Integrative Genomic and Transcriptomic Analysis of White-Rot Fungi Ganoderma tsugae Growing on Both Coniferous and Broad-Leaved Trees
Abstract
1. Introduction
2. Materials and Methods
2.1. Fungal Strains and Culture Conditions
2.2. Genome Sequencing, Assembly, and Annotation
2.3. Transcriptome Sequencing and Annotation
3. Results
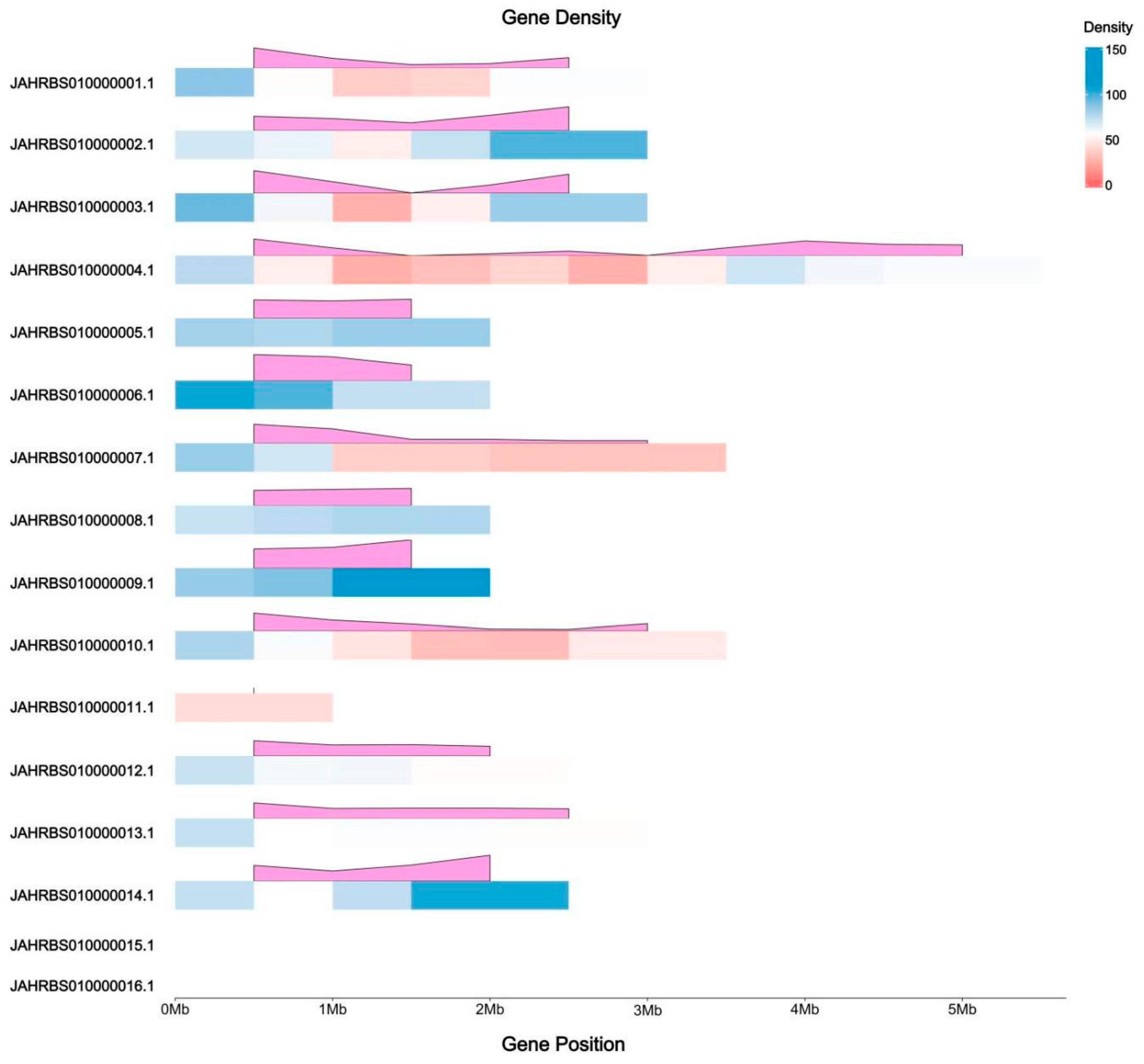
3.1. Whole-Genome Sequencing and Assembly
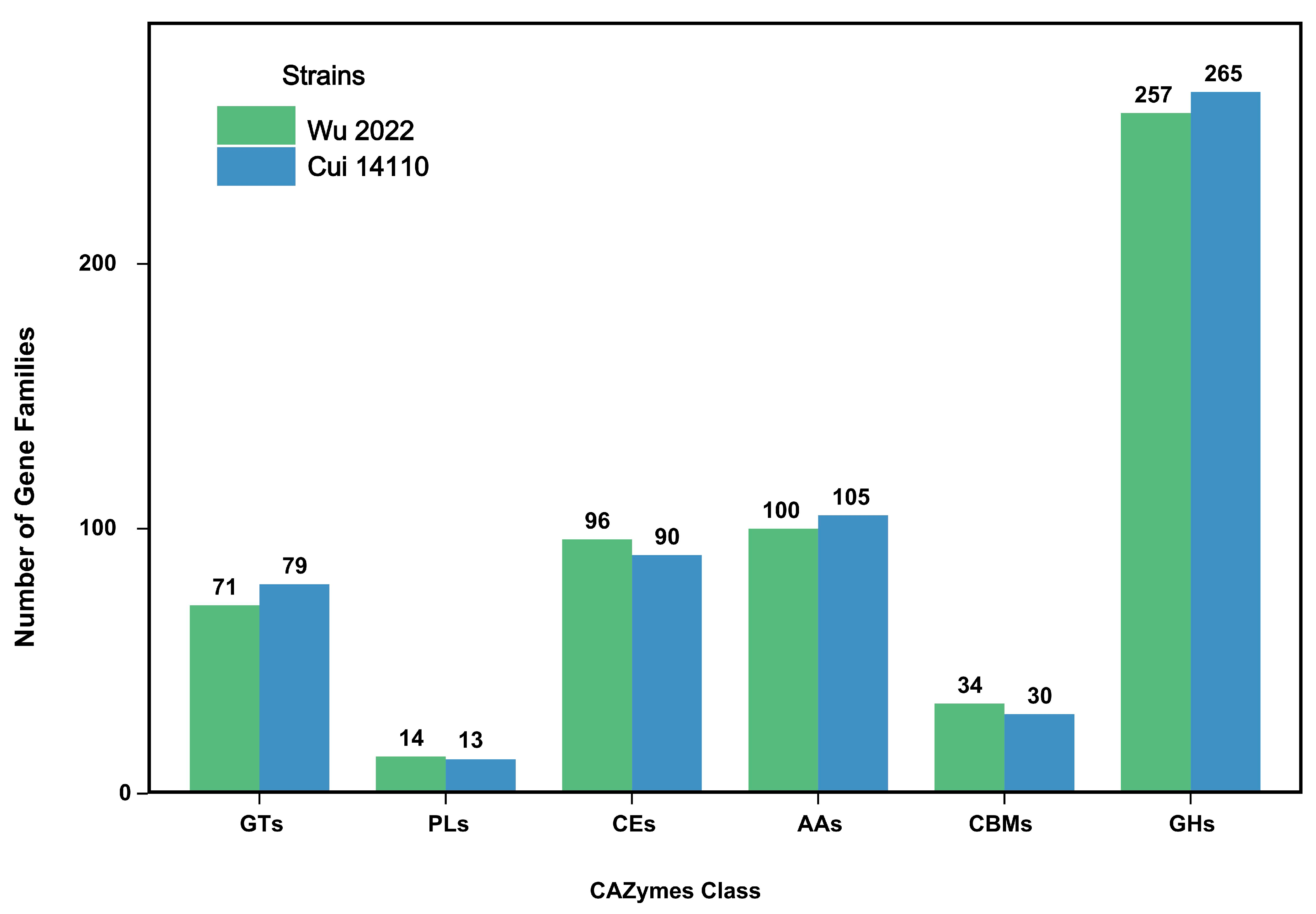
3.2. Genome Functional Annotation
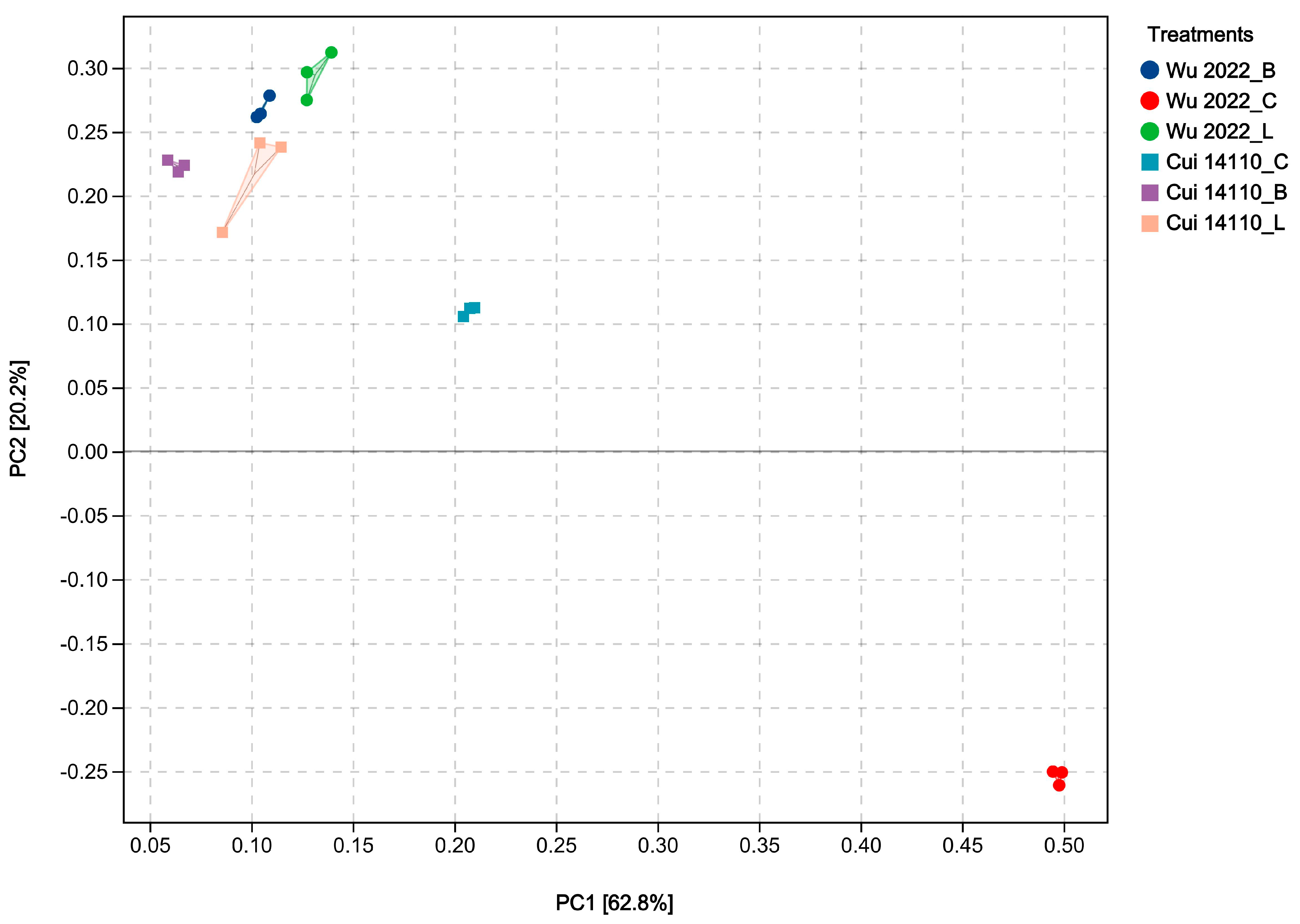
3.3. Transcriptome Sequencing of G. Tsugae Cultured on Different Substrates
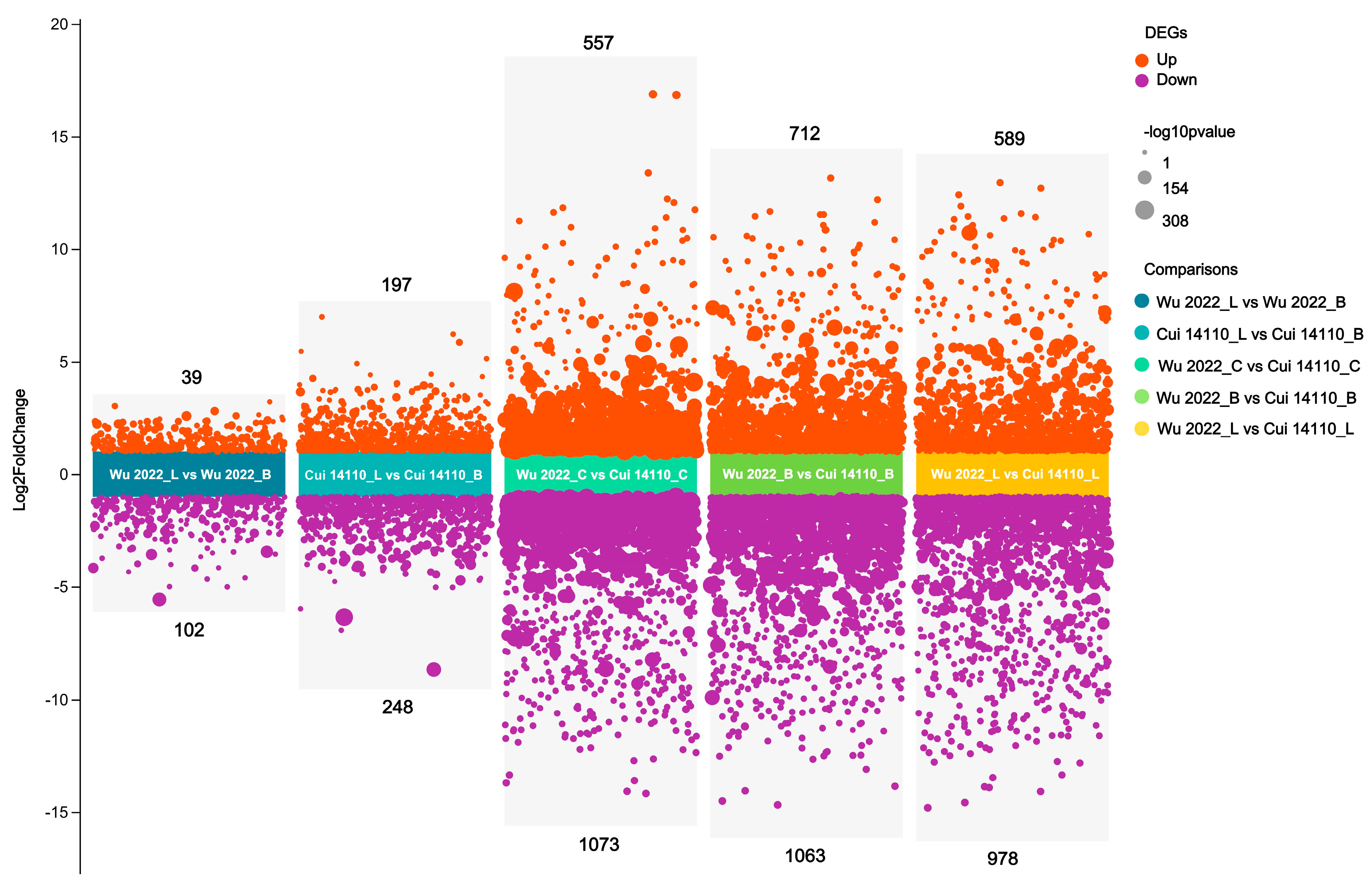
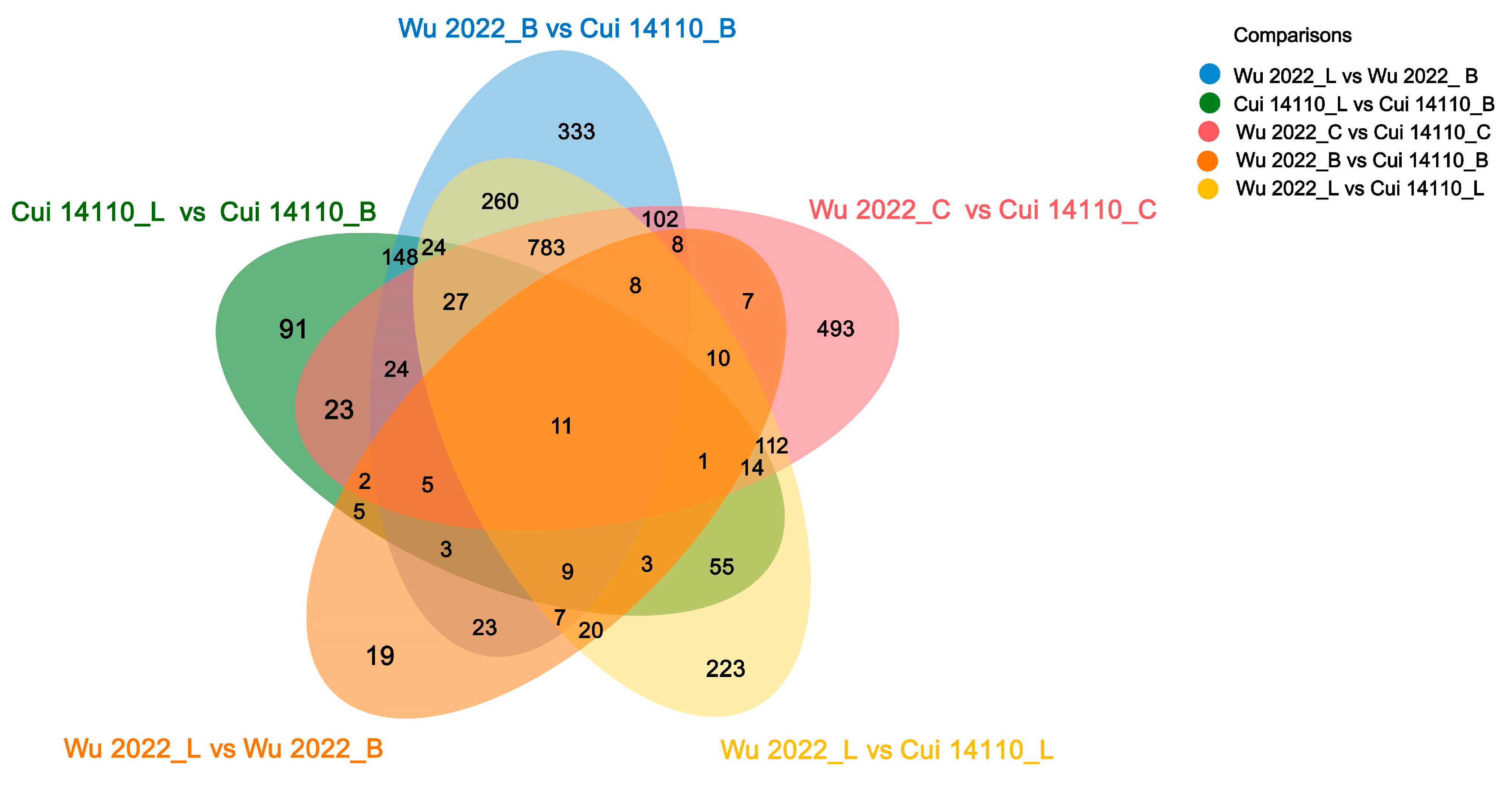
3.4. Differential Expressed Genes Among Six Treatments
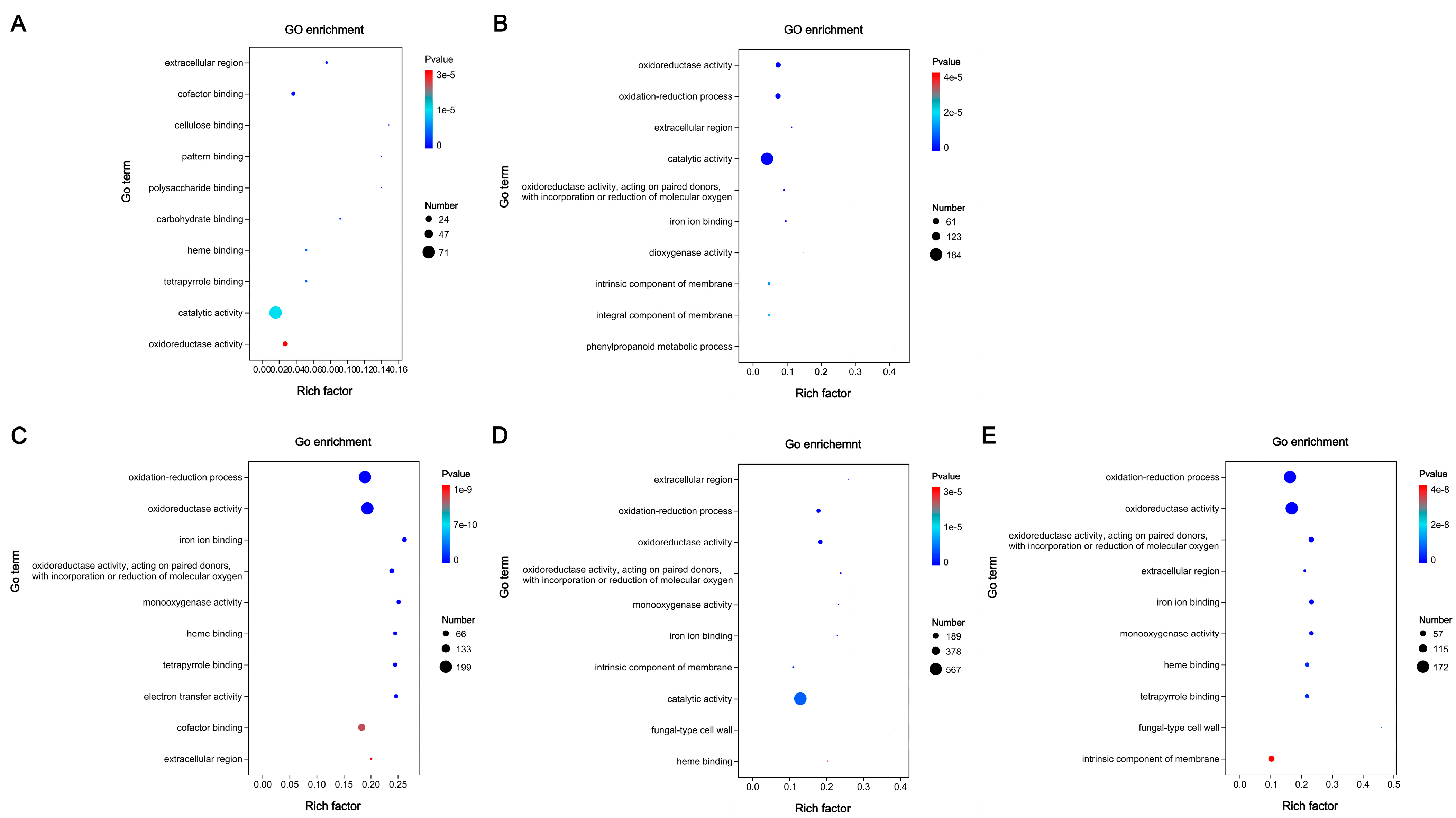
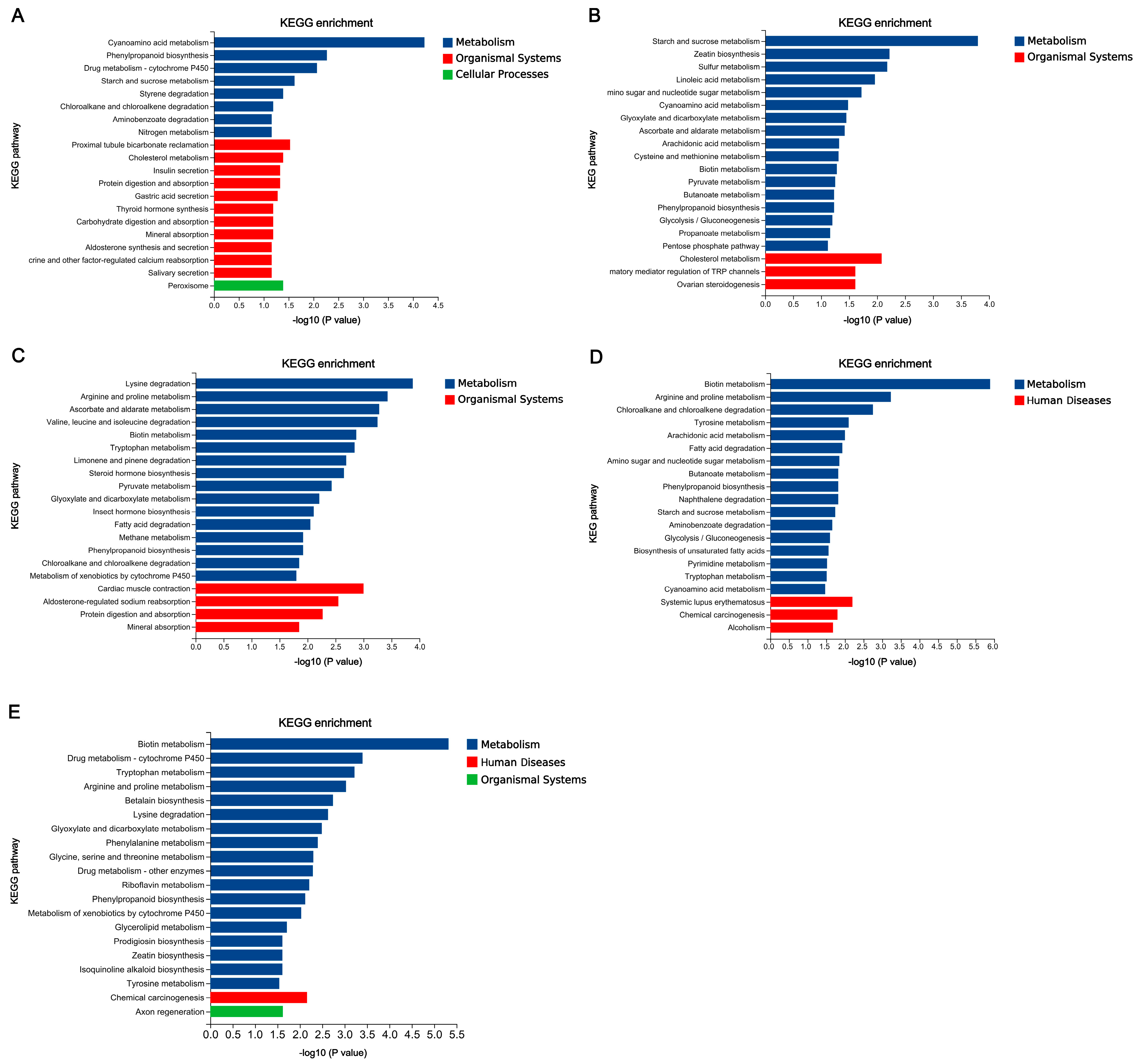
3.5. Functional Annotation of DEGs
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Sun, Y.F.; Xing, J.H.; He, X.L.; Wu, D.M.; Song, C.G.; Liu, S.; Vlasák, J.; Gates, G.; Gibertoni, T.B.; Cui, B.K. Species diversity, systematic revision and molecular phylogeny of Ganodermataceae (Polyporales, Basidiomycota) with an emphasis on Chinese collections. Stud. Mycol. 2016, 101, 287–415. [Google Scholar] [CrossRef]
- Annele, H. Lignin-modifying enzymes from selected white-rot fungi: Production and role from in lignin degradation. FEMS Microbiol. Rev. 1994, 13, 125–135. [Google Scholar] [CrossRef]
- Xu, S.D. Isolation and Purification of Laccase from the White Rot Fungus Leiotrametes lactinea and the Enzymatic Activity in Dye Degradation. Master’s Thesis, Shandong Agricultural University, Tai’An, China, 2021; pp. 1–82. (In Chinese). [Google Scholar]
- Hu, S. Functional Genomics Analysis and Terpene Synthase-Related Gene Mining of Ganoderma tsugae. Master’s Thesis, Jilin Agricultural University, Changchun, China, 2023; pp. 1–69. (In Chinese). [Google Scholar]
- Deng, W.; Zhao, W.; Yang, Y. Laccase purified from white-rot fungus Ganoderma lucidum. Int. J. Environ. Res. Public Health 2022, 19, 8150. [Google Scholar] [CrossRef]
- Li, Q.; Wu, X.Q.; Zhang, X.J. Research progress in microbial degradation of straw lignin. Acta Microbiol. Sin. 2014, 63, 4118–4132. (In Chinese) [Google Scholar]
- Łebkowska, M.; Załęska-Radziwiłł, M. Application of white-rot fungi for biodegradation of refractory organic compounds—A review. Desalination Water Treat. 2014, 52, 3708–3713. [Google Scholar] [CrossRef]
