.; Hube, B.; Gow, N.A.R.; Flores-Carreon, A. Kex2 protease converts the endoplasmic reticulum alpha1,2-mannosidase of Candida albicans into a soluble cytosolic form. Microbiology 2008, 154, 3782–3794. [Google Scholar] [CrossRef][Green Version]
- Mora-Montes, H.M.; Lopez-Romero, E.; Zinker, S.; Ponce-Noyola, P.; Flores-Carreon, A. Hydrolysis of Man9GlcNAc2 and Man8GlcNAc2 oligosaccharides by a purified alpha-mannosidase from Candida albicans. Glycobiology 2004, 14, 593–598. [Google Scholar] [CrossRef]
- Gonzalez-Hernandez, R.J.; Jin, K.; Hernandez-Chavez, M.J.; Diaz-Jimenez, D.F.; Trujillo-Esquivel, E.; Clavijo-Giraldo, D.M.; Tamez-Castrellon, A.K.; Franco, B.; Gow, N.A.R.; Mora-Montes, H.M. Phosphomannosylation and the functional analysis of the extended Candida albicans MNN4-like gene family. Front. Microbiol. 2017, 8, 2156. [Google Scholar] [CrossRef]
- Hernandez-Chavez, M.J.; Franco, B.; Clavijo-Giraldo, D.M.; Hernandez, N.V.; Estrada-Mata, E.; Mora-Montes, H.M. Role of protein phosphomannosylation in the Candida tropicalis-macrophage interaction. FEMS Yeast Res. 2018, 18. [Google Scholar] [CrossRef]
- Mora-Montes, H.M.; Bates, S.; Netea, M.G.; Castillo, L.; Brand, A.; Buurman, E.T.; Diaz-Jimenez, D.F.; Jan Kullberg, B.; Brown, A.J.; Odds, F.C.; et al. A multifunctional mannosyltransferase family in Candida albicans determines cell wall mannan structure and host-fungus interactions. J. Biol. Chem. 2010, 285, 12087–12095. [Google Scholar] [CrossRef]
- Mukaremera, L.; Lee, K.K.; Mora-Montes, H.M.; Gow, N.A.R. Candida albicans yeast, pseudohyphal, and hyphal morphogenesis differentially affects immune recognition. Front. Immunol. 2017, 8, 629. [Google Scholar] [CrossRef]
- Cheng, S.C.; van de Veerdonk, F.L.; Lenardon, M.; Stoffels, M.; Plantinga, T.; Smeekens, S.; Rizzetto, L.; Mukaremera, L.; Preechasuth, K.; Cavalieri, D.; et al. The dectin-1/inflammasome pathway is responsible for the induction of protective T-helper 17 responses that discriminate between yeasts and hyphae of Candida albicans. J. Leukoc. Biol. 2011, 90, 357–366. [Google Scholar] [CrossRef] [PubMed]
- Hernandez-Chavez, M.J.; Clavijo-Giraldo, D.M.; Novak, A.; Lozoya-Perez, N.E.; Martinez-Alvarez, J.A.; Salinas-Marin, R.; Hernandez, N.V.; Martinez-Duncker, I.; Gacser, A.; Mora-Montes, H.M. Role of protein mannosylation in the Candida tropicalis-host Interaction. Front. Microbiol. 2019, 10, 2743. [Google Scholar] [CrossRef] [PubMed]
- Munro, C.A.; Bates, S.; Buurman, E.T.; Hughes, H.B.; Maccallum, D.M.; Bertram, G.; Atrih, A.; Ferguson, M.A.; Bain, J.M.; Brand, A.; et al. Mnt1p and Mnt2p of Candida albicans are partially redundant alpha-1,2-mannosyltransferases that participate in O-linked mannosylation and are required for adhesion and virulence. J. Biol. Chem. 2005, 280, 1051–1060. [Google Scholar] [CrossRef] [PubMed]
- Gow, N.A.R.; Netea, M.G.; Munro, C.A.; Ferwerda, G.; Bates, S.; Mora-Montes, H.M.; Walker, L.; Jansen, T.; Jacobs, L.; Tsoni, V.; et al. Immune recognition of Candida albicans beta-glucan by dectin-1. J. Infect. Dis. 2007, 196, 1565–1571. [Google Scholar] [CrossRef]
- Kopecka, M.; Gabriel, M. The influence of congo red on the cell wall and (1----3)-beta-D-glucan microfibril biogenesis in Saccharomyces cerevisiae. Arch. Microbiol. 1992, 158, 115–126. [Google Scholar] [CrossRef]
- Dean, N. Yeast glycosylation mutants are sensitive to aminoglycosides. Proc. Natl. Acad. Sci. USA 1995, 92, 1287–1291. [Google Scholar] [CrossRef]
