The Candida albicans Biofilm Matrix: Composition, Structure and Function
Abstract
:1. Introduction
2. Composition of the C. albicans Biofilm Matrix
2.1. Extraction Techniques for the Isolation of the C. albicans Biofilm Matrix
2.2. Compositional Analyses of the C. albicans Biofilm Matrix
2.2.1. Proteins
2.2.2. Carbohydrates
2.2.3. Lipids
2.2.4. Nucleic Acids
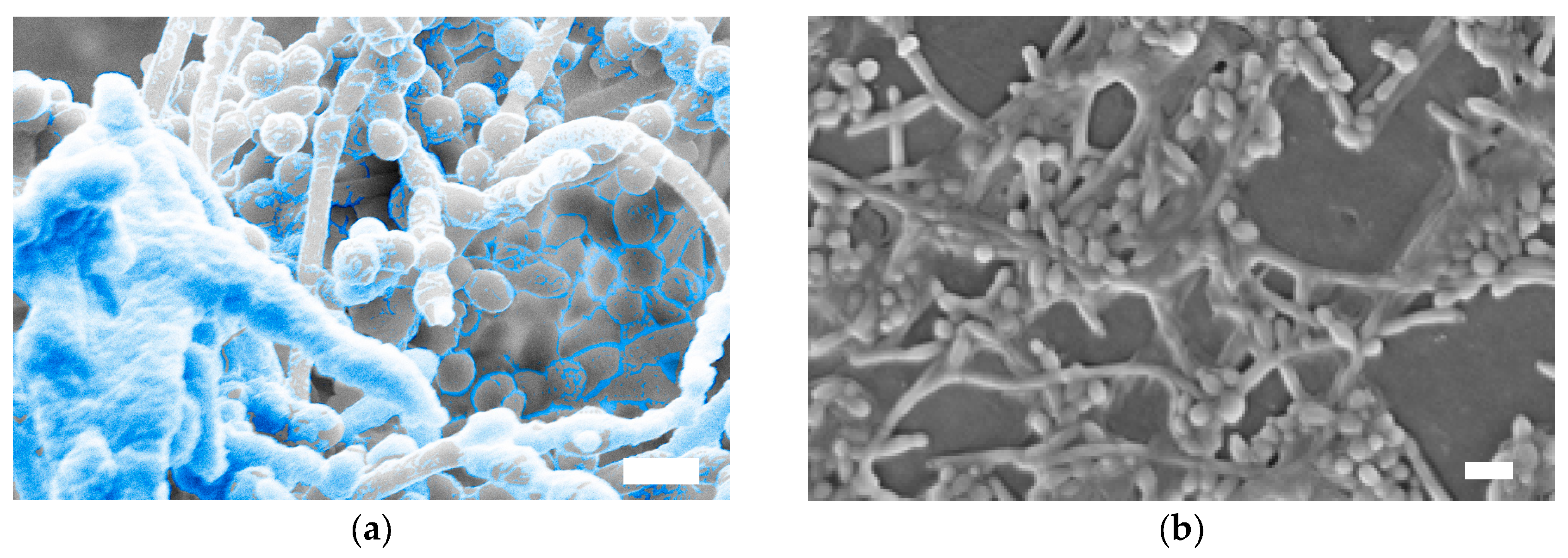
3. Structure of the C. albicans Biofilm Matrix
4. Functions of the C. albicans Biofilm Matrix
4.1. Mechanical Stability, Adhesive and Cohesive Interactions
4.2. Antifungal Drug Resistance
4.3. Protection from the Host Immune System
4.4. Role of the C. albicans Biofilm Matrix in Mixed Fungal/Bacterial Biofilms
5. Conclusions and Future Outlook
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Pfaller, M.A.; Diekema, D.J. Epidemiology of invasive candidiasis: A persistent public health problem. Clin. Microbiol. Rev. 2007, 20, 133–163. [Google Scholar] [CrossRef] [PubMed]
- Ramage, G.; Martinez, J.P.; Lopez-Ribot, J.L. Candida biofilms on implanted biomaterials: A clinically significant problem. FEMS Yeast Res. 2006, 6, 979–986. [Google Scholar] [CrossRef] [PubMed]
- Ramage, G.; Mowat, E.; Jones, B.; Williams, C.; Lopez-Ribot, J. Our current understanding of fungal biofilms. Crit. Rev. Microbiol. 2009, 35, 340–355. [Google Scholar] [CrossRef] [PubMed]
- Ramage, G.; Saville, S.P.; Thomas, D.P.; Lopez-Ribot, J.L. Candida biofilms: An update. Eukaryot. Cell 2005, 4, 633–638. [Google Scholar] [CrossRef] [PubMed]
- Mitchell, K.F.; Zarnowski, R.; Andes, D.R. Fungal super glue: The biofilm matrix and its composition, assembly, and functions. PLoS Pathog. 2016, 12, e1005828. [Google Scholar] [CrossRef] [PubMed]
- Al-Fattani, M.A.; Douglas, L.J. Biofilm matrix of Candida albicans and Candida tropicalis: Chemical composition and role in drug resistance. J. Med. Microbiol. 2006, 55, 999–1008. [Google Scholar] [CrossRef] [PubMed]
- Baillie, G.S.; Douglas, L.J. Matrix polymers of Candida biofilms and their possible role in biofilm resistance to antifungal agents. J. Antimicrob. Chemother. 2000, 46, 397–403. [Google Scholar] [CrossRef] [PubMed]
- Thomas, D.P.; Bachmann, S.P.; Lopez-Ribot, J.L. Proteomics for the analysis of the Candida albicans biofilm lifestyle. Proteomics 2006, 6, 5795–5804. [Google Scholar] [CrossRef] [PubMed]
- Zarnowski, R.; Sanchez, H.; Andes, D.R. Large-scale production and isolation of Candida biofilm extracellular matrix. Nat. Protoc. 2016, 11, 2320–2327. [Google Scholar] [CrossRef] [PubMed]
