PaPro1 and IDC4, Two Genes Controlling Stationary Phase, Sexual Development and Cell Degeneration in Podospora anserina
Abstract
1. Introduction
2. Materials and Methods
2.1. Strains and Culture Conditions
2.2. Mutation Identifications
2.3. Gene Deletions
2.4. Gene Complementation
2.5. Plasmid Constructions for mCherry-Tagging of IDC4
2.6. RT-qPCR Experiments
2.7. Microscopy
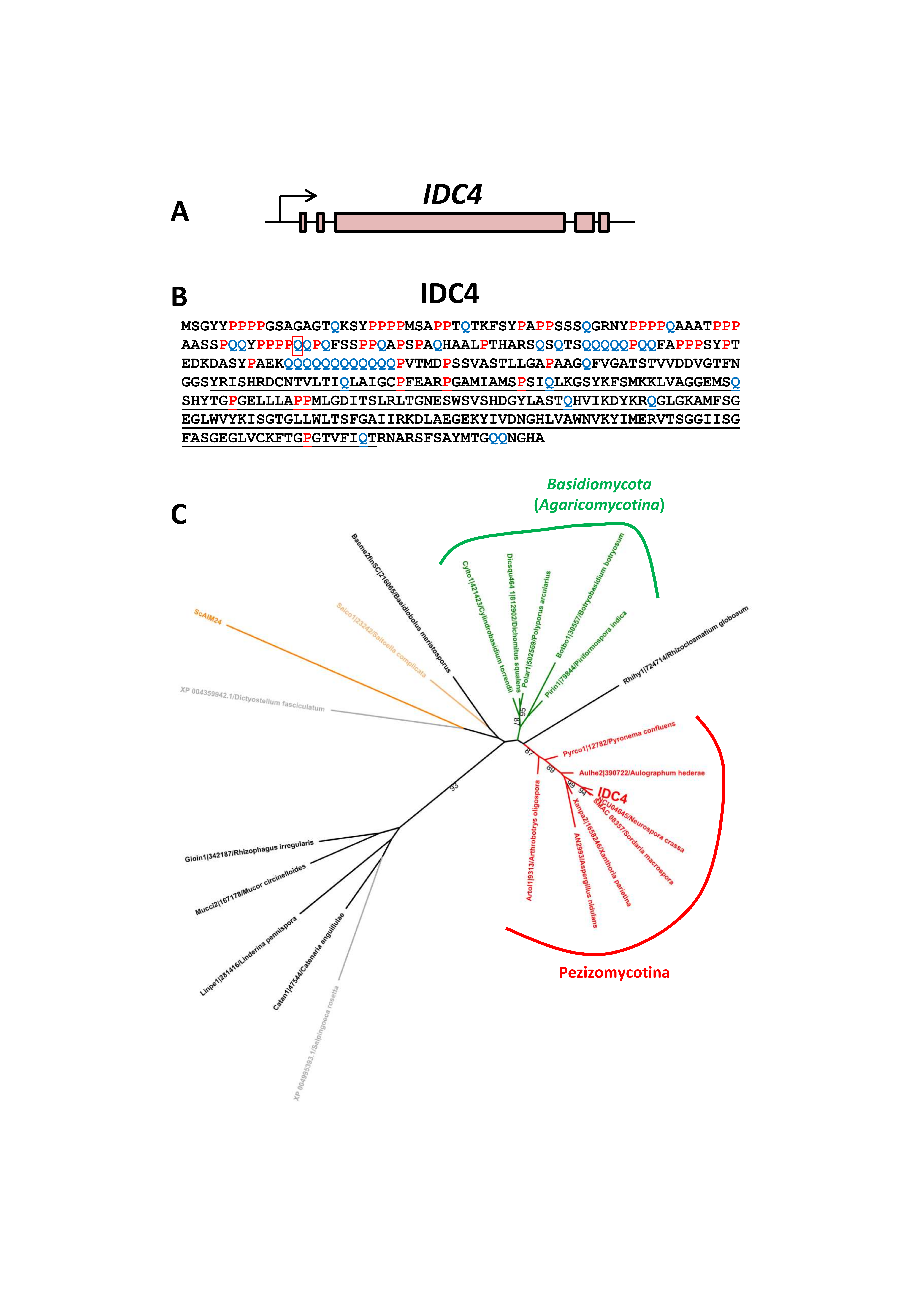
2.8. Phylogenetic Analysis
2.9. FIMO Analysis
2.10. Protein Extraction and Western Blot Analysis
3. Results
3.1. Identification of the Genes Affected in IDC508, IDC510 and IDC511
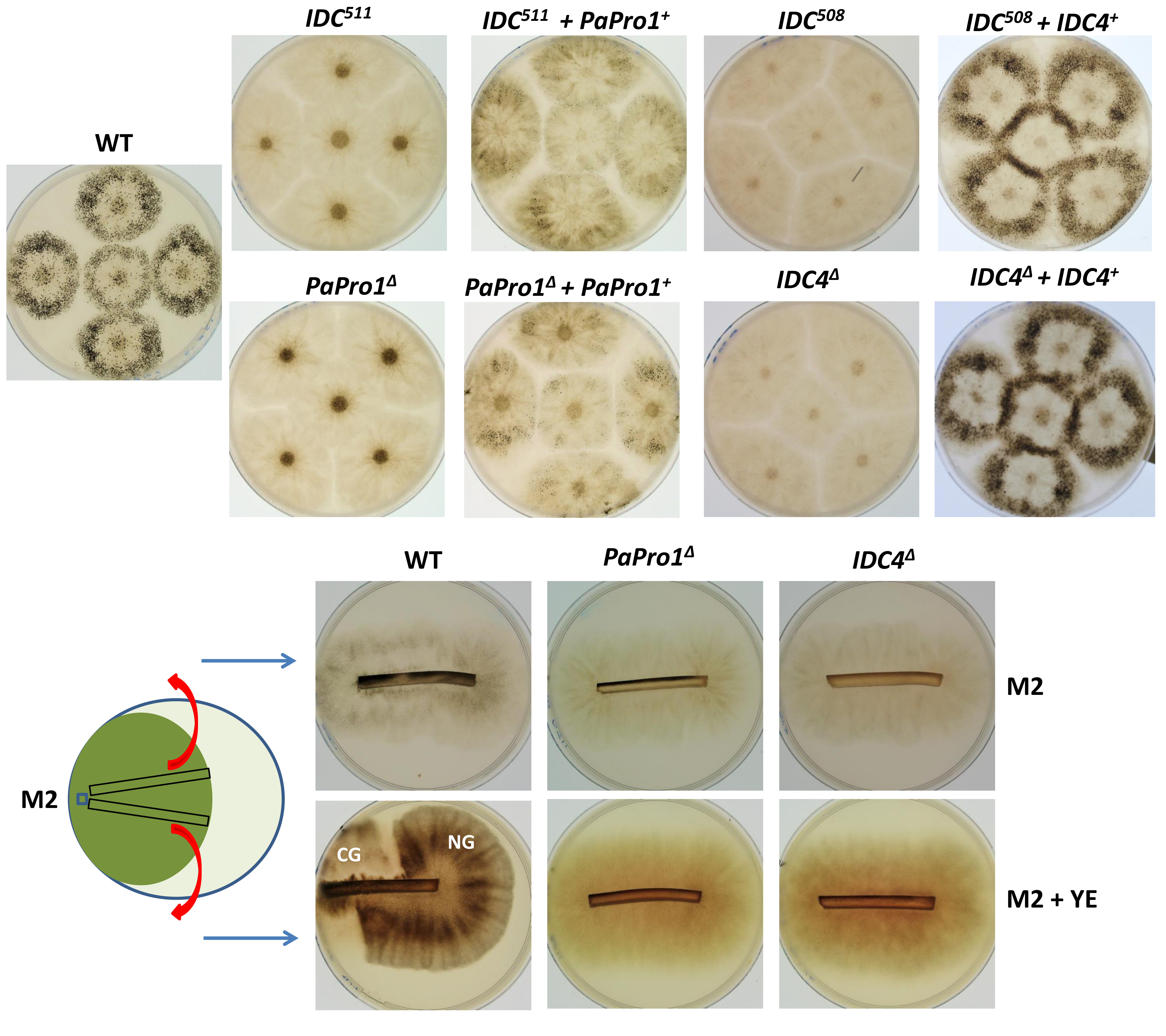
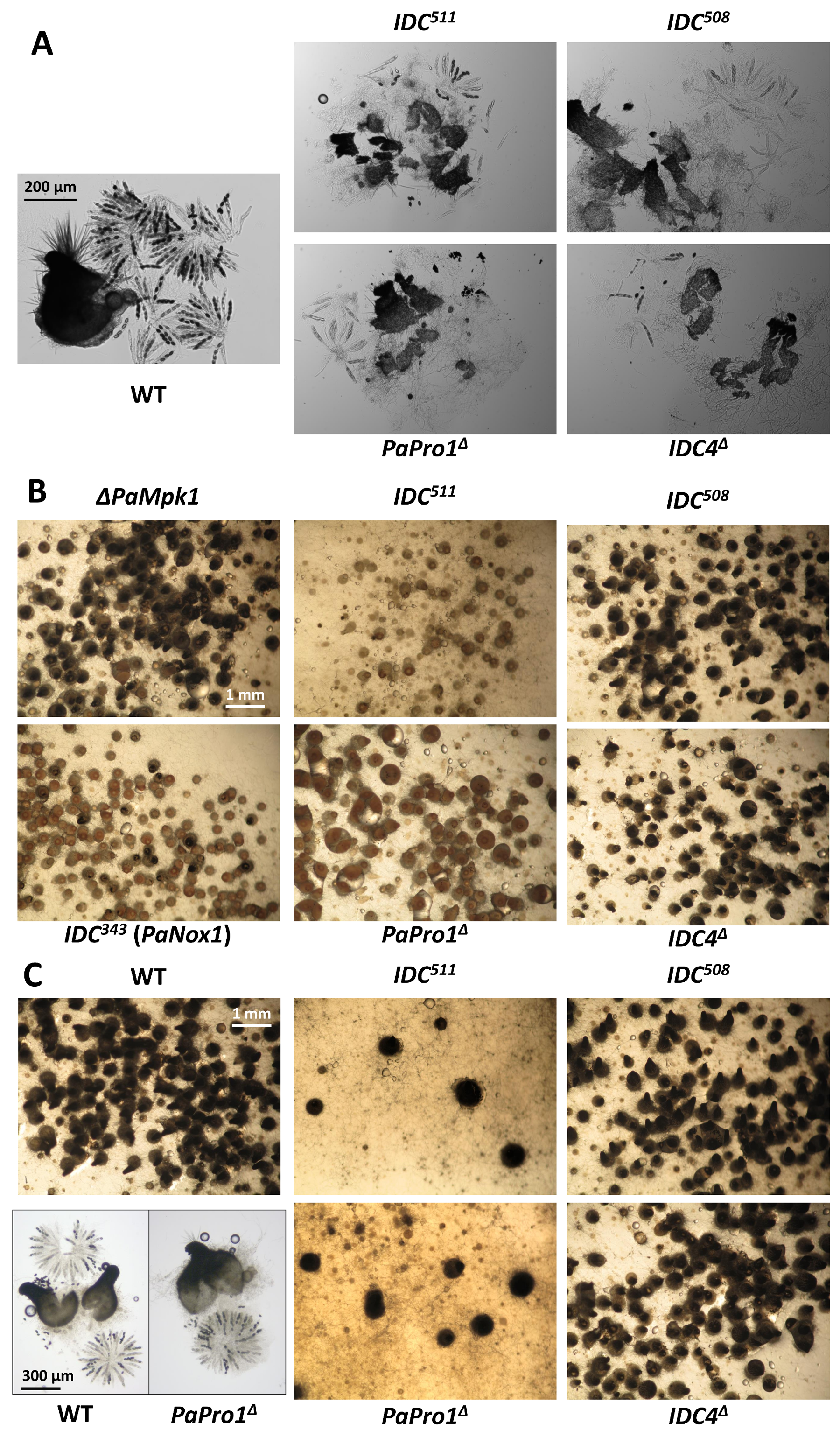
3.2. Inactivation of PaPro1 in IDC511 Creates the IDC Phenotype
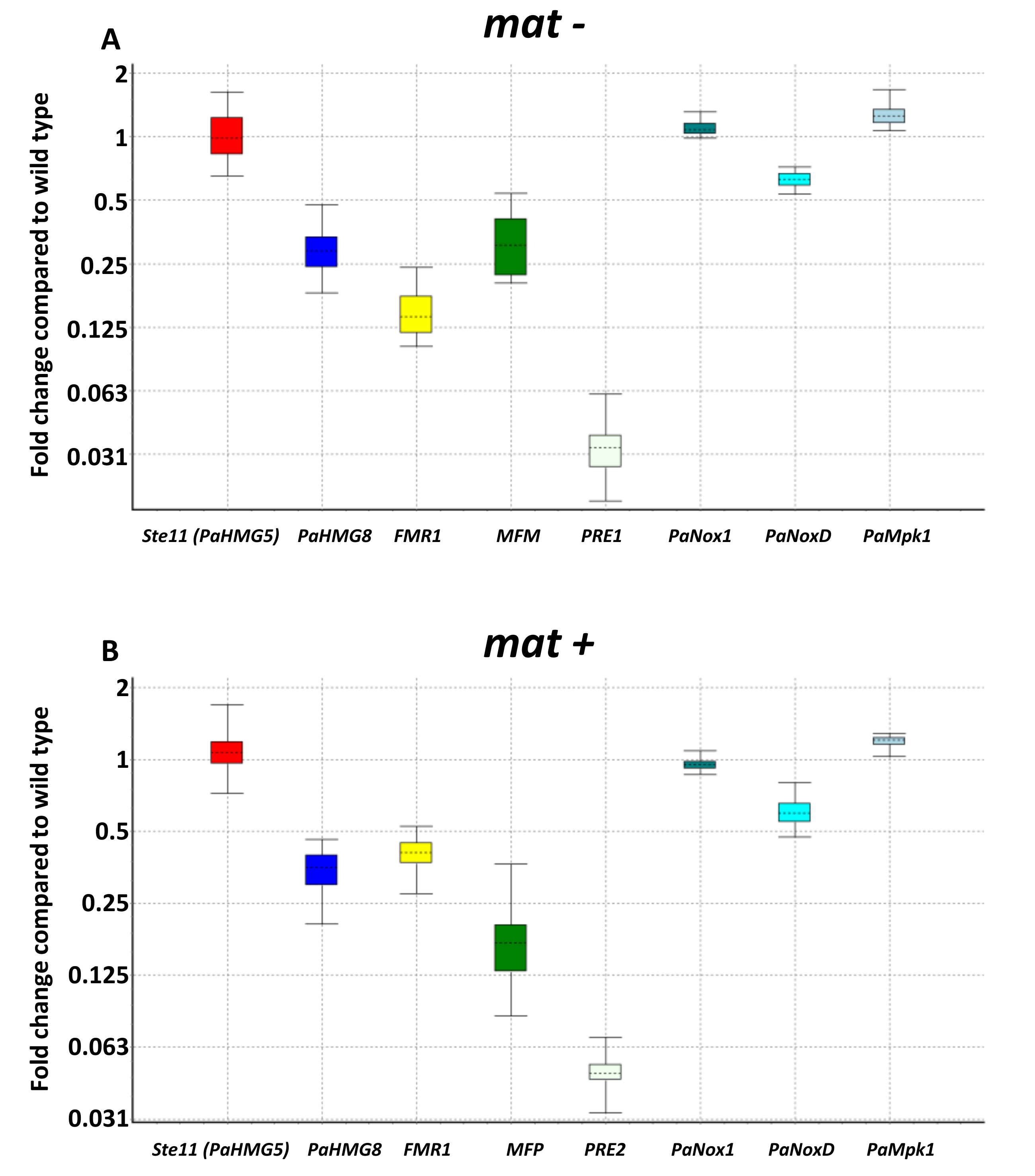
3.3. Transcription of PaPro1 Is Not Regulated by the MAPK Pathways
3.4. Expression of Selected Genes in the PaPro1Δ Mutant
3.5. Inactivation of IDC4, a Gene Coding for a Protein with an AIM24 Domain, Creates an IDC Phenotype
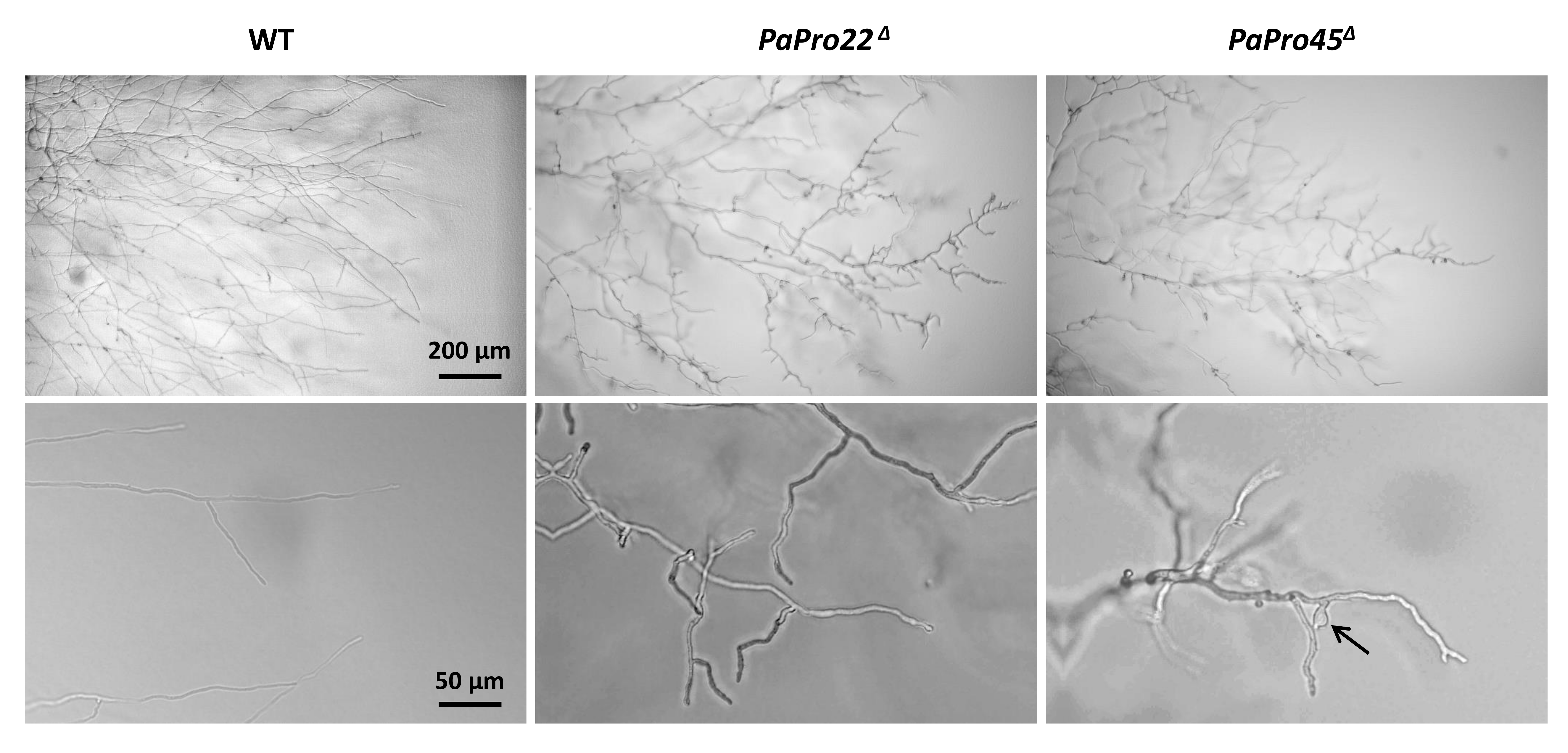
3.6. Fine Analysis of the IDC Phenotype of the IDC4 Mutants
3.7. Expression of IDC4
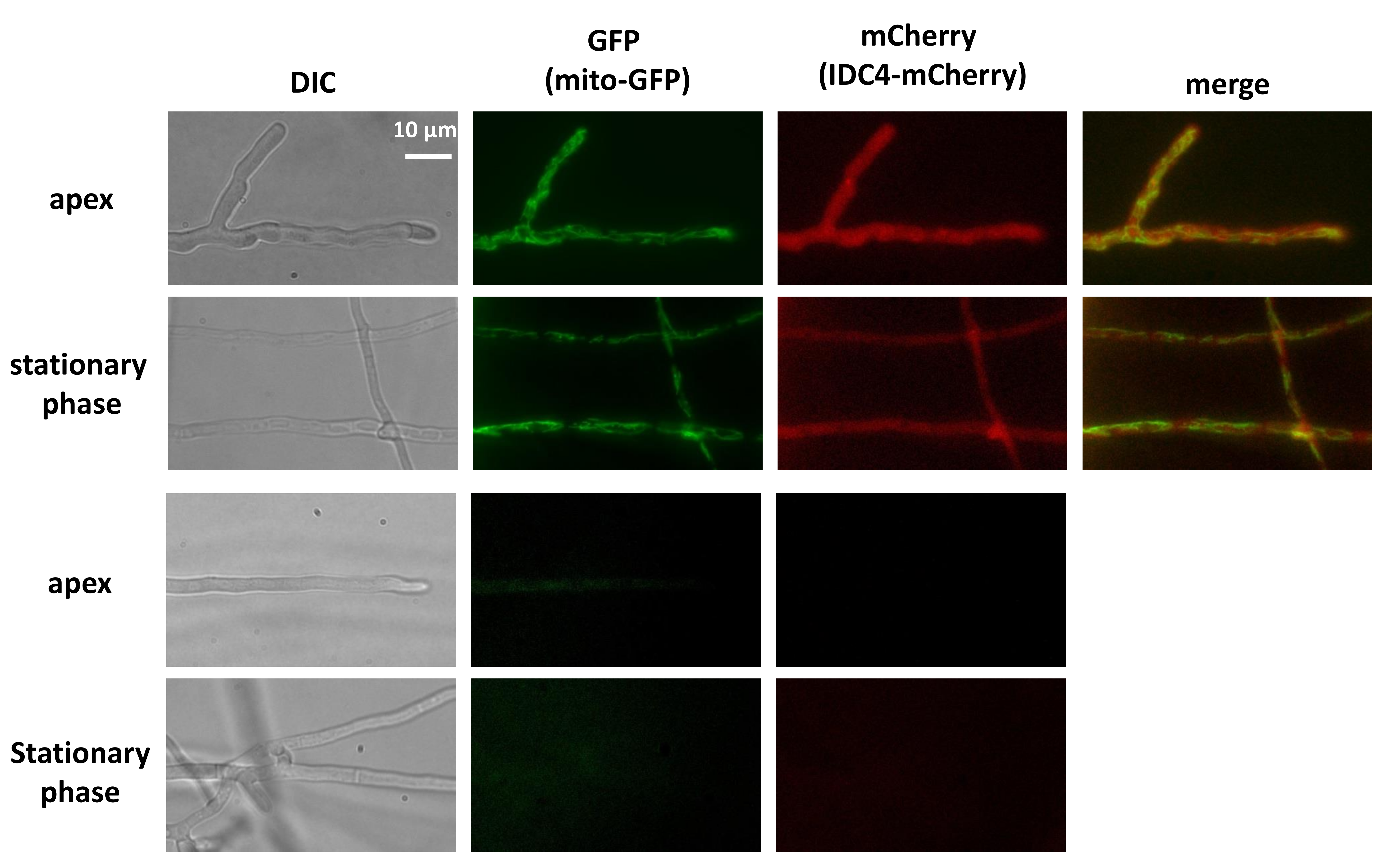
3.8. IDC4 is Localized in the Cytosol
3.9. Double Mutant Analyses Enable to Place PaPro1 and IDC4 Upstream of PDC1
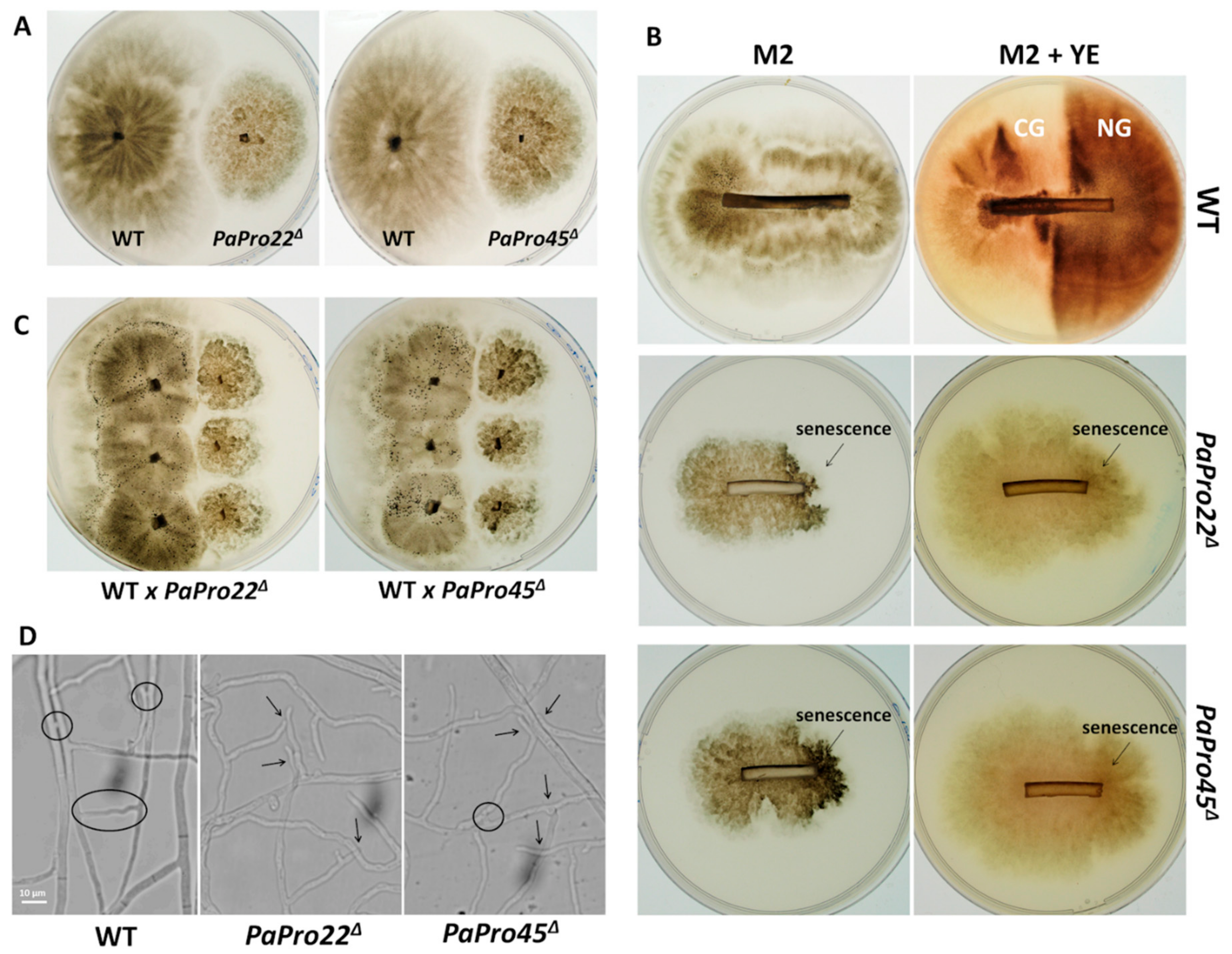
3.10. Mutants of the STRIPAK Complex Do Not Have a Typical “Pink” IDC Phenotype
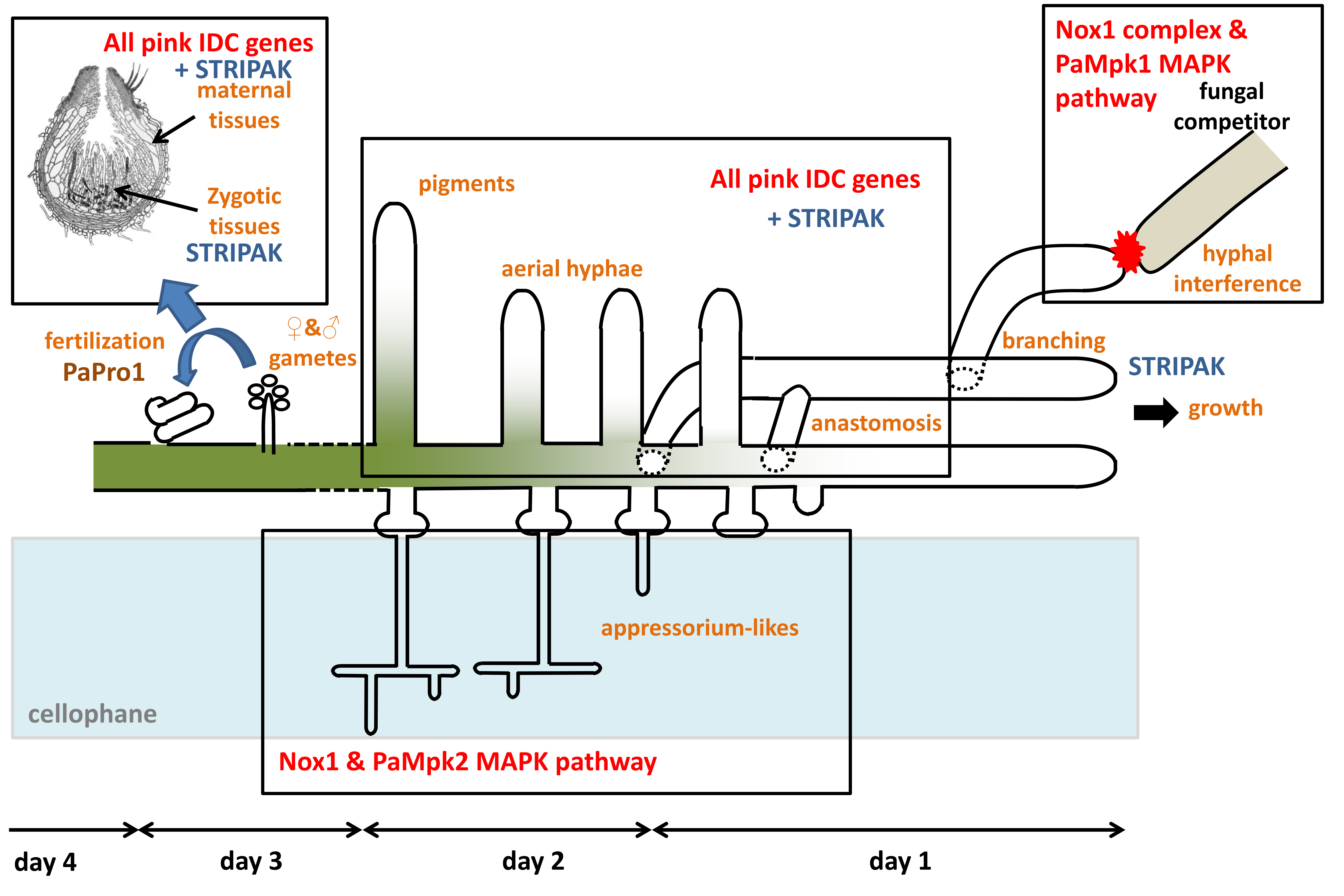
4. Discussion
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Silar, P.; Daboussi, M.J. Non-conventional infectious elements in filamentous fungi. Trends Genet. 1999, 15, 141–145. [Google Scholar] [CrossRef]
- Haedens, V.; Malagnac, F.; Silar, P. Genetic control of an epigenetic cell degeneration syndrome in Podospora anserina. Fungal Genet. Biol. 2005, 42, 564–577. [Google Scholar] [CrossRef] [PubMed]
- Silar, P.; Haedens, V.; Rossignol, M.; Lalucque, H. Propagation of a novel cytoplasmic, infectious and deleterious determinant is controlled by translational accuracy in Podospora anserina. Genetics 1999, 151, 87–95. [Google Scholar] [PubMed]
- Kicka, S.; Silar, P. PaASK1, a mitogen-activated protein kinase kinase kinase that controls cell degeneration and cell differentiation in Podospora anserina. Genetics 2004, 166, 1241–1252. [Google Scholar] [CrossRef] [PubMed]
- Malagnac, F.; Lalucque, H.; Lepere, G.; Silar, P. Two nadph oxidase isoforms are required for sexual reproduction and ascospore germination in the filamentous fungus Podospora anserina. Fungal Genet. Biol. 2004, 41, 982–997. [Google Scholar] [CrossRef] [PubMed]
- Kicka, S.; Bonnet, C.; Sobering, A.K.; Ganesan, L.P.; Silar, P. A mitotically inheritable unit containing a MAP kinase module. Proc. Natl. Acad. Sci. USA 2006, 103, 13445–13450. [Google Scholar] [CrossRef] [PubMed]
- Chan Ho Tong, L.; Silar, P.; Lalucque, H. Genetic control of anastomosis in Podospora anserina. Fungal Genet. Biol. 2014, 70C, 94–103. [Google Scholar] [CrossRef] [PubMed]