- Rollenhagen, C.; Mamtani, S.; Ma, D.; Dixit, R.; Eszterhas, S.; Lee, S.A. The role of secretory pathways in Candida albicans pathogenesis. J. Fungi 2020, 6, 26. [Google Scholar] [CrossRef]
- de Nobel, H.; Ruiz, C.; Martin, H.; Morris, W.; Brul, S.; Molina, M.; Klis, F.M. Cell wall perturbation in yeast results in dual phosphorylation of the Slt2/Mpk1 MAP kinase and in an Slt2-mediated increase in FKS2-lacZ expression, glucanase resistance and thermotolerance. Microbiology 2000, 146, 2121–2132. [Google Scholar] [CrossRef]
- Prill, S.K.; Klinkert, B.; Timpel, C.; Gale, C.A.; Schröppel, K.; Ernst, J.F. PMT family of Candida albicans: Five protein mannosyltransferase isoforms affect growth, morphogenesis and antifungal resistance. Mol. Microbiol. 2005, 55, 546–560. [Google Scholar] [CrossRef]
- Karhinen, L.; Makarow, M. Activity of recycling Golgi mannosyltransferases in the yeast endoplasmic reticulum. J. Cell Sci. 2004, 117, 351–358. [Google Scholar] [CrossRef]
- Harris, S.L.; Waters, M.G. Localization of a yeast early Golgi mannosyltransferase, Och1p, involves retrograde transport. J. Cell Biol. 1996, 132, 985–998. [Google Scholar] [CrossRef]
- Bruinsma, P.; Spelbrink, R.G.; Nothwehr, S.F. Retrograde transport of the mannosyltransferase Och1p to the early Golgi requires a component of the COG transport complex. J. Biol. Chem 2004, 279, 39814–39823. [Google Scholar] [CrossRef]
- Costanzo, M.; VanderSluis, B.; Koch, E.N.; Baryshnikova, A.; Pons, C.; Tan, G.; Wang, W.; Usaj, M.; Hanchard, J.; Lee, S.D.; et al. A global genetic interaction network maps a wiring diagram of cellular function. Science 2016, 353. [Google Scholar] [CrossRef]
- Surma, M.A.; Klose, C.; Peng, D.; Shales, M.; Mrejen, C.; Stefanko, A.; Braberg, H.; Gordon, D.E.; Vorkel, D.; Ejsing, C.S.; et al. A lipid E-MAP identifies Ubx2 as a critical regulator of lipid saturation and lipid bilayer stress. Mol. Cell 2013, 51, 519–530. [Google Scholar] [CrossRef]
- Schuldiner, M.; Collins, S.R.; Thompson, N.J.; Denic, V.; Bhamidipati, A.; Punna, T.; Ihmels, J.; Andrews, B.; Boone, C.; Greenblatt, J.F.; et al. Exploration of the function and organization of the yeast early secretory pathway through an epistatic miniarray profile. Cell 2005, 123, 507–519. [Google Scholar] [CrossRef]
- Cunningham, K.W.; Wickner, W.T. Yeast KEX2 protease and mannosyltransferase I are localized to distinct compartments of the secretory pathway. Yeast 1989, 5, 25–33. [Google Scholar] [CrossRef]
- Gantner, B.N.; Simmons, R.M.; Canavera, S.J.; Akira, S.; Underhill, D.M. Collaborative induction of inflammatory responses by dectin-1 and Toll-like receptor 2. J. Exp. Med. 2003, 197, 1107–1117. [Google Scholar] [CrossRef]
- West, L.; Lowman, D.W.; Mora-Montes, H.M.; Grubb, S.; Murdoch, C.; Thornhill, M.H.; Gow, N.A.; Williams, D.; Haynes, K. Differential virulence of Candida glabrata glycosylation mutants. J. Biol. Chem. 2013, 288, 22006–22018. [Google Scholar] [CrossRef]
- Wheeler, R.T.; Fink, G.R. A drug-sensitive genetic network masks fungi from the immune system. PLoS Pathog. 2006, 2, e35. [Google Scholar] [CrossRef]
- Heinsbroek, S.E.M.; Taylor, P.R.; Martinez, F.O.; Martinez-Pomares, L.; Brown, G.D.; Gordon, S. Stage-specific sampling by pattern recognition receptors during Candida albicans phagocytosis. PLoS Pathog. 2008, 4, e1000218. [Google Scholar] [CrossRef]
- Reid, D.M.; Gow, N.A.R.; Brown, G.D. Pattern recognition: Recent insights from Dectin-1. Curr. Opin. Immunol. 2009, 21, 30–37. [Google Scholar] [CrossRef] [PubMed]