- Zarnowski, R.; Westler, W.M.; Lacmbouh, G.A.; Marita, J.M.; Bothe, J.R.; Bernhardt, J.; Lounes-Hadj Sahraoui, A.; Fontaine, J.; Sanchez, H.; Hatfield, R.D.; et al. Novel entries in a fungal biofilm matrix encyclopedia. mBio 2014, 5, e01333-14. [Google Scholar] [CrossRef] [PubMed]
- Lattif, A.A.; Mukherjee, P.K.; Chandra, J.; Roth, M.R.; Welti, R.; Rouabhia, M.; Ghannoum, M.A. Lipidomics of Candida albicans biofilms reveals phase-dependent production of phospholipid molecular classes and role for lipid rafts in biofilm formation. Microbiology 2011, 157, 3232–3242. [Google Scholar] [CrossRef] [PubMed]
- Chaffin, W.L.; Lopez-Ribot, J.L.; Casanova, M.; Gozalbo, D.; Martinez, J.P. Cell wall and secreted proteins of Candida albicans: Identification, function, and expression. Microbiol. Mol. Biol. Rev. 1998, 62, 130–180. [Google Scholar] [PubMed]
- Nombela, C.; Gil, C.; Chaffin, W.L. Non-conventional protein secretion in yeast. Trends Microbiol. 2006, 14, 15–21. [Google Scholar] [CrossRef] [PubMed]
- Uppuluri, P.; Chaturvedi, A.K.; Srinivasan, A.; Banerjee, M.; Ramasubramaniam, A.K.; Kohler, J.R.; Kadosh, D.; Lopez-Ribot, J.L. Dispersion as an important step in the Candida albicans biofilm developmental cycle. PLoS Pathog. 2010, 6, e1000828. [Google Scholar] [CrossRef] [PubMed]
- Nett, J.; Lincoln, L.; Marchillo, K.; Massey, R.; Holoyda, K.; Hoff, B.; VanHandel, M.; Andes, D. Putative role of β-1,3 glucans in Candida albicans biofilm resistance. Antimicrob. Agents Chemother. 2007, 51, 510–520. [Google Scholar] [CrossRef] [PubMed]
- Nett, J.E.; Crawford, K.; Marchillo, K.; Andes, D.R. Role of fks1p and matrix glucan in Candida albicans biofilm resistance to an echinocandin, pyrimidine, and polyene. Antimicrob. Agents Chemother. 2010, 54, 3505–3508. [Google Scholar] [CrossRef] [PubMed]
- Nett, J.E.; Sanchez, H.; Cain, M.T.; Andes, D.R. Genetic basis of Candida biofilm resistance due to drug-sequestering matrix glucan. J. Infect. Dis. 2010, 202, 171–175. [Google Scholar] [CrossRef] [PubMed]
- Taff, H.T.; Nett, J.E.; Zarnowski, R.; Ross, K.M.; Sanchez, H.; Cain, M.T.; Hamaker, J.; Mitchell, A.P.; Andes, D.R. A Candida biofilm-induced pathway for matrix glucan delivery: Implications for drug resistance. PLoS Pathog. 2012, 8, e1002848. [Google Scholar] [CrossRef] [PubMed]
- Alem, M.A.; Douglas, L.J. Effects of aspirin and other nonsteroidal anti-inflammatory drugs on biofilms and planktonic cells of Candida albicans. Antimicrob. Agents Chemother. 2004, 48, 41–47. [Google Scholar] [CrossRef] [PubMed]
