Abstract
Mucor species are fast-growing filamentous fungi, widespread in natural ecosystems. As opportunistic pathogens, some species can cause mucormycoses in humans and animals, while others hold significant economic value in food fermentation and bioengineering. In this study, four novel species were identified from soil samples collected in Xizang Autonomous Region and Yunnan Province, China, and their establishment as new species was supported by morphological characteristics and molecular data (ITS-LSU-RPB1), with phylogenetic analyses conducted using the Maximum Likelihood (ML) and Bayesian Inference (BI) methods. M. globosporus sp. nov. is characterized by producing globose chlamydospores. M. multimorphus sp. nov. is distinguished by swelling in the sporangiophores. M. polymorphus sp. nov. is differentiated by polymorphic chlamydospores. And M. xizangensis sp. nov. reflects its geographical origin in the Xizang Autonomous Region. Comprehensive descriptions of each novel taxon are presented herein. This study constitutes the ninth segment in an ongoing series elucidating early-diverging fungal diversity in China, expanding the understanding of the phylogeny of Mucor fungi and extending the worldwide number of known Mucor species to 137.
1. Introduction
The genus Mucor, a group of fast-growing, early-diverging fungi, is species-rich and distributed widely in natural ecosystems [1,2]. It is commonly found in soil, air, herbivore dung, insects, necromass of animals and plants, and other damp environments [3,4,5,6]. As coprophilic fungi, some species serve as pioneer decomposers during fecal decomposition, significantly contributing to material cycling in ecosystems through mediating the biogeochemical cycles of carbon and nitrogen [7,8]. Some species are opportunistic pathogens of animals, causing cutaneous mucormycoses in humans, especially in immunocompromised individuals [9,10,11]. Several Mucor species can also induce the spoilage of natural and artificial foods [3]. However, certain Mucor species exhibit significant application value in food industries, frequently used for fermenting soybean products, cheeses, and other foods [12,13]. And certain species play an important role in bioengineering by producing various enzymes such as lipases, proteases, phytases, cellulases, and uricases, which are vital for biocatalytic processes and industrial applications [14,15,16,17,18,19,20].
The genus was originally described by Fresenius in 1850 [21] and characterized by simple or branched sporangiophores arising directly from substrates, non-apophysate and globose sporangia with persistent or deliquescent, incrusted sporangial walls, and zygospores borne on opposed suspensors [22,23]. However, the morphological classification of Mucor remains contentious. For example, the traditional taxonomic literature historically differentiated Rhizomucor from Mucor based on rhizoids [24,25], but recent molecular studies demonstrate that certain Mucor species also produce rhizoids (e.g., Mucor changshaensis) [5,26,27]. This explains why some Mucor species were misclassified into the genus Rhizomucor. Most Mucor species are mesophilic, exhibiting optimal growth at 20–30 °C and survival within 10–42 °C [28]. By contrast, a minority are psychrophilic, characterized by an optimal growth temperature of approximately 15 °C, a minimum growth temperature capable of reaching 0 °C, and a maximum growth temperature in the vicinity of 20 °C [29,30].
Since its formal inclusion in Linnaeus’ Species Plantarum (1753) [31], the genus Mucor has witnessed continuous taxonomic refinements, from the initial morphological delineation to the contemporary phylogenetic reclassification. In 2018, Wijayawardene et al. stated that among more than 300 literature-recorded Mucor species, only approximately 60 species were valid or could be validated [32]. Recent taxonomic revisions integrating phylogenetic analyses and morphological characters have led to the systematic reclassification of multiple species previously assigned to Mucor into other genera [11]. Nevertheless, the discovery and formal description of novel species in recent years have further enriched the taxonomic diversity of the genus Mucor [5,27,33,34,35]. Currently, 133 species are accepted (https://www.catalogueoflife.org/, accessed on 30 June 2025).
During the investigation of soil fungal diversity in China, eight strains were classified into four new Mucor species based on ITS-LSU-RPB1 molecular data, morphological characteristics, and maximum growth temperatures. Phylogenetic trees, detailed descriptions, and photographs of these new taxa are presented herein. This is the ninth report of a serial work on the diversity of early-diverging Chinese fungi [36,37,38,39,40,41,42,43].
2. Materials and Methods
2.1. Sample Collection and Strain Isolation
Soil samples were collected from Xizang Autonomous Region and Yunnan Province, China, between 2022 and 2024—with two sampling expeditions in Xizang Autonomous Region and eight in Yunnan Province. Sampling was carried out during the rainy season or the vigorous vegetation growth period in both regions. Yunnan sampling sites were located in subtropical evergreen broad-leaved forests, coniferous-broad-leaved mixed forests, and alpine meadows, with dominant red and yellow soils and high vegetation coverage. Xizang Autonomous Region sites focused on alpine meadows, shrubs, and sparse valley forests, with alpine meadow soils as the main soil type. All samples were collected from 5 to 10 cm depth, labeled with a waterproof tag, indicating the collection date, administrative division, GPS coordinates, and altitude. These samples were then temporarily stored in a 4 °C incubator in the laboratory.