- Lacaze, I.; Lalucque, H.; Siegmund, U.; Silar, P.; Brun, S. Identification of NoxD/Pro41 as the homologue of the p22phoxNADPH oxidase subunit in fungi. Mol. Microbiol. 2015, 95, 1006–1024. [Google Scholar] [CrossRef] [PubMed]
- Brun, S.; Malagnac, F.; Bidard, F.; Lalucque, H.; Silar, P. Functions and regulation of the nox family in the filamentous fungus Podospora anserina: A new role in cellulose degradation. Mol. Microbiol. 2009, 74, 480–496. [Google Scholar] [CrossRef] [PubMed]
- Lalucque, H.; Malagnac, F.; Green, K.; Gautier, V.; Grognet, P.; Chan Ho Tong, L.; Scott, B.; Silar, P. IDC2 and IDC3, two genes involved in cell non-autonomous signaling of fruiting body development in the model fungus Podospora anserina. Dev. Biol. 2017, 421, 126–138. [Google Scholar] [CrossRef] [PubMed]
- Timpano, H.; Chan Ho Tong, L.; Gautier, V.; Lalucque, H.; Silar, P. The papsr1 and pawhi2 genes are members of the regulatory network that connect stationary phase to mycelium differentiation and reproduction in Podospora anserina. Fungal Genet. Biol. 2016, 94, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Lalucque, H.; Malagnac, F.; Brun, S.; Kicka, S.; Silar, P. A non-mendelian mapk-generated hereditary unit controlled by a second MAPK pathway in Podospora anserina. Genetics 2012, 191, 419–433. [Google Scholar] [CrossRef] [PubMed]
- Jamet-Vierny, C.; Debuchy, R.; Prigent, M.; Silar, P. IDC1, a pezizomycotina-specific gene that belongs to the PaMpk1 MAP kinase transduction cascade of the filamentous fungus Podospora anserina. Fungal Genet. Biol. 2007, 44, 1219–1230. [Google Scholar] [CrossRef] [PubMed]
- Jonkers, W.; Leeder, A.C.; Ansong, C.; Wang, Y.; Yang, F.; Starr, T.L.; Camp, D.G., II; Smith, R.D.; Glass, N.L. HAM-5 functions as a map kinase scaffold during cell fusion in Neurospora crassa. PLoS Genet. 2014, 10, e1004783. [Google Scholar] [CrossRef] [PubMed]
- Dettmann, A.; Heilig, Y.; Valerius, O.; Ludwig, S.; Seiler, S. Fungal communication requires the MAK-2 pathway elements STE-20 and RAS-2, the NRC-1 adapter STE-50 and the MAP kinase scaffold HAM-5. PLoS Genet. 2014, 10, e1004762. [Google Scholar] [CrossRef] [PubMed]
- Fleissner, A.; Glass, N.L. So, a protein involved in hyphal fusion in Neurospora crassa, localizes to septal plugs. Eukaryot. Cell 2007, 6, 84–94. [Google Scholar] [CrossRef] [PubMed]
- Fleissner, A.; Sarkar, S.; Jacobson, D.J.; Roca, M.G.; Read, N.D.; Glass, N.L. The so locus is required for vegetative cell fusion and postfertilization events in Neurospora crassa. Eukaryot. Cell 2005, 4, 920–930. [Google Scholar] [CrossRef] [PubMed]
- Teichert, I.; Steffens, E.K.; Schnaß, N.; Fränzel, B.; Krisp, C.; Wolters, D.A.; Kück, U. Pro40 is a scaffold protein of the cell wall integrity pathway, linking the MAP kinase module to the upstream activator protein kinase c. PLoS Genet. 2014, 10, e1004582. [Google Scholar] [CrossRef] [PubMed]