- Wang, H.; Jin, C.; Li, X.; Ma, J.X.; Ye, F.; Tang, L.X.; Si, J.; Cui, B.K. A green biocatalyst fabricated by fungal laccase immobilized onto Fe3O4@polyaniline-chitosan nanofibrous composites for the removal of phenolic compounds. Chem. Eng. J. 2014, 507, 160486. [Google Scholar] [CrossRef]
- Si, J.; Wu, Y.; Ma, H.F.; Cao, Y.J.; Sun, Y.F.; Cui, B.K. Selection of a pH- and temperature-stable laccase from Ganoderma australe and its application for bioremediation of textile dyes. J. Environ. Manag. 2021, 299, 113619. [Google Scholar] [CrossRef]
- Yang, J.; Jiang, J.C.; Zhang, N.; Xu, H.; Xie, J.C.; Zhao, J. Research progress on lignin degradation by microorganism. Biomass Chem. Eng. 2021, 55, 62–70. (In Chinese) [Google Scholar]
- Yan, L.H.; Chen, X.Q.; Wang, L.; Yan, Y.S.; Hu, S.H. Medicinal value and artificial cultivation technology of Ganoderma tsugae in pine fir. Inn. Mong. Agric. Sci. Technol. 2009, 1, 123. (In Chinese) [Google Scholar]
- Xu, T.M.; Wu, D.M.; Gao, N.; Liu, S.; Sun, Y.F.; Cui, B.K. Species diversity, taxonomic classification and ecological habits of polypore fungi in China. Mycology 2025, 16, 419–544. [Google Scholar] [CrossRef]
- Qin, G.F.; Xu, M.Q.; Dai, Y.C. The forest pathogens of root and butt rot in Northeast China. For. Res. 2000, 13, 15–22. (In Chinese) [Google Scholar]
- Jiang, N.; Hu, S.; Pang, B.; Li, Z.H.; Yuan, X.H.; Xiao, S.J.; Fu, Y.P. Genome of Ganoderma species provides insights into the evolution, conifers substrate utilization, and terpene synthesis for Ganoderma tsugae. Front. Microbiol. 2021, 12, 724451. [Google Scholar] [CrossRef] [PubMed]
- Wu, T.H.; Cai, M.J.; Hu, H.P.; Jiao, C.W.; Zhang, Z.; Liu, Y.C.; Chen, J.; Xiao, C.; Li, X.M.; Gao, X.; et al. Whole-genome sequencing and transcriptome analysis of Ganoderma lucidum Strain Yw-1-5 provides new insights into the enhanced effect of Tween80 on exopolysaccharide production. J. Fungi 2022, 8, 1081. [Google Scholar] [CrossRef] [PubMed]
- Wu, Q.; Liu, H.; Shi, Y.X.; Li, W.T.; Huang, J.; Xue, F.F.; Liu, Y.N.; Liu, G.Q. Characteristics of the genome, transcriptome and ganoderic acid of the medicinal fungus Ganoderma lingzhi. J. Fungi 2022, 8, 1257. [Google Scholar] [CrossRef]