Subsequently, strains were isolated following the plate dilution method and single spore isolation method described in previous studies [44,45]. In short, 1 g of soil sample was weighed and suspended in 10 mL of sterile water to prepare a soil suspension with a concentration of 10−1. Accurate 1 mL of this suspension was transferred to 9 mL sterile water and mixed thoroughly to obtain a soil suspension with a concentration of 10−2. Serial dilutions were continued to prepare soil suspensions at concentrations of 10−3 and 10−4. About 200 μL of 10−3 and 10−4 soil suspensions were drawn by a sterile pipette and spread evenly onto the surface of Rose Bengal Chloramphenicol agar [46] (RBC: peptone 5.00 g/L, glucose 10.00 g/L, MgSO4·7H2O 0.50 g/L, KH2PO4 1.00 g/L rose bengal 0.05 g/L, chloramphenicol 0.10 g/L, agar 15.00 g/L) containing 0.03% streptomycin sulfate. The coated plates were incubated in the dark at 25 °C for 3–5 d. After sporangia produced, individual spores were picked under a stereomicroscope (Olympus SZX10, OLYMPUS, Tokyo, Japan) using a sterile inoculation loop and inoculated onto Potato Dextrose Agar (PDA: 200 g potato, 20 g dextrose, 20 g agar, 1000 mL distilled water, pH 7.0). The inoculated PDA plates were incubated at 25 °C in the dark. Strains were purified and stored with 10% glycerol at 4 °C.
The type strains were deposited at the China General Microbiological Culture Collection Center, Beijing, China (CGMCC) and duplicated at Shandong Normal University, Jinan, China (XG). The dried specimens were stored at the Herbarium Mycologicum Academiae Sinicae, Beijing, China (Fungarium, HMAS). Taxonomic information of these new taxa was submitted to the fungal name database (https://nmdc.cn/fungalnames/, accessed on 30 June 2025).
2.2. Morphology and Maximum Growth Temperature
Colonies were cultured on PDA medium at 25 °C for 2–5 days. Macro- and microscopic structures were observed daily. Tape-stripping and wet-mount methods were adopted in the slide preparation process.
A high-definition color digital camera (DP80, Olympus, Tokyo, Japan) was used to photograph both the obverse and reverse sides of the colonies. A stereomicroscope (Olympus SZX10, Olympus, Tokyo, Japan) and an optical microscope (BX53, Olympus, Tokyo, Japan) were used to observe microscopic structures (including hyphae, rhizoids, stolons, sporangiophores, sporangia, collars, columellae, apophyses, sporangiospores, chlamydospores, and zygospores). After that, Adobe Photoshop CC 2019 was used to layout of different microstructures images. Then, Digimizer software (https://www.digimizer.com/, accessed on 30 June 2025) was systematically utilized to measure various dimensions of the microstructures, with the statistical data covering 20 measurements.
The maximum growth temperature was performed following the methodology described by previous study [47,48,49]. To determine the maximum growth temperature of the target fungal strain, the strain was initially cultured at 25 °C for 3 d, followed by a controlled daily temperature increase of 1 °C until colony formation halted. Three independent parallel replicates were incorporated in the design for statistical reliability.
2.3. DNA Extraction, PCR Amplification, and Sequencing
After the target strains were incubated at 25 °C for 5–7 d on PDA solid medium plates, cell total DNAs were extracted from the mycelia using the BeaverBeads Plant DNA Kit [50] (Cat. No.: 70409–20; BEAVER Biomedical Engineering Co., Ltd., Suzhou, China). For strains with inefficient genomic DNA amplification, reverse transcription was performed to synthesize cDNA from total RNA as an alternative template using SPARKscript II All-in-one RT SuperMix for qPCR (With gDNA Eraser) (Cat# AG0305-B; Shandong Sparkjade Biotechnology Co., Ltd., Jinan, China). Genomic loci ITS, LSU, and RPB1 were amplified by polymerase chain reaction (PCR) using the primer pairs ITS4/ITS5 [51], LR0R/LR7 [52], and RPB1-Af/RPB1-Cr [53], respectively (Table 1). PCR amplification was carried out using a 25μL reaction system, including 12.5μL of 2 × Hieff Canace®Plus PCR Master Mix (Yeasen Biotechnology, Shanghai, China, Cat No. 10154ES03), 10 µL ddH2O, 1 μL of each of the forward and reverse primers (10 µM) (TsingKe, Beijing, China), and 1 μL fungal genomic DNA template. The amplification products were detected by 1% agarose gel electrophoresis, and the band specificity was observed after staining with TS-GelRed Nucleic Acid Gel Stain (10,000 × in water; TSJ002; Beijing Tsingke Biotech Co., Ltd., Beijing, China). Gel recovery was carried out using a Gel Extraction Kit (Cat# AE0101-C; Shandong Sparkjade Biotechnology Co., Ltd., Jinan, China). DNA sequencing was performed by the Biosune Company Limited (Shanghai, China). Consensus sequences were obtained through MAFFT v.7.0 alignment, and assembled with MEGA v.7.0. Then all sequences were uploaded to the GenBank database, with the accession numbers provided in Table S1.