- Espagne, E.; Lespinet, O.; Malagnac, F.; Da Silva, C.; Jaillon, O.; Porcel, B.M.; Couloux, A.; Aury, J.M.; Segurens, B.; Poulain, J.; et al. The genome sequence of the model ascomycete fungus Podospora anserina. Genome Biol. 2008, 9, R77. [Google Scholar] [CrossRef] [PubMed]
- Grognet, P.; Bidard, F.; Kuchly, C.; Tong, L.C.; Coppin, E.; Benkhali, J.A.; Couloux, A.; Wincker, P.; Debuchy, R.; Silar, P. Maintaining two mating types: Structure of the mating type locus and its role in heterokaryosis in Podospora anserina. Genetics 2014, 197, 421–432. [Google Scholar] [CrossRef] [PubMed]
- Rizet, G.; Engelmann, C. Contribution à l'étude génétique d'un ascomycète tétrasporé : Podospora anserina (ces.) rehm. Rev. Cytol. Biol. Vég. 1949, 11, 201–304. [Google Scholar]
- Silar, P. Podospora anserina: From laboratory to biotechnology. In Genomics of Soil- and Plant-Associated Fungi; Horwitz, B., Mukherjee, P., Mukherjee, M., Kubicek, C., Eds.; Springer: Heidelberg, Germany; New York, NY, USA; Dordrecht, The Netherlands; London, UK, 2013; pp. 283–309. [Google Scholar]
- Silar, P. Two new easy-to-use vectors for transformations. Fungal Genet. Newsl. 1995, 42, 73. [Google Scholar] [CrossRef]
- Sellem, C.H.; Marsy, S.; Boivin, A.; Lemaire, C.; Sainsard-Chanet, A. A mutation in the gene encoding cytochrome c1 leads to a decreased ros content and to a long-lived phenotype in the filamentous fungus Podospora anserina. Fungal Genet. Biol. 2007, 44, 648–658. [Google Scholar] [CrossRef] [PubMed]
- Bidard, F.; Ait Benkhali, J.; Coppin, E.; Imbeaud, S.; Grognet, P.; Delacroix, H.; Debuchy, R. Genome-wide gene expression profiling of fertilization competent mycelium in opposite mating types in the heterothallic fungus Podospora anserina. PLoS ONE 2011, 6, e21476. [Google Scholar] [CrossRef] [PubMed]
- Ait Benkhali, J.; Coppin, E.; Brun, S.; Peraza-Reyes, L.; Martin, T.; Dixelius, C.; Lazar, N.; van Tilbeurgh, H.; Debuchy, R. A network of hmg-box transcription factors regulates sexual cycle in the fungus Podospora anserina. PLoS Genet. 2013, 9, e1003642. [Google Scholar] [CrossRef] [PubMed]
- Vandesompele, J.; De Preter, K.; Pattyn, F.; Poppe, B.; Van Roy, N.; De Paepe, A.; Speleman, F. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002, 3, R0034. [Google Scholar] [CrossRef]
- Pfaffl, M.W.; Horgan, G.W.; Dempfle, L. Relative expression software tool (rest) for group-wise comparison and statistical analysis of relative expression results in real-time PCR. Nucleic Acids Res. 2002, 30, e36. [Google Scholar] [CrossRef] [PubMed]
- Bustin, S.A.; Benes, V.; Garson, J.A.; Hellemans, J.; Huggett, J.; Kubista, M.; Mueller, R.; Nolan, T.; Pfaffl, M.W.; Shipley, G.L.; et al. The miqe guidelines: Minimum information for publication of quantitative real-time PCR experiments. Clin. Chem. 2009, 55, 611–622. [Google Scholar] [CrossRef] [PubMed]
- Katoh, K.; Kuma, K.-I.; Toh, H.; Miyata, T. Mafft version 5: Improvement in accuracy of multiple sequence alignment. Nucleic Acids Res. 2005, 33, 511–518. [Google Scholar] [CrossRef] [PubMed]