- Gaskell, J.; Blanchette, R.A.; Stewart, P.E.; BonDurant, S.S.; Adams, M.; Sabat, G.; Kersten, P.; Cullen, D. Transcriptome and secretome analyses of the wood decay fungus Wolfiporia cocos support alternative mechanisms of lignocellulose conversion. Appl. Environ. Microbiol. 2016, 82, 3979–3987. [Google Scholar] [CrossRef]
- Martinez, D.; Challacombe, J.; Morgenstern, I.; Hibbett, D.; Schmoll, M.; Kubicek, C.P.; Ferreira, P.; Ruiz-Duenas, F.J.; Martinez, A.T.; Kersten, P.; et al. Genome, transcriptome, and secretome analysis of wood decay fungus Postia placenta supports unique mechanisms of lignocellulose conversion. Proc. Natl. Acad. Sci. 2009, 106, 1954–1959. [Google Scholar] [CrossRef]
- Vanden Wymelenberg, A.; Gaskell, J.; Mozuch, M.; Sabat, G.; Ralph, J.; Skyba, O.; Mansfield, S.D.; Blanchette, R.A.; Martinez, D.; Grigoriev, I.; et al. Comparative transcriptome and secretome analysis of wood decay fungi Postia placenta and Phanerochaete chrysosporium. Appl. Environ. Microbiol. 2010, 76, 3599–3610. [Google Scholar] [CrossRef] [PubMed]
- Tan, Y.; Yu, X.L.; Zhang, Z.G.; Tian, J.L.; Feng, N.; Tang, C.H.; Zou, G.; Zhang, J.S. An efficient CRISPR/Cas9 genome editing system for a Ganoderma lucidum cultivated strain by ribonucleoprotein method. J. Fungi 2023, 9, 1170. [Google Scholar] [CrossRef]
- Lv, D.M.; Xu, Y.; Wang, Z.X.; Zhang, Q.L.; Yan, J.P.; Xu, J.W. CRISPR/Cas9-mediated genome editing in Ganoderma lucidum: Recent advances and biotechnological opportunities. World J. Microbiol. Biotechnol. 2025, 41, 223. [Google Scholar] [CrossRef]
- Walker, B.J.; Abeel, T.; Shea, T.; Priest, M.; Abouelliel, A.; Sakthikumar, S.; Cuomo, C.A.; Zeng, Q.; Wortman, J.; Young, S.K.; et al. Pilon: An integrated tool for comprehensive microbial variant detection and genome assembly improvement. PLoS ONE 2014, 9, e112963. [Google Scholar] [CrossRef]
- Simão, F.A.; Waterhouse, R.M.; Ioannidis, P.; Kriventseva, E.V.; Zdobnov, E.M. BUSCO: Assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics 2015, 31, 3210–3212. [Google Scholar] [CrossRef] [PubMed]
- Majoros, W.H.; Pertea, M.; Salzberg, S.L. TigrScan and GlimmerHMM: Two open source ab initio eukaryotic gene-finders. Bioinformatics 2004, 20, 2878–2879. [Google Scholar] [CrossRef] [PubMed]
- Stanke, M.; Morgenstern, B. AUGUSTUS: A web server for gene prediction in eukaryotes that allows user-defined constraints. Nucleic Acids Res. 2005, 33, W465–W467. [Google Scholar] [CrossRef]