Table 1.
PCR primers and programs used in this study.
2.4. Phylogenetic Analyses
The phylogenetic analyses were constructed based on concatenated ITS-LSU-RPB1 sequences, using both the Maximum Likelihood (ML) and Bayesian Inference (BI) methods. The optimal evolutionary model of each locus was determined by MrModelTest v2.3 [54] and subsequently applied in the Bayesian inference (BI) analysis. The ML analysis was conducted using RAxML-HPC2 on XSEDE v.8.2.12 on the CIPRES Science Gateway platform (https://www.phylo.org/, accessed on 30 June 2025), with 1000 bootstrap replicates [55]. The BI analysis was conducted on a Linux system server, with a quick start configured with an automatic stop option. Bayesian inference consisted of five million generations with four parallel runs, employing stopping rules and sampling frequencies of 100 generations [56]. The burn-in score was set to 0.25, and the posterior probability (PP) was determined based on the remaining trees. The evolutionary trees were uploaded to the iTOL website (https://itol.embl.de, accessed on 30 June 2025) for layout and adjustment. The final refinements were carried out using Adobe Illustrator CC 2019.
3. Results
3.1. Phylogeny
The molecular dataset included 83 strains in total, consisting of 46 Mucor species, with Backusella lamprospora (CBS 118.08) as an outgroup. The dataset consisted of 3290 characters, covering ITS rDNA (1–1172), LSU rDNA (1173–2283), and RPB1 (2284–3290). Among them, 1808 characters were constant, 246 variable characters were parsimony-uninformative, and 1236 characters were parsimony-informative. The results of the MrModelTest analysis indicated that the Dirichlet base frequencies and the GTR + I + G evolutionary model were suitable for the two partitions in the Bayesian inference. The topology of the Maximum Likelihood (ML) tree, consistent with that of the Bayesian Inference (BI) tree, was chosen as a representative for detailed illustration (Figure 1). Eight strains of Mucor isolated in this study were divided into four independent clades: M. globosporus (MLBV = 100, BIPP = 1.00), M. multimorphus (MLBV = 100, BIPP = 1.00), M. polymorphus (MLBV = 100, BIPP = 1.00), and M. xizangensis (MLBV = 100, BIPP = 1.00). As for phylogenetic relationship, M. globosporus is closely related with M. moniliformis, M. multimorphus and M. xizangensis are sisters to each other, and M. polymorphus is basal to M. multimorphus and M. xizangensis.
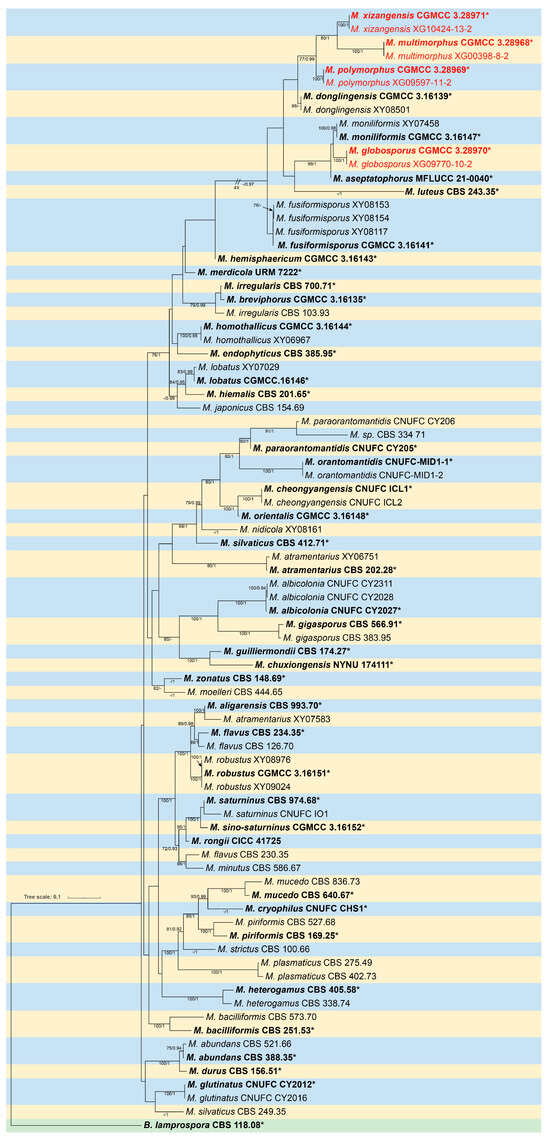
Figure 1.