- Guindon, S.; Gascuel, O. Simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst. Biol. 2003, 52, 696–704. [Google Scholar] [CrossRef] [PubMed]
- Letunic, I.; Bork, P. Interactive tree of life (itol): An online tool for phylogenetic tree display and annotation. Bioinformatics 2007, 23, 127–128. [Google Scholar] [CrossRef] [PubMed]
- Masloff, S.; Pöggeler, S.; Kück, U. The pro1 gene from Sordaria macrospora encodes a C6 zinc finger transcription factor required for fruiting body development. Genetics 1999, 152, 191–199. [Google Scholar] [PubMed]
- Masloff, S.; Jacobsen, S.; Pöggeler, S.; Kück, U. Functional analysis of the C6 zinc finger gene pro1 involved in fungal sexual development. Fungal Genet. Biol. 2002, 36, 107–116. [Google Scholar] [CrossRef]
- Silar, P. Simple genetic tools to study fruiting body development in fungi. Open Mycol. 2014, 8, 148–155. [Google Scholar] [CrossRef]
- Silar, P. Peroxide accumulation and cell death in filamentous fungi induced by contact with a contestant. Mycol. Res. 2005, 109, 137–149. [Google Scholar] [CrossRef] [PubMed]
- Silar, P. Hyphal interference: Self versus non-self fungal recognition and hyphal death. In Biocommunication of Fungi; Witzany, G., Ed.; Springer: Dordrecht, The Netherlands, 2012. [Google Scholar]
- Bidard, F.; Coppin, E.; Silar, P. The transcriptional response to the inactivation of the PaMpk1 and PaMpk2 MAP kinase pathways in Podospora anserina. Fungal Genet. Biol. 2012, 49, 643–652. [Google Scholar] [CrossRef] [PubMed]
- Steffens, E.K.; Becker, K.; Krevet, S.; Teichert, I.; Kück, U. Transcription factor pro1 targets genes encoding conserved components of fungal developmental signaling pathways. Mol. Microbiol. 2016, 102, 792–809. [Google Scholar] [CrossRef] [PubMed]
- Teichert, I.; Wolff, G.; Kück, U.; Nowrousian, M. Combining laser microdissection and RNA-seq to chart the transcriptional landscape of fungal development. BMC Genom. 2012, 13, 511–511. [Google Scholar] [CrossRef] [PubMed]
- Nowrousian, M.; Frank, S.; Koers, S.; Strauch, P.; Weitner, T.; Ringelberg, C.; Dunlap, J.C.; Loros, J.J.; Kück, U. The novel er membrane protein pro41 is essential for sexual development in the filamentous fungus Sordaria macrospora. Mol. Microbiol. 2007, 64, 923–937. [Google Scholar] [CrossRef] [PubMed]
- Grant, C.E.; Bailey, T.L.; Noble, W.S. Fimo: Scanning for occurrences of a given motif. Bioinformatics 2011, 27, 1017–1018. [Google Scholar] [CrossRef] [PubMed]
- Fischer, M.S.; Wu, V.W.; Lee, J.E.; O’Malley, R.C.; Glass, N.L. Regulation of cell-to-cell communication and cell wall integrity by a network of map kinase pathways and transcription factors in Neurospora crassa. Genetics 2018, 209, 489. [Google Scholar] [CrossRef] [PubMed]