- Ter-Hovhannisyan, V.; Lomsadze, A.; Chernoff, Y.; Borodovsky, M. Gene prediction in novel fungal genomes using an abinitio algorithm with unsupervised training. Genome Res. 2008, 18, 1979–1990. [Google Scholar] [CrossRef]
- Benjamin, B.; Chao, X.; Huson, D.H. Fast and sensitive protein alignment using DIAMOND. Nat. Methods 2015, 12, 59–60. [Google Scholar] [CrossRef]
- Camacho, C.; Coulouris, G.; Avagyan, V.; Ma, N.; Papadopoulos, J.; Bealer, K.; Madden, T.L. BLAST+: Architecture and applications. BMC Bioinform. 2009, 10, 421. [Google Scholar] [CrossRef]
- Conesa, A.; Götz, S.; García-Gómez, J.M.; Terol, J.; Talón, M.; Robles, M. Blast2GO: A universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics 2005, 21, 3674–3676. [Google Scholar] [CrossRef] [PubMed]
- Cantalapiedra, C.P.; Hernandez-Plaza, A.; Letunic, I.; Bork, P.; Huerta-Cepas, J. eggNOG-mapper v2: Functional annotation, orthology assignments, and domain prediction at the metagenomic scale. Mol. Biol. Evol. 2021, 38, 5825–5829. [Google Scholar] [CrossRef] [PubMed]
- Moriya, Y.; Itoh, M.; Okuda, S.; Yoshizawa, A.C.; Kanehisa, M. KAAS: An automatic genome annotation and pathway reconstruction server. Nucleic Acids Res. 2007, 35, W182–W185. [Google Scholar] [CrossRef]
- Chen, S.; Zhou, Y.; Chen, Y.; Gu, J. fastp: An ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 2018, 34, 884–890. [Google Scholar] [CrossRef]
- Wang, L.; Feng, Z.; Wang, X.; Wang, X.; Zhang, X. DEGseq: An R package for identifying differentially expressed genes from RNA-seq data. Bioinformatics 2010, 26, 136–138. [Google Scholar] [CrossRef]
- Fehér, Z.; Szirák, K. Signal transduction in fungi—The role of protein phosphorylation. Acta Microbiol. Immunol. Hung. 1999, 46, 269. [Google Scholar]
- Pawson, T. Specificity in signal transduction: From phosphotyrosine-SH2 domain interactions to complex cellular systems. Cell 2004, 116, 191–203. [Google Scholar] [CrossRef]
- Goldberg, A.L. Protein degradation and protection against misfolded or damaged proteins. Nature 2003, 426, 895–899. [Google Scholar] [CrossRef] [PubMed]
- Chu, X.L.; Feng, M.G.; Ying, S.H. Qualitative ubiquitome unveils the potential significances of protein lysine ubiquitination in hyphal growth of Aspergillus nidulans. Curr. Genet. 2016, 62, 191–201. [Google Scholar] [CrossRef]