Maximum Likelihood (ML) phylogenetic tree of Mucor based on ITS, LSU, and RPB1 sequences, with Backusella lamprospora (CBS 118.08) as outgroup. Nodes are labeled with ML bootstrap values (MLBV ≥ 70%) and Bayesian inference posterior probabilities (BIPP ≥ 0.9), separated by a slash “/”. Ex-type or ex-holotype strains are indicated in bold black and marked with an asterisk “*”. Strains isolated in this study are shown in red. The scale bar in the lower left represents 0.1 substitutions per site.
3.2. Taxonomy
3.2.1. Mucor globosporus Z.Y. Ding, H. Zhao & X.Y. Liu, sp. Nov., Figure 2
Fungal Names—FN 573008.
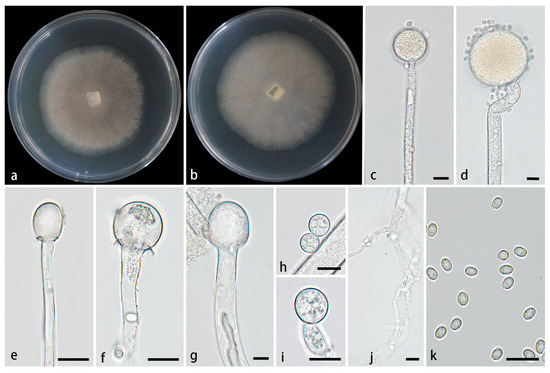
Figure 2.
Mucor globosporus ex-holotype CGMCC 3.28970. (a,b) Colonies cultured on PDA at 25 °C for 3 days, (a) obverse, (b) reverse; (c,d) sporangia; (e–g) columellae; (h,i) chlamydospores; (j) rhizoids; (k) sporangiospores. Scale bars: (c–k) 10 μm.
Type—China, Yunnan Province, Xishuangbanna Dai Autonomous Prefecture, Menghai County, Mengman Town, G219 (West Scenic Line) (22°08′01″ N, 100°18′87″ E, altitude 1367.38 m), from soil, 6 July 2024, Z.Y. Ding, holotype HMAS 354077, ex-type living culture CGMCC 3.28970 (=XG09777-10-1).
Etymology—The epithet globosporus (Lat.) refers to globose chlamydospores in this species.
Description—Colonies on PDA at 25 °C for 3 d, reaching 59 mm in diameter, rapidly growing with a growth rate of 19.7 mm/d, initially white, gradually becoming black-brown, floccose. Hyphae flourishing, occasionally branched, hyaline, aseptate when young, septate with age, radial growth. Rhizoids present, but rare. Stolons absent. Sporangiophores arising from substrate and aerial hyphae, erect or few slightly bent, unbranched, hyaline, occasionally with a swelling, 4.8–14.3 µm wide. Sporangia globose, pale yellow to light brown, 16.6–76.1 μm in diameter. Collars present or absent, if present usually distinct and large. Columellae subglobose, globose, ellipsoidal, hyaline or subhyaline, smooth-walled, 5.2–30.2 µm long and 4.5–27 µm wide. Apophyses absent. Sporangiospores usually ovoid, rarely globose, 3.0–5.1 µm long and 2.6–3.7 µm wide. Chlamydospores rare, globose, 6.6–11.3 µm long and 6.6–11.2 µm wide. Zygospores unknown.
Cultured characteristics and maximum growth temperature: Under the same culture conditions, the colonies grow faster on PDA than on MEA (Figure 3). On PDA, the colonies reach 73 mm in diameter for 5 d at 25 °C. On MEA, the colonies reach 58 mm in diameter for 5 d at 25 °C. No growth was obversed at 33 °C.