- Harner, M.E.; Unger, A.-K.; Izawa, T.; Walther, D.M.; Özbalci, C.; Geimer, S.; Reggiori, F.; Brügger, B.; Mann, M.; Westermann, B.; et al. Aim24 and MICOS modulate respiratory function, tafazzin-related cardiolipin modification and mitochondrial architecture. eLife 2014, 3, e01684. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, T.S.; Lalucque, H.; Silar, P. Identification and characterization of PDC1, a novel protein involved in the epigenetic cell degeneration crippled growth in Podospora anserina. Mol. Microbiol. 2018. in revision. [Google Scholar]
- Kück, U.; Beier, A.M.; Teichert, I. The composition and function of the striatin-interacting phosphatases and kinases (stripak) complex in fungi. Fungal Genet. Biol. 2016, 90, 31–38. [Google Scholar] [CrossRef] [PubMed]
- Bloemendal, S.; Lord, K.M.; Rech, C.; Hoff, B.; Engh, I.; Read, N.D.; Kück, U. A mutant defective in sexual development produces aseptate ascogonia. Eukaryot. Cell 2010, 9, 1856–1866. [Google Scholar] [CrossRef] [PubMed]
- Nordzieke, S.; Zobel, T.; Fränzel, B.; Wolters, D.A.; Kück, U.; Teichert, I. A fungal sarcolemmal membrane-associated protein (SLMAP) homolog plays a fundamental role in development and localizes to the nuclear envelope, endoplasmic reticulum, and mitochondria. Eukaryot. Cell 2015, 14, 345–358. [Google Scholar] [CrossRef] [PubMed]
- Fu, C.; Iyer, P.; Herkal, A.; Abdullah, J.; Stout, A.; Free, S.J. Identification and characterization of genes required for cell-to-cell fusion in Neurospora crassa. Eukaryot. Cell 2011, 10, 1100–1109. [Google Scholar] [CrossRef] [PubMed]
- Green, K.A.; Becker, Y.; Fitzsimons, H.L.; Scott, B. An Epichloë festucae homologue of MOB3, a component of the stripak complex, is required for the establishment of a mutualistic symbiotic interaction with lolium perenne. Mol. Plant Pathol. 2016, 17, 1480–1492. [Google Scholar] [CrossRef] [PubMed]
- Coppin, E.; Silar, P. Identification of PaPKS1, a polyketide synthase involved in melanin formation and its use as a genetic tool in Podospora anserina. Mycol. Res. 2007, 111, 901–908. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, T.S.; Lalucque, H.; Malagnac, F.; Silar, P. Prions and prion-like phenomena in epigenetic inheritance. In Handbook of Epigenetics, 2nd ed.; Tollefsbol, T., Ed.; Elsevier: New York, NY, USA, 2017; pp. 61–72. [Google Scholar]
- Colot, H.V.; Park, G.; Turner, G.E.; Ringelberg, C.; Crew, C.M.; Litvinkova, L.; Weiss, R.L.; Borkovich, K.A.; Dunlap, J.C. A high-throughput gene knockout procedure for Neurospora reveals functions for multiple transcription factors. Proc. Natl. Acad. Sci. USA 2006, 103, 10352–10357. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, A.; Cartwright, G.M.; Saikia, S.; Kayano, Y.; Takemoto, D.; Kato, M.; Tsuge, T.; Scott, B. Proa, a transcriptional regulator of fungal fruiting body development, regulates leaf hyphal network development in the Epichloë festucae-lolium perenne symbiosis. Mol. Microbiol. 2013, 90, 551–568. [Google Scholar] [CrossRef] [PubMed]
- Eaton, C.J.; Dupont, P.-Y.; Solomon, P.; Clayton, W.; Scott, B.; Cox, M.P. A core gene set describes the molecular basis of mutualism and antagonism in Epichloë spp. Mol. Plant-Microbe Interact. 2014, 28, 218–231. [Google Scholar] [CrossRef] [PubMed]