- Kuppuraj, S.P.; Venkidasamy, B.; Selvaraj, D.; Ramalingam, S. Comprehensive in silico and gene expression profiles of MnP family genes in Phanerochaete chrysosporium towards lignin biodegradation. Int. Biodeterior. Biodegrad. 2021, 157, 105143. [Google Scholar] [CrossRef]
- Cajnko, M.M.; Oblak, J.; Grilc, M.; Likozar, B. Enzymatic bioconversion process of lignin: Mechanisms, reactions and kinetics. Bioresour. Technol. 2021, 340, 125655. [Google Scholar] [CrossRef]
- Kanehisa, M.; Furumichi, M.; Tanabe, M.; Sato, Y.; Morishima, K. KEGG: New perspectives on genomes, pathways, diseases and drugs. Nucleic Acids Res. 2017, 45, D353–D361. [Google Scholar] [CrossRef] [PubMed]
- Reis, V.C.B.; Torres, F.A.G.; Poças-Fonseca, M.J.; De-Souza, M.T.; De-Souza, D.P.; Almeida, J.R.M.; Marinho-Silva, C.; Parachin, N.S.; da Silva Dantas, A.; Mello-de-Sousa, T.M.; et al. Cell cycle, DNA replication, repair, and recombination in the dimorphic human pathogenic fungus Paracoccidioides brasiliensis. Genet. Mol. Res. 2005, 4, 232–250. [Google Scholar]
- Wood, W.A.; Kellogg, S.T. Biomass Part A: Cellulose and Hemicellulose; Methods in Enzymology; Elsevier: Amsterdam, The Netherlands, 1988; pp. 1–774. [Google Scholar]
- Holyavka, M.G.; Artyukhov, V.G. Comparative analysis of the primary structures of glycoside hydrolases. Biochem. Suppl. Ser. B Biomed. Chem. 2023, 17, 1–5. [Google Scholar] [CrossRef]
- Liu, Y.; Li, T.; Zhu, H.; Cao, L.H.; Liang, L.B.; Liu, D.Y.; Shen, Q.R. Methionine inducing carbohydrate esterase secretion of Trichoderma harzianum enhances the accessibility of substrate glycosidic bonds. Microb. Cell Factories 2024, 23, 120. [Google Scholar] [CrossRef]
- Vladimír, P.; Peter, B. Microbial xylanolytic carbohydrate esterases. Essays Biochem. 2023, 67, 479–491. [Google Scholar] [CrossRef] [PubMed]
- Bradley, E.L.; Ökmen, B.; Doehlemann, G.; Henrissat, B.; Bradshaw, R.E.; Mesarich, C.H. Secreted glycoside hydrolase proteins as effectors and invasion patterns of plant-associated fungi and Oomycetes. Front. Plant Sci. 2022, 13, 853106. [Google Scholar] [CrossRef] [PubMed]
- Xiao, Y.; Lu, H.Q.; Liu, Y.; Sang, Y.X.; Sun, J.L. Whole-genome sequencing and functional analysis of a novel chitin-degrading strain Rhodococcus sp. 11-3. J. Biosci. Bioeng. 2022, 134, 167–173. [Google Scholar] [CrossRef] [PubMed]