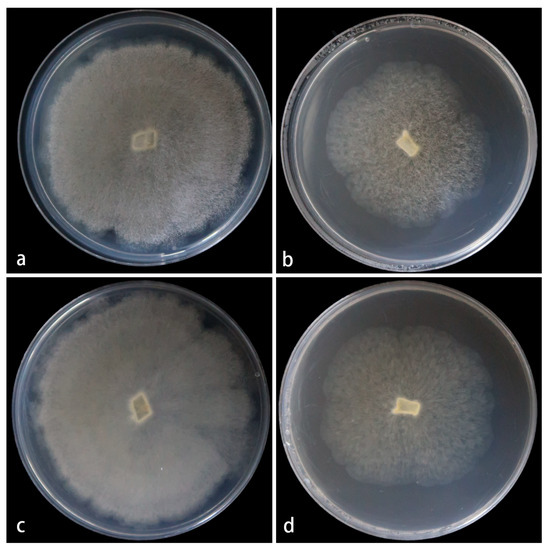
Figure 3.
Mucor globosporus ex-holotype CGMCC 3.28970. Colonies cultured on PDA and on MEA at 25 °C for 5 days, (a,c) Colony on PDA; (b,d) Colony on MEA.
Additional strains examined—China, Yunnan Province, Xishuangbanna Dai Autonomous Prefecture, Menghai County, Mengman Town (22°11′02″ N, 100°17′22″ E, altitude 1492.96 m), from soil, 6 July 2024, Z.Y. Ding, living culture XG09770-10-2.
GenBank accession numbers—CGMCC 3.28970 (ITS, PV819211; LSU, PV833754; RPB1, PX048331), XG09770-10-2 (ITS, PV819212; LSU, PV833755; RPB1, PX048332).
Notes—Based on the ITS-LSU-RPB1 phylogenetic tree, two strains of the Mucor globosporus sp. nov. formed a fully supported lineage (MLBV = 100, BIPP = 1.00; Figure 1), which is sister to M. moniliformis. Morphologically, the new species is distinguished from M. inflatus in sporangia, columellae, chlamydospores and rhizoids. The new species produces subglobose, globose and ellipsoidal columellae, while M. moniliformis only has the first two shapes. The chlamydospores of the new species are only globose, while M. moniliformis exhibits various shapes including ellipsoidal, ovoid, subglobose, globose or irregular. Additionally, rhizoids are present in the new species, whereas they are absent in M. moniliformis [27].
3.2.2. Mucor multimorphus Z.Y. Ding, H. Zhao & X.Y. Liu, sp. Nov., Figure 4
Fungal Names—FN 573010.
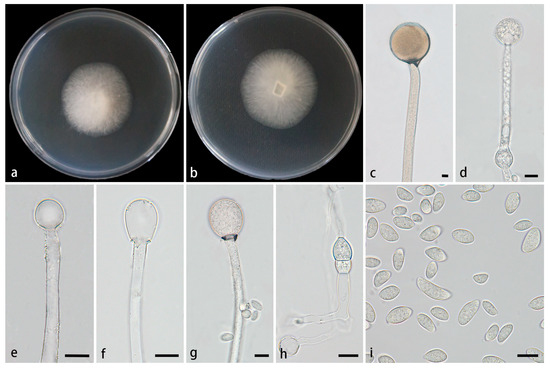
Figure 4.
Mucor multimorphus ex-holotype CGMCC 3.28968. (a,b) Colonies cultured on PDA at 25 °C for 3 days, (a) obverse, (b) reverse; (c,d) sporangia; (e–g) columellae; (h) chlamydospores; (i) sporangiospores. Scale bars: (c–i) 10 μm.
Type—China, Xizang Autonomous Region, Xigaze City, Jilong County (The latitude and longitude are not clear, altitude 3040 m), from soil, 1 August 2022, Z.Y. Ding, holotype HMAS 354075, ex-type living culture CGMCC 3.28968 (=XG00398-8-1).
Etymology—The epithet multimorphus (Lat.) refers to producing multiple shapes of sporangiospores.
Description—Colonies on PDA at 25 °C for 9 d, reaching 77 mm in diameter, slowly growing with a growth rate of 8.56 mm/d, initially white, gradually becoming Cream yellow with age, floccose. Hyphae flourishing, usually unbranched, hyaline, occasionally septate, radial growth. Rhizoids absent. Stolons absent. Sporangiophores arising from substrate and aerial hyphae, erect or few slightly bent, unbranched, hyaline, sometimes accompanied by a swelling, 5.0–15.8 µm wide. Septa sometimes present in sporangiophores. Sporangia globose, pale yellow to pale brown, 33.8–70.0 μm in diameter. Collars present, usually small. Columellae globose, ellipsoidal, pyriform, hyaline or subhyaline, smooth-walled, 10.1–40.0 µm long and 7.5–39.7 µm wide. Apophyses absent. Sporangiospores multiple shaped, mainly fusiform and ellipsoidal, occasionally irregular, 5.4–18.0 µm long and 3.3–7.8 µm wide. Chlamydospores produced in substrate hyphae, ellipsoidal or irregular, occasionally present, 6.3–16.8 µm long and 8.1–12.5 µm wide. Zygospores unknown.