- Flemming, H.C.; Wingender, J. The biofilm matrix. Nat. Rev. Microbiol. 2010, 8, 623–633. [Google Scholar] [CrossRef] [PubMed]
- Martins, M.; Uppuluri, P.; Thomas, D.P.; Cleary, I.A.; Henriques, M.; Lopez-Ribot, J.L.; Oliveira, R. Presence of extracellular DNA in the Candida albicans biofilm matrix and its contribution to biofilms. Mycopathologia 2010, 169, 323–331. [Google Scholar] [CrossRef] [PubMed]
- Mitchell, K.F.; Zarnowski, R.; Sanchez, H.; Edward, J.A.; Reinicke, E.L.; Nett, J.E.; Mitchell, A.P.; Andes, D.R. Community participation in biofilm matrix assembly and function. Proc. Natl. Acad. Sci. USA 2015, 112, 4092–4097. [Google Scholar] [CrossRef] [PubMed]
- Ramage, G.; Wickes, B.L.; Lopez-Ribot, J.L. A seed and feed model for the formation of Candida albicans biofilms under flow conditions using an improved modified robbins device. Rev. Iberoam. Micol. 2008, 25, 37–40. [Google Scholar] [CrossRef]
- Uppuluri, P.; Chaturvedi, A.K.; Lopez-Ribot, J.L. Design of a simple model of Candida albicans biofilms formed under conditions of flow: Development, architecture, and drug resistance. Mycopathologia 2009, 168, 101–109. [Google Scholar] [CrossRef] [PubMed]
- Ramage, G.; Rajendran, R.; Sherry, L.; Williams, C. Fungal biofilm resistance. Int. J. Microbiol. 2012, 2012, 528521. [Google Scholar] [CrossRef] [PubMed]
- Taff, H.T.; Mitchell, K.F.; Edward, J.A.; Andes, D.R. Mechanisms of Candida biofilm drug resistance. Future Microbiol. 2013, 8, 1325–1337. [Google Scholar] [CrossRef] [PubMed]
- Ramage, G.; Bachmann, S.; Patterson, T.F.; Wickes, B.L.; Lopez-Ribot, J.L. Investigation of multidrug efflux pumps in relation to fluconazole resistance in Candida albicans biofilms. J. Antimicrob. Chemother. 2002, 49, 973–980. [Google Scholar] [CrossRef] [PubMed]
- Vediyappan, G.; Rossignol, T.; d’Enfert, C. Interaction of Candida albicans biofilms with antifungals: Transcriptional response and binding of antifungals to beta-glucans. Antimicrob. Agents Chemother. 2010, 54, 2096–2111. [Google Scholar] [CrossRef] [PubMed]
- Martins, M.; Henriques, M.; Lopez-Ribot, J.L.; Oliveira, R. Addition of dnase improves the in vitro activity of antifungal drugs against Candida albicans biofilms. Mycoses 2012, 55, 80–85. [Google Scholar] [CrossRef] [PubMed]
- Srikantha, T.; Daniels, K.J.; Pujol, C.; Kim, E.; Soll, D.R. Identification of genes upregulated by the transcription factor bcr1 that are involved in impermeability, impenetrability, and drug resistance of Candida albicans a/α biofilms. Eukaryot. Cell 2013, 12, 875–888. [Google Scholar] [CrossRef] [PubMed]