- Biely, P. Microbial glucuronoyl esterases: 10 years after discovery. Appl. Environ. Microbiol. 2016, 82, 7014–7018. [Google Scholar] [CrossRef]
- Masahiro, W.; Harumi, F.; Hiroyuki, I.; Kazuhiko, I. Crystal structure of an acetylesterase from Talaromyces cellulolyticus and the importance of a disulfide bond near the active site. FEBS Lett. 2015, 589, 1200–1206. [Google Scholar] [CrossRef]
- Pentari, C.; Katsimpouras, C.; Haon, M.; Berrin, J.; Zerva, A.; Topakas, E. Exploring the synergy between fungal CE15 glucuronoyl esterases and xylanases for lignocellulose saccharification. Biotechnol. Biofuels Bioprod. 2025, 18, 38. [Google Scholar] [CrossRef] [PubMed]
- Selim, S.; Harun-Ur-Rashid, M.; Hamoud, Y.A.; Shaghaleh, H. Utilization of bacterial enzymes for cellulose and hemicelluloses degradations: Medical and industrial benefits. BioResources 2025, 20, 8135–8148. [Google Scholar] [CrossRef]
- Adamczyk, S.; Latvala, S.; Poimala, A.; Adamczyk, B.; Hytönen, T.; Pennanen, T. Diterpenes and triterpenes show potential as biocides against pathogenic fungi and oomycetes: A screening study. Biotechnol. Lett. 2023, 45, 1555–1563. [Google Scholar] [CrossRef]
- Chauhan, P.S. Role of various bacterial enzymes in complete depolymerization of lignin: A review. Biocatal. Agric. Biotechnol. 2020, 23, 101498. [Google Scholar] [CrossRef]
- Dong, C.D.; Tiwari, A.; Anisha, G.S.; Chen, C.W.; Singh, A.; Haldar, D.; Patel, A.K.; Singhania, R.R. Laccase: A potential biocatalyst for pollutant degradation. Environ. Pollut. 2023, 319, 120999. [Google Scholar] [CrossRef] [PubMed]
- Varnaite, R.; Raudoniene, V.; Bridziuviene, D. Enzymatic biodegradation of lignin-cellulose complex in plant origin material. Mater. Sci.-Medzg. 2011, 17, 99–103. [Google Scholar] [CrossRef]
- Prieto, R.; Woloshuk, C.P. Ord1, an oxidoreductase gene responsible for conversion of O-methylsterigmatocystin to aflatoxin in Aspergillus flavus. Appl. Environ. Microbiol. 1997, 63, 1661–1666. [Google Scholar] [CrossRef]
- Yu, J.; Chang, P.K.; Ehrlich, K.C.; Cary, J.W.; Montalbano, B.; Dyer, J.M.; Bhatnagar, D.; Cleveland, T.E. Characterization of the critical amino acids of an Aspergillus parasiticus Cytochrome P-450 monooxygenase encoded by ordA that is involved in the biosynthesis of aflatoxins B1, G1, B2, and G2. Appl. Environ. Microbiol. 1999, 64, 4834–4841. [Google Scholar] [CrossRef]