Cultured characteristics and maximum growth temperature: Under the same culture conditions, the colonies grow faster on PDA than on MEA (Figure 5). On PDA, the colonies reaching 55 mm in diameter for 5 d at 25 °C. On MEA, the colonies reaching 45 mm in diameter for 5 d at 25 °C. No growth was obversed at 31 °C.
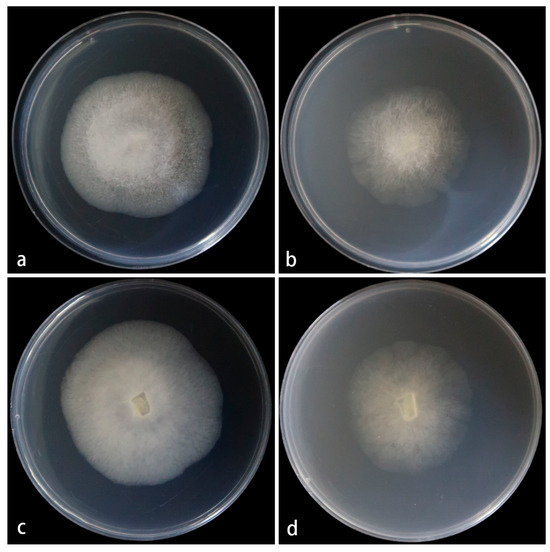
Figure 5.
Mucor multimorphus ex-holotype CGMCC 3.28968. Colonies cultured on PDA and on MEA at 25 °C for 5 days, (a,c) Colony on PDA; (b,d) Colony on MEA.
Additional strains examined—China, Xizang Autonomous Region, Xigaze City, Jilong County (The latitude and longitude are not clear, altitude 3040 m), from a soil sample, 1 August 2022, Z.Y. Ding, living culture XG00398-8-2.
GenBank accession numbers—CGMCC 3.28968 (ITS, PV819207; LSU, PV833750; RPB1, PV889321), XG00398-8-2 (ITS, PV819208; LSU, PV833751; RPB1, PV889322).
Notes—In the phylogenetic tree of ITS-LSU-RPB1, two strains of the Mucor multimorphus sp. nov. formed a fully supported independent clade (MLBV = 100, BIPP = 1.00; Figure 1), which is closely related to M. xizangensis. Morphologically, the new species differs from M. xizangensis in sporangiophores, sporangia, sporangiospores, and chlamydospores. It occasionally forms a swelling on sporangiophores, while M. xizangensis does not. It is larger than M. xizangensis in sporangia (33.8–70 μm vs. 23.3–58.6 μm). In sporangiospores and chlamydospores, it differs from M. xizangensis by larger size and more shapes. Specifically, it produces predominantly fusiform and ellipsoidal sporangiospores (5.4–18.0 × 3.3–7.8 μm) and ellipsoidal or irregular chlamydospores (6.3–16.8 μm × 8.1–12.5 μm), whereas M. xizangensis forms mainly ellipsoidal sporangiospores (3.9–8.4 × 2.4–4.9 μm) and globose chlamydospores (4.0–11.1 μm × 3.9–10.7 μm). Physiologically, the maximum growth temperature of the new species is 1 °C lower than that of M. xizangensis (30 °C vs. 31 °C).
3.2.3. Mucor polymorphus Z.Y. Ding, H. Zhao & X.Y. Liu, sp. Nov., Figure 6
Fungal Names—FN 573009.
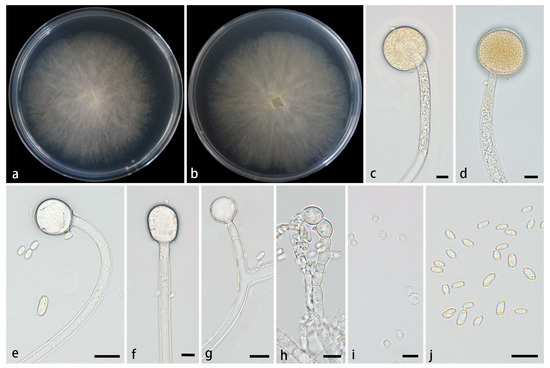
Figure 6.
Mucor polymorphus ex-holotype CGMCC 3.28969. (a,b) Colonies cultured on PDA at 25 °C for 3 days, (a) obverse, (b) reverse; (c,d) sporangia; (e–g) columellae; (h,i) chlamydospores; (j) sporangiospores. Scale bars: (c–j) 10 μm.