- Cho, Y.; Kim, K.-H.; La Rota, M.; Scott, D.; Santopietro, G.; Callihan, M.; Mitchell, T.K.; Lawrence, C.B. Identification of novel virulence factors associated with signal transduction pathways in Alternaria brassicicola. Mol. Microbiol. 2009, 72, 1316–1333. [Google Scholar] [CrossRef] [PubMed]
- Sun, Q.; Choi, G.H.; Nuss, D.L. Hypovirus-responsive transcription factor gene pro1 of the chestnut blight fungus Cryphonectria parasitica is required for female fertility, asexual spore development, and stable maintenance of hypovirus infection. Eukaryot. Cell 2009, 8, 262–270. [Google Scholar] [CrossRef] [PubMed]
- Son, H.; Seo, Y.-S.; Min, K.; Park, A.R.; Lee, J.; Jin, J.-M.; Lin, Y.; Cao, P.; Hong, S.-Y.; Kim, E.-K.; et al. A phenome-based functional analysis of transcription factors in the cereal head blight fungus, Fusarium graminearum. PLoS Pathog. 2011, 7, e1002310. [Google Scholar] [CrossRef] [PubMed]
- Vienken, K.; Scherer, M.; Fischer, R. The Zn(II)Cys6 putative Aspergillus nidulans transcription factor repressor of sexual development inhibits sexual development under low-carbon conditions and in submersed culture. Genetics 2005, 169, 619–630. [Google Scholar] [CrossRef] [PubMed]
- Vienken, K.; Fischer, R. The Zn(II)2Cys6 putative transcription factor nosa controls fruiting body formation in Aspergillus nidulans. Mol. Microbiol. 2006, 61, 544–554. [Google Scholar] [CrossRef] [PubMed]
- Soukup, A.A.; Farnoodian, M.; Berthier, E.; Keller, N.P. Nosa, a transcription factor important in Aspergillus fumigatus stress and developmental response, rescues the germination defect of a laea deletion. Fungal Genet. Biol. 2012, 49, 857–865. [Google Scholar] [CrossRef] [PubMed]
- Nowrousian, M.; Ringelberg, C.; Dunlap, J.C.; Loros, J.J.; Kück, U. Cross-species microarray hybridization to identify developmentally regulated genes in the filamentous fungus Sordaria macrospora. Mol. Genet. Genom. 2005, 273, 137–149. [Google Scholar] [CrossRef] [PubMed]
- Deckers, M.; Balleininger, M.; Vukotic, M.; Römpler, K.; Bareth, B.; Juris, L.; Dudek, J. Aim24 stabilizes respiratory chain supercomplexes and is required for efficient respiration. FEBS Lett. 2014, 588, 2985–2992. [Google Scholar] [CrossRef] [PubMed]
- Daskalov, A.; Heller, J.; Herzog, S.; Fleissner, A.; Glass, N.L. Molecular mechanisms regulating cell fusion and heterokaryon formation in filamentous fungi. Microbiol. Spectr. 2017, 5. [Google Scholar] [CrossRef] [PubMed]
- Teichert, I.; Nowrousian, M.; Pöggeler, S.; Kück, U. Chapter four—The filamentous fungus Sordaria macrospora as a genetic model to study fruiting body development. In Advances in Genetics; Theodore Friedmann, J.C.D., Stephen, F.G., Eds.; Academic Press: Cambridge, MA, USA, 2014; Volume 87, pp. 199–244. [Google Scholar]