- Reading, N.S.; Welch, K.D.; Aust, S.D. Free radical reactions of wood-degrading fungi. Wood Deterior. Preserv. 2003, 845, 16–31. [Google Scholar]
- Martínez, A.T.; Speranza, M.; Ruiz-Dueñas, F.J.; Ferreira, P.; Camarero, S.; Guillén, F.; Martínez, M.J.; Gutiérrez, A.; del Río, J.C. Biodegradation of lignocellulosics: Microbial, chemical, and enzymatic aspects of the fungal attack of lignin. Int. Microbiol. 2005, 8, 195–204. [Google Scholar] [PubMed]
- Daâssi, D.; Zouari-Mechichi, H.; Belbahri, L.; Barriuso, J.; Martínez, M.J.; Nasri, M.; Tahar, M.T. Phylogenetic and metabolic diversity of Tunisian forest wood-degrading fungi: A wealth of novelties and opportunities for biotechnology. 3 Biotech 2016, 6, 46. [Google Scholar] [CrossRef]
- Yao, T.; Feng, K.; Xie, M.; Barros, J.; Tschaplinski, T.J.; Tuskan, G.A.; Muchero, W.; Chen, J.G. Phylogenetic occurrence of the phenylpropanoid pathway and lignin biosynthesis in plants. Front. Plant Sci. 2021, 12, 704697. [Google Scholar] [CrossRef]
- Mori, T.; Ohno, H.; Ichinose, H.; Hirokazu, K.; Hirofumi, H. White-rot fungus Phanerochaete chrysosporium metabolizes chloropyridinyl-type neonicotinoid insecticides by an N-dealkylation reaction catalyzed by two cytochrome P450s. J. Hazard. Mater. 2021, 402, 123831. [Google Scholar] [CrossRef]
- Morel, M.; Meux, E.; Mathieu, Y.; Thuillier, A.; Chibani, K.; Harvengt, L.; Jacquot, J.P.; Gelhaye, E. Xenomic networks variability and adaptation traits in wood decaying fungi. Microb. Biotechnol. 2013, 6, 248–263. [Google Scholar] [CrossRef]
- Young, D.; Rice, J.; Martin, R.; Lindquist, E.; Lipzen, A.; Grigoriev, I.; Hibbett, D. Degradation of bunker C fuel oil by white-rot fungi in sawdust cultures suggests potential applications in bioremediation. PLoS ONE 2015, 10, e0130381. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.S.; Chen, X.; Jin, E.H.; Wang, A.K.; Chen, T.T.; Zhang, X.L.; Zhu, J.W.; Dong, L.L.; Sun, Y.L.; Yu, C.X.; et al. The GSA Family in 2025: A broadened sharing platform for multi-omics and multimodal data. Genom. Proteom. Bioinform. 2025, 23, qzaf072. [Google Scholar] [CrossRef] [PubMed]
- CNCB-NGDC Members and Partners. Database Resources of the National Genomics Data Center, China National Center for Bioinformation in 2025. Nucleic Acids Res. 2025, 53, D30–D44. [Google Scholar] [CrossRef] [PubMed]