Type—China, Yunnan Province, Puer City, Simao District, Yixiang Town, Yutang Section, Longlongba Jinyu Tea Estate (22°68′48″ N, 101°07′32″ E, altitude 1549.97 m), from soil, 5 July 2024, Z.Y. Ding, holotype HMAS 354076, ex-type living culture CGMCC 3.28969 (=XG09597-11-1).
Etymology—The epithet polymorphus (Lat.) refers to the polymorphic chlamydospores in this species.
Description—Colonies on PDA at 25 °C for 3 d, reaching 69 mm in diameter, rapidly growing with a growth rate of 23 mm/d, initially white, gradually becoming yellowish-brown with age, floccose. Hyphae flourishing, unbranched, hyaline, aseptate when juvenile, septate with age, radial growth. Rhizoids absent. Stolons absent. Sporangiophores arising from substrate and aerial hyphae, erect or few slightly bent, occasionally branched, hyaline, 2.2–14.4 µm wide. Septa sometimes present in sporangiophores. Sporangia globose, pale yellow to light brown, 36.7–49.0 μm in diameter. Collars present or absent, usually small. Columellae globose, ovoid, ellipsoidal, hyaline or subhyaline, smooth-walled, 5.1–28.3 µm long and 5.8–24.1 µm wide. Apophyses absent. Sporangiospores usually fusiform, 3.7–7.0 µm long and 1.9–3.6 µm wide. Chlamydospores produced in substrate hyphae, in chains, globose, ovoid, cylindrical or irregular, 4.5–17.2 µm long and 3.9–13.9 µm wide. Zygospores unknown.
Cultured characteristics and maximum growth temperature: Under the same culture conditions, the colonies grow faster on PDA than on MEA (Figure 7). On PDA, the colonies reaching 82 mm in diameter for 5 d at 25 °C. On MEA, the colonies reaching 80 mm in diameter for 5 d at 25 °C. No growth was obversed at 32 °C.
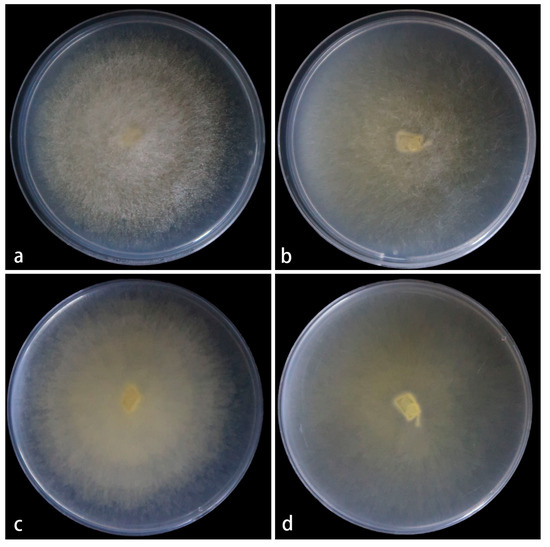
Figure 7.
Mucor polymorphus ex-holotype CGMCC 3.28969. Colonies cultured on PDA and on MEA at 25 °C for 5 days, (a,c) Colony on PDA; (b,d) Colony on MEA.
Additional strains examined—China, Yunnan Province, Puer City, Simao District, Yixiang Town, Yutang Section, Longlongba Jinyu Tea Estate (22°68′48″ N, 101°07′32″ E, altitude 1549.97 m), from soil, 5 July 2024, Z.Y. Ding living culture XG09597-11-2.
GenBank accession numbers—CGMCC 3.28969 (ITS, PV819209; LSU, PV833752; RPB1, PV948857), XG09597-11-2 (ITS, PV819210; LSU, PV833753; RPB1, PV948858).