- Turrà, D.; Segorbe, D.; Pietro, A.D. Protein kinases in plant-pathogenic fungi: Conserved regulators of infection. Annu. Rev. Phytopathol. 2014, 52, 267–288. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Zhu, J.; Ying, S.-H.; Feng, M.-G. Three mitogen-activated protein kinases required for cell wall integrity contribute greatly to biocontrol potential of a fungal entomopathogen. PLoS ONE 2014, 9, e87948. [Google Scholar] [CrossRef] [PubMed]
- Becker, Y.; Eaton, C.J.; Brasell, E.; May, K.J.; Becker, M.; Hassing, B.; Cartwright, G.M.; Reinhold, L.; Scott, B. The fungal cell-wall integrity mapk cascade is crucial for hyphal network formation and maintenance of restrictive growth of Epichloë festucae in symbiosis with lolium perenne. Mol. Plant-Microbe Interact. 2014, 28, 69–85. [Google Scholar] [CrossRef] [PubMed]
- Green, K.A.; Becker, Y.; Tanaka, A.; Takemoto, D.; Fitzsimons, H.L.; Seiler, S.; Lalucque, H.; Silar, P.; Scott, B. SymB and SymC, two membrane associated proteins, are required for Epichloë festucae hyphal cell–cell fusion and maintenance of a mutualistic interaction with lolium perenne. Mol. Microbiol. 2017, 103, 657–677. [Google Scholar] [CrossRef] [PubMed]
- Egan, M.J.; Wang, Z.Y.; Jones, M.A.; Smirnoff, N.; Talbot, N.J. Generation of reactive oxygen species by fungal nadph oxidases is required for rice blast disease. Proc. Natl. Acad. Sci. USA 2007, 104, 11772–11777. [Google Scholar] [CrossRef] [PubMed]
- Siegmund, U.; Heller, J.; van Kann, J.A.; Tudzynski, P. The nadph oxidase complexes in Botrytis cinerea: Evidence for a close association with the er and the tetraspanin Pls1. PLoS ONE 2013, 8, e55879. [Google Scholar] [CrossRef] [PubMed]
- Herzog, S.; Schumann, M.R.; Fleißner, A. Cell fusion in Neurospora crassa. Curr. Opin. Microbiol. 2015, 28, 53–59. [Google Scholar] [CrossRef] [PubMed]
- Beier, A.; Teichert, I.; Krisp, C.; Wolters, D.A.; Kück, U. Catalytic subunit 1 of protein phosphatase 2A is a subunit of the stripak complex and governs fungal sexual development. mBio 2016, 7. [Google Scholar] [CrossRef] [PubMed]
- Dettmann, A.; Heilig, Y.; Ludwig, S.; Schmitt, K.; Illgen, J.; Fleissner, A.; Valerius, O.; Seiler, S. HAM-2 and HAM-3 are central for the assembly of the Neurospora stripak complex at the nuclear envelope and regulate nuclear accumulation of the MAP kinase MAK-1 in a MAK-2-dependent manner. Mol. Microbiol. 2013, 90, 796–812. [Google Scholar] [CrossRef] [PubMed]




| Genes | Promoter Size | p = 0.001 | p = 0.0001 |
|---|---|---|---|
| Ste11 | 3216 | 2 (0) | 0 |
| PaHGM8 | 1506 | 4 (2) | 2 (1) |
| FMR1 | 480 | 0 | 0 |
| MFM | 1974 | 1 (1) | 0 |
| PRE1 | 1054 | 5 (5) | 0 |
| FPR1 | 3945 | 4 (4) | 1 (1) |
| MFP | 1011 | 0 | 0 |
| PRE2 | 1236 | 1 (1) | 0 |
| PaNoxD | 3774 | 12 (3) | 0 |
| PaNox1 | 3630 | 10 (3) | 3 (1) |
| PaMpk1 | 1704 | 5 (3) | 1 (1) |
| Gene | Id from the Genome Project | Available Mutants | Function | Ref. | N. crassa (S. macrospora) Orthologue | Site of Action during Perithecium Development | Hyphal Interference | Appressorium-Like | Fertility on Paper | Additional Phenotype |