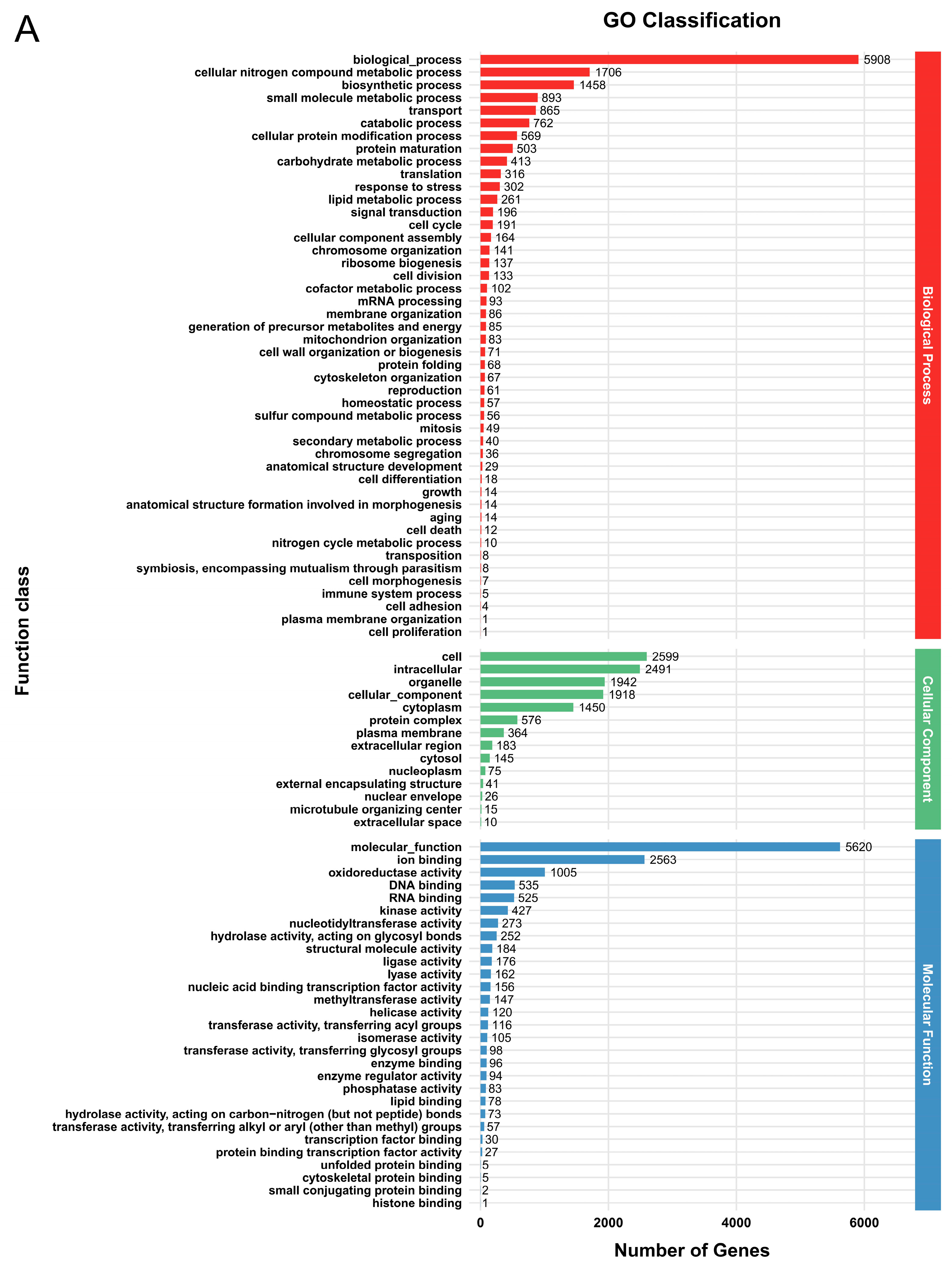
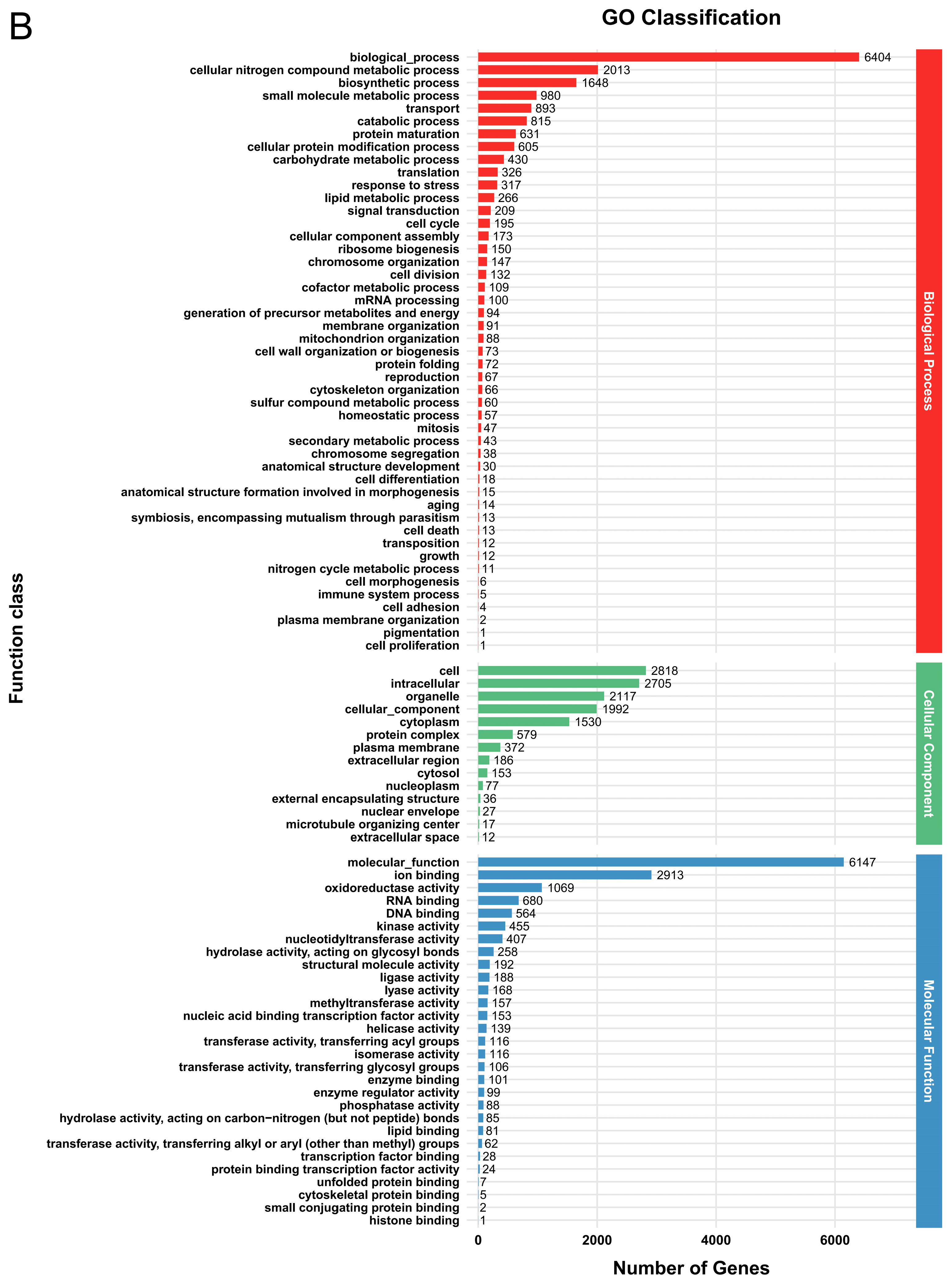
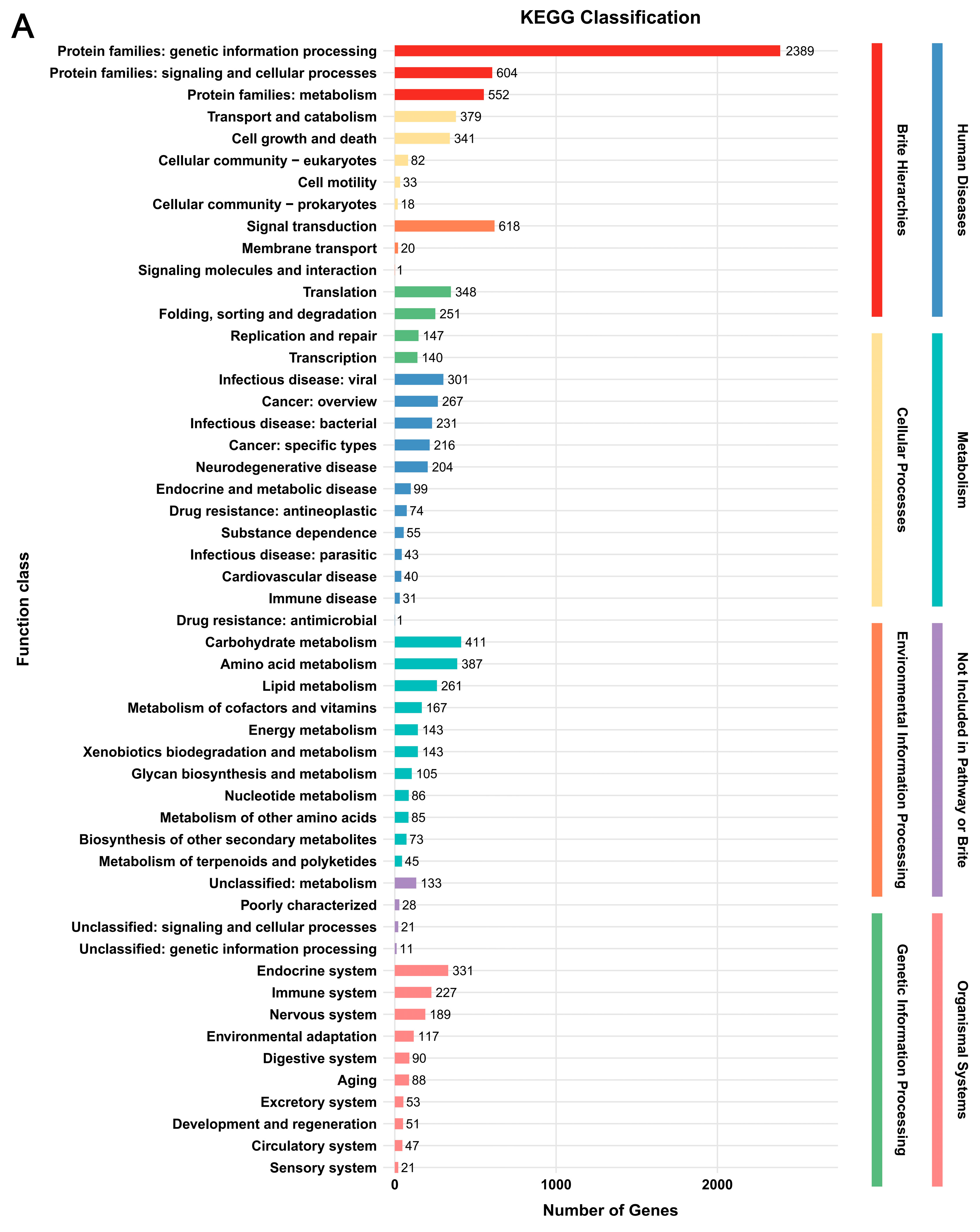
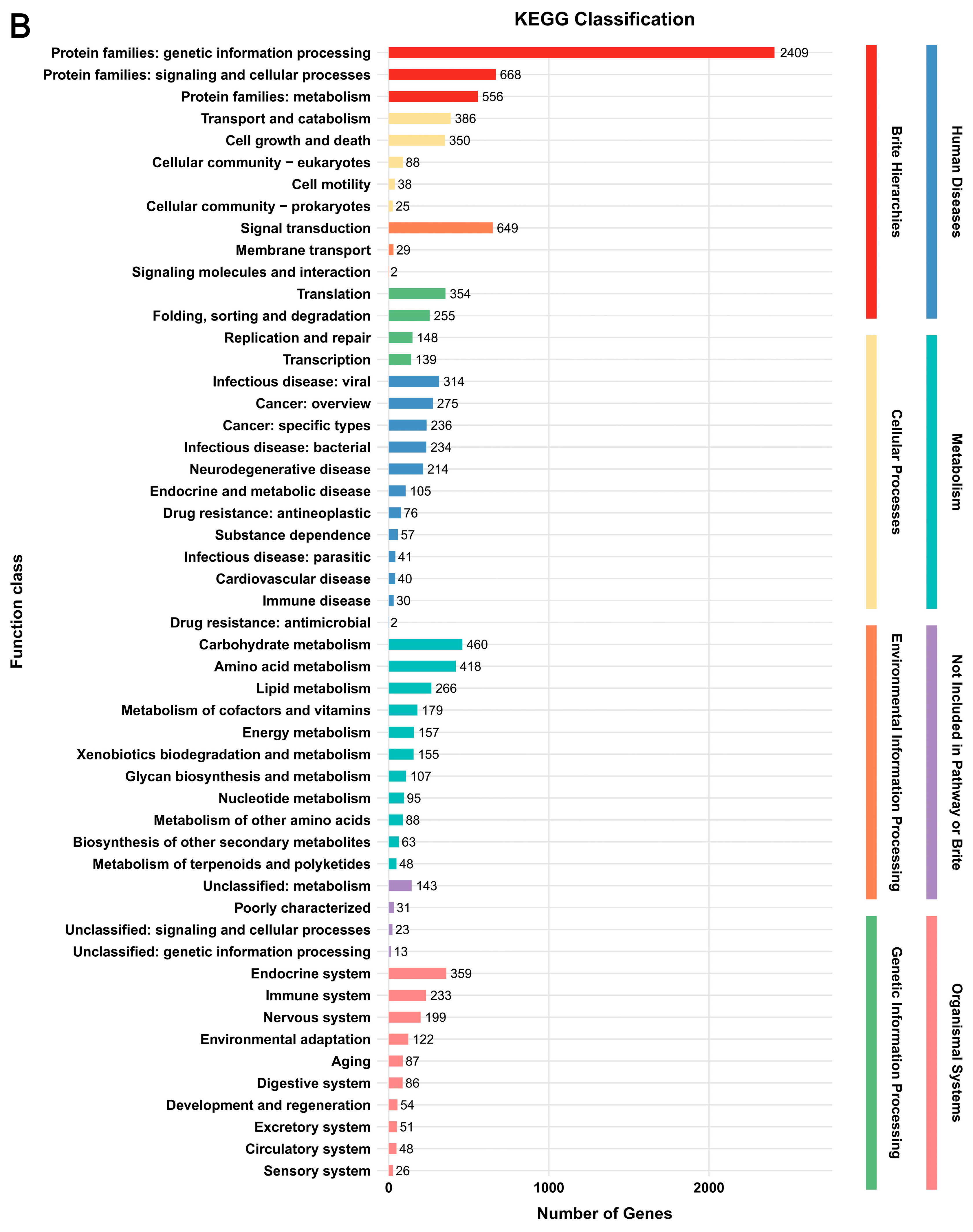
| Parameter | G. tsugae Wu 2022 | G. tsugae Cui 14110 |
|---|---|---|
| Sequencing platform | Illumina NovaSeq | Illumina NovaSeq pacbio Sequel |
| Genome size (Mb) | 40.8 | 45.6 |
| Number of contigs | 1134 | 660 |
| Contig N50 (bp) | 100,812 | 168,103 |
| Number of scaffolds | 1099 | 270 |
| Scaffold N50 (bp) | 103,400 | 3,119,812 |
| Number of genes | 12,496 | 13,450 |
| Average gene length (bp) | 1772 | 1702 |
| Average exons per gene | 5.6 | 5.2 |
| GC content (%) | 56.03 | 55.85 |
| BUSCO (% complete) | 98.5 | 98.1 |
| Database | G. tsugae Wu 2022 | G. tsugae Cui 14110 |
|---|---|---|
| NR | 12,220 (97.8%) | 13,281 (98.7%) |
| eggNOG | 9203 (73.6%) | 9835 (73.1%) |
| GO | 6616 (52.9%) | 7176 (53.4%) |
| KEGG | 3685 (29.4%) | 3824 (28.4%) |
| CAZymes | 572 (4.58%) | 582 (4.38%) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2026 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license.
Share and Cite
Sun, Y.; Lv, M.; Luo, M.; Yao, Z.; Zhou, M.; Fang, Y.; Wu, D.; Gao, N.; Cui, B. Integrative Genomic and Transcriptomic Analysis of White-Rot Fungi Ganoderma tsugae Growing on Both Coniferous and Broad-Leaved Trees. J. Fungi 2026, 12, 35. https://doi.org/10.3390/jof12010035
Sun Y, Lv M, Luo M, Yao Z, Zhou M, Fang Y, Wu D, Gao N, Cui B. Integrative Genomic and Transcriptomic Analysis of White-Rot Fungi Ganoderma tsugae Growing on Both Coniferous and Broad-Leaved Trees. Journal of Fungi. 2026; 12(1):35. https://doi.org/10.3390/jof12010035
Chicago/Turabian StyleSun, Yifei, Mengxue Lv, Meiqin Luo, Ziqi Yao, Miao Zhou, Yuxuan Fang, Dongmei Wu, Neng Gao, and Baokai Cui. 2026. "Integrative Genomic and Transcriptomic Analysis of White-Rot Fungi Ganoderma tsugae Growing on Both Coniferous and Broad-Leaved Trees" Journal of Fungi 12, no. 1: 35. https://doi.org/10.3390/jof12010035
APA StyleSun, Y., Lv, M., Luo, M., Yao, Z., Zhou, M., Fang, Y., Wu, D., Gao, N., & Cui, B. (2026). Integrative Genomic and Transcriptomic Analysis of White-Rot Fungi Ganoderma tsugae Growing on Both Coniferous and Broad-Leaved Trees. Journal of Fungi, 12(1), 35. https://doi.org/10.3390/jof12010035