- McKenzie, C.G.; Koser, U.; Lewis, L.E.; Bain, J.M.; Mora-Montes, H.M.; Barker, R.N.; Gow, N.A.; Erwig, L.P. Contribution of Candida albicans cell wall components to recognition by and escape from murine macrophages. Infect. Immun. 2010, 78, 1650–1658. [Google Scholar] [CrossRef] [PubMed]

| Strain | Origin | Genotype | Reference |
|---|---|---|---|
| NGY152 | Derived from CAI-4 | ura3Δ-iro1Δ::imm434/ura3Δ-iro1Δ::imm434; RPSI/rps1Δ::CIp10 | [28] |
| kex2Δ/kex2Δ | Derived from CAI-4 | ura3Δ-iro1Δ::imm434/ura3Δ-iro1Δ::imm434; kex2Δ::dp1200/kex2Δ::dp1200 | [26] |
| HMY209 | Derived from kex2Δ/kex2Δ | As kex2Δ/kex2Δ, but RPSI/rps1Δ::CIp10 | This work |
| HMY210 | Derived from kex2Δ/kex2Δ | As kex2Δ/kex2Δ, but RPSI/rps1Δ:: CIp10-KEX2 | This work |
| Cell Wall Abundance | Phosphomannan Content (μg) a | Porosity (%) b | Protein Content (μg) c | |||
|---|---|---|---|---|---|---|
| Strain | Chitin (%) | Mannan (%) | Glucan (%) | |||
| WT (NGY152) | 1.8 ± 0.5 | 43.7 ± 1.0 | 54.4 ± 0.7 | 95.7 ± 9.1 | 22.4 ± 7.9 | 62.4 ± 4.7 |
| kex2Δ (HMY209) | 8.8 ± 1.0 * | 28.1 ± 0.5 * | 63.0 ± 0.8 * | 45.9 ± 15.2 * | 70.5 ± 12.4 * | 96.8 ± 10.0 * |
| kex2Δ + KEX2 (HMY210) | 1.9 ± 0.5 | 41.6 ± 0.5 | 56.5 ± 0.6 | 89.1 ± 12.5 | 27.1 ± 8.8 | 55.0 ± 4.0 |
| Enzyme Activity | Strain | |||
|---|---|---|---|---|
| WT (NGY152) | kex2Δ (HMY209) | kex2Δ + KEX2 (HMY210) | ||
| α-1,2-Mannosidase a | Cytosolic compartment | 6.9 ± 1.5 | 0.02 ± 0.01 * | 6.3 ± 1.1 |
| Golgi complex | 9.5 ± 1.3 | 8.9 ± 2.0 | 9.0 ± 1.8 | |
| Endoplasmic reticulum | 18.7 ± 1.9 | 20.7 ± 2.7 | 19.1 ± 1.6 | |
| α-1,2-Mannosyltransferase b | Cytosolic compartment | 5.0 × 102 ± 2.0 × 102 | 4.0 × 102 ± 3.0 × 102 | 3.0 × 102 ± 2.0 × 102 |
| Golgi complex | 8.0 × 106 ± 9.0 × 105 | 14.0 × 106 ± 1.1 ×10 * | 9.0 × 106 ± 1.2 × 106 | |
| Endoplasmic reticulum | 6.0 × 103 ± 1.5 × 103 | 4.0 × 103 ± 1.1 × 103 | 2.0 × 103 ± 1.5 × 103 | |
| α-1,3-Mannosyltransferase b | Cytosolic compartment | ND | ND | ND |
| Golgi complex | 3.0 × 106 ± 1.0 × 106 | 3.0 × 106 ± 1.8 × 106 | 2.0 × 106 ± 0.8 × 106 | |
| Endoplasmic reticulum | 8.0 × 102 ± 2.4 × 102 | 7.0 × 102 ± 1.1 × 102 | 9.0 × 102 ± 3.5 × 102 | |
| α-1,6-Mannosyltransferase b | Cytosolic compartment | ND | ND | ND |
| Golgi complex | 9.0 × 105 ± 1.8 × 105 | 2.0 × 102 ± 0.9 × 102 * | 7.0 × 105 ± 2.4 × 105 | |
| Endoplasmic reticulum | 3.0 × 102 ± 0.6 × 102 | 2.4 × 102 ± 1.0 × 102 | 2.6 × 102 ± 0.9 × 102 | |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gómez-Gaviria, M.; Lozoya-Pérez, N.E.; Staniszewska, M.; Franco, B.; Niño-Vega, G.A.; Mora-Montes, H.M. Loss of Kex2 Affects the Candida albicans Cell Wall and Interaction with Innate Immune Cells. J. Fungi 2020, 6, 57. https://doi.org/10.3390/jof6020057
Gómez-Gaviria M, Lozoya-Pérez NE, Staniszewska M, Franco B, Niño-Vega GA, Mora-Montes HM. Loss of Kex2 Affects the Candida albicans Cell Wall and Interaction with Innate Immune Cells. Journal of Fungi. 2020; 6(2):57. https://doi.org/10.3390/jof6020057
Chicago/Turabian StyleGómez-Gaviria, Manuela, Nancy E. Lozoya-Pérez, Monika Staniszewska, Bernardo Franco, Gustavo A. Niño-Vega, and Hector M. Mora-Montes. 2020. "Loss of Kex2 Affects the Candida albicans Cell Wall and Interaction with Innate Immune Cells" Journal of Fungi 6, no. 2: 57. https://doi.org/10.3390/jof6020057
APA StyleGómez-Gaviria, M., Lozoya-Pérez, N. E., Staniszewska, M., Franco, B., Niño-Vega, G. A., & Mora-Montes, H. M. (2020). Loss of Kex2 Affects the Candida albicans Cell Wall and Interaction with Innate Immune Cells. Journal of Fungi, 6(2), 57. https://doi.org/10.3390/jof6020057