|---|---|---|---|---|---|---|---|---|---|---|
| PaASK1 | Pa_5_9370 | IDC118, IDC172, IDC507 | MAPKKK | [4] | mik1 (mik1) | mycelium | − | + | − | |
| PaMKK1 | Pa_7_10270 | IDC404, IDC505 | MAPKK | [6] | mek-1 (mek1) | mycelium | − | + | − | |
| PaMpk1 | Pa_2_13340 | ΔPaMpk1 | MAPK | [6] | mak-1 (mak1) | mycelium | − | + | − | |
| PaSo | Pa_1_7440 | IDC821, PaSoΔ | MAPK1 scaffold? | [7] | So = Ham-1 (pro40) | peridium | +/− | + | − | |
| PaTLK2 | Pa_7_8030 | IDC510, ΔPaTLK2 | MAPKKK | [12], this paper | NRC-1 | mycelium | + | − | − | no ascospore germination |
| PaMKK2 | Pa_2_820 | ΔPaMKK2 | MAPKK | [12] | mek-2 | mycelium | + | − | − | no ascospore germination |
| PaMpk2 | Pa_5_5680 | ΔPaMpk2 | MAPK | [12] | mak-2 | mycelium | + | − | − | no ascospore germination |
| IDC1 | Pa_3_8520 | IDC1, IDC318, IDC502 | MAPK2 scaffold? | [13] | Ham-5 | mycelium and peridium | +/− | + | − | |
| PaNox1 | Pa_1_2410 | IDC343 | Nox catalytic | [5] | nox-1 (nox1) | peridium | +/− | − | +/− | |
| PaNoxD | Pa_1_7250 | IDC509, PaNoxDΔ | Nox docking | [8] | Ham-6 (pro41) | peridium | +/− | − | +/− | |
| PaNoxR | Pa_7_11300 | IDC524, ΔPaNoxR | Nox regulator | [9] | NOR-1 (nor1) | peridium | +/− | − | - | no ascospore germination |
| IDC2 | Pa_1_16080 | IDC506, IDC519, IDC3Δ | ? GPI anchored | [10] | Ham-7 | diffusible | + | + | +/− | |
| IDC3 | Pa_1_1990 | IDC522, IDC3Δ | ? transmembrane | [10] | Not yet studied | diffusible | + | + | +/− | |
| IDC4 | Pa_2_230 | IDC508, IDC4Δ | ? cytosolic | this paper | NCU04645 | mycelium | +/− | + | − | |
| PaPsr1 | Pa_1_3870 | scle1, PaPsr1Δ | ? | [11] | psr-1 | peridium | + | + | − | |
| PaWhi2 | Pa_4_7330 | IDC815, PaWhi2Δ | Phosphatase? | [11] | whi-2 | peridium | + | + | − | |
| PaPro1 | Pa_1_10140 | IDC511, PaPro1Δ | Transcription factor | this paper | ADV-1 (pro1) | mycelium and peridium (+ fertilization) | +/− | + | − |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gautier, V.; Tong, L.C.H.; Nguyen, T.-S.; Debuchy, R.; Silar, P. PaPro1 and IDC4, Two Genes Controlling Stationary Phase, Sexual Development and Cell Degeneration in Podospora anserina. J. Fungi 2018, 4, 85. https://doi.org/10.3390/jof4030085
Gautier V, Tong LCH, Nguyen T-S, Debuchy R, Silar P. PaPro1 and IDC4, Two Genes Controlling Stationary Phase, Sexual Development and Cell Degeneration in Podospora anserina. Journal of Fungi. 2018; 4(3):85. https://doi.org/10.3390/jof4030085
Chicago/Turabian StyleGautier, Valérie, Laetitia Chan Ho Tong, Tinh-Suong Nguyen, Robert Debuchy, and Philippe Silar. 2018. "PaPro1 and IDC4, Two Genes Controlling Stationary Phase, Sexual Development and Cell Degeneration in Podospora anserina" Journal of Fungi 4, no. 3: 85. https://doi.org/10.3390/jof4030085
APA StyleGautier, V., Tong, L. C. H., Nguyen, T.-S., Debuchy, R., & Silar, P. (2018). PaPro1 and IDC4, Two Genes Controlling Stationary Phase, Sexual Development and Cell Degeneration in Podospora anserina. Journal of Fungi, 4(3), 85. https://doi.org/10.3390/jof4030085