- Xie, Z.; Thompson, A.; Sobue, T.; Kashleva, H.; Xu, H.; Vasilakos, J.; Dongari-Bagtzoglou, A. Candida albicans biofilms do not trigger reactive oxygen species and evade neutrophil killing. J. Infect. Dise. 2012, 206, 1936–1945. [Google Scholar] [CrossRef] [PubMed]
- Johnson, C.J.; Cabezas-Olcoz, J.; Kernien, J.F.; Wang, S.X.; Beebe, D.J.; Huttenlocher, A.; Ansari, H.; Nett, J.E. The extracellular matrix of Candida albicans biofilms impairs formation of neutrophil extracellular traps. PLoS Pathog. 2016, 12, e1005884. [Google Scholar] [CrossRef] [PubMed]
- Fox, E.P.; Cowley, E.S.; Nobile, C.J.; Hartooni, N.; Newman, D.K.; Johnson, A.D. Anaerobic bacteria grow within Candida albicans biofilms and induce biofilm formation in suspension cultures. Curr. Biol. 2014, 24, 2411–2416. [Google Scholar] [CrossRef] [PubMed]
- Kong, E.F.; Tsui, C.; Kucharikova, S.; Andes, D.; van Dijck, P.; Jabra-Rizk, M.A. Commensal protection of Staphylococcus aureus against antimicrobials by Candida albicans biofilm matrix. mBio 2016, 7. [Google Scholar] [CrossRef] [PubMed]
- De Brucker, K.; Tan, Y.; Vints, K.; De Cremer, K.; Braem, A.; Verstraeten, N.; Michiels, J.; Vleugels, J.; Cammue, B.P.; Thevissen, K. Fungal β-1,3-glucan increases ofloxacin tolerance of Escherichia coli in a polymicrobial E. coli/Candida albicans biofilm. Antimicrob. Agents Chemother. 2015, 59, 3052–3058. [Google Scholar] [CrossRef] [PubMed]
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license ( http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pierce, C.G.; Vila, T.; Romo, J.A.; Montelongo-Jauregui, D.; Wall, G.; Ramasubramanian, A.; Lopez-Ribot, J.L. The Candida albicans Biofilm Matrix: Composition, Structure and Function. J. Fungi 2017, 3, 14. https://doi.org/10.3390/jof3010014
Pierce CG, Vila T, Romo JA, Montelongo-Jauregui D, Wall G, Ramasubramanian A, Lopez-Ribot JL. The Candida albicans Biofilm Matrix: Composition, Structure and Function. Journal of Fungi. 2017; 3(1):14. https://doi.org/10.3390/jof3010014
Chicago/Turabian StylePierce, Christopher G., Taissa Vila, Jesus A. Romo, Daniel Montelongo-Jauregui, Gina Wall, Anand Ramasubramanian, and Jose L. Lopez-Ribot. 2017. "The Candida albicans Biofilm Matrix: Composition, Structure and Function" Journal of Fungi 3, no. 1: 14. https://doi.org/10.3390/jof3010014
APA StylePierce, C. G., Vila, T., Romo, J. A., Montelongo-Jauregui, D., Wall, G., Ramasubramanian, A., & Lopez-Ribot, J. L. (2017). The Candida albicans Biofilm Matrix: Composition, Structure and Function. Journal of Fungi, 3(1), 14. https://doi.org/10.3390/jof3010014