Notes—Based on the ITS-LSU-RPB1 phylogenetic tree, two strains of the Mucor polymorphus sp. nov. formed a fully supported clade (MLBV = 100, BIPP = 1.00; Figure 1), which is closely related to M. multimorphus and M. xizangensis. Morphologically, the new species is distinguished from M. multimorphus and M. xizangensis by sporangiophores, columellae, sporangiospores, and chlamydospores. Compared with M. multimorphus, the new species exhibits thinner sporangiophores (2.2–14.4 μm vs. 5.0–15.8 μm), smaller columellae (5.1–28.3 × 5.8–24.1 μm vs. 10.1–40.0 × 7.5–39.7 μm), and smaller sporangiospores (5.1–28.3 × 1.9–3.6 μm vs. 5.4–18.0 × 3.3–7.8 μm). Moreover, the new species produces globose, ovoid, and ellipsoidal columellae and fusiform sporangiospores, while M. multimorphus forms globose, ellipsoidal, and pear-shaped columellae and fusiform or ellipsoidal sporangiospores. Regarding chlamydospores, the new species produces chain-like with globose, ovoid, cylindrical or irregular ones, while M. multimorphus forms ellipsoidal or irregular ones. In contrast to M. xizangensis, the new species features thinner sporangiophores (2.2–14.4 μm vs. 5.0–17.8 μm), smaller columellae (5.1–28.3 × 5.8–24.1 μm vs. 7.9–30.1 × 7.7–29.4 μm), smaller sporangiospores (5.1–28.3 × 1.9–3.6 μm vs. 3.9–8.4 × 2.4–4.9 μm). And M. xizangensis produces globose, ellipsoidal, ovoid, and pyriform columellae, ellipsoidal sporangiospores and globose chlamydospores. Physiologically, the maximum growth temperature of the new species is 1 °C higher than those of M. multimorphus and the same as those of M. xizangensis.
3.2.4. Mucor xizangensis Z.Y. Ding, H. Zhao & X.Y. Liu, sp. Nov., Figure 8
Fungal Names—FN 572032.
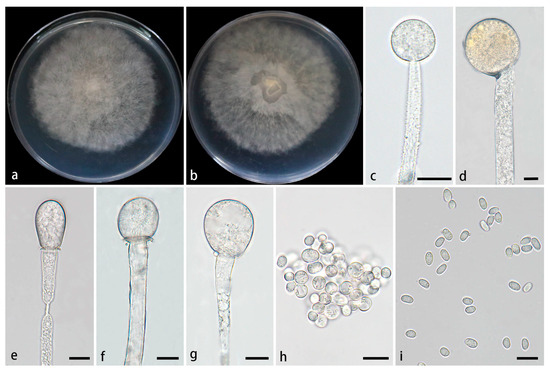
Figure 8.
Mucor xizangensis ex-holotype CGMCC 3.28971. (a,b) Colonies cultured on PDA at 25 °C for 5 days, (a) obverse, (b) reverse; (c,d) sporangia; (e–g) columellae; (h) chlamydospores; (i) sporangiospores. Scale bars: (c–i) 10 μm.
Type—China, Xizang Autonomous Region, Nyingchi City and Milin City, close to the Yarlung Zangbo Grand Canyon (29°64′36″ N, 94°88′53″ E, altitude 2779.72 m), from soil, 29 August 2024, X.Y. Liu, holotype HMAS 354078, ex-type living culture CGMCC 3.28971 (=XG10424-13-1).
Etymology—The epithet xizangensis (Lat.) refers to the location, Xizang Autonomous Region, China, where the ex-holotype was collected.
Description—Colonies on PDA at 25 °C for 5 d, reaching 80 mm in diameter, rapidly growing with a growth rate of 16 mm/d, initially white, gradually becoming grayish-white, floccose. Hyphae flourishing, unbranched, hyaline, radial growth. Rhizoids absent. Stolons absent. Sporangiophores arising from substrate and aerial hyphae, erect or few slightly bent, unbranched, hyaline, 5.0–17.8 µm wide. Sporangia globose, white to light grayish-brown, 23.3–58.6 μm in diameter. Collars present or absent, if present usually distinct and large. Columellae globose, ellipsoidal, ovoid, sometimes pyriform, hyaline or subhyaline, smooth-walled, 7.9–30.1 µm long and 7.7–29.4 µm wide. Apophyses absent. Sporangiospores usually ellipsoidal, 3.9–8.4 µm long and 2.4–4.9 µm wide. Chlamydospores produced in substrate hyphae, mainly globose, 4.0–11.1 µm long and 3.9–10.7 µm wide. Zygospores unknown.
Cultured characteristics and maximum growth temperature: Under the same culture conditions, the colonies grow faster on PDA than on MEA (Figure 9). On PDA, the colonies reaching 80 mm in diameter for 5 d at 25 °C. On MEA, the colonies reaching 75 mm in diameter for 5 d at 25 °C. No growth was obversed at 32 °C.
Figure 9.
Mucor xizangensis ex-holotype CGMCC 3.28971. Colonies cultured on PDA and on MEA at 25 °C for 5 days, (a,c) Colony on PDA; (b,d) Colony on MEA.
Additional strains examined—China, Xizang Autonomous Region, Nyingchi City and Milin City, close to the Yarlung Zangbo Grand Canyon (29°64′36″ N, 94°88′53″ E, altitude 2779.72 m), from soil, 29 August 2024, Z.Y. Ding, living culture XG10424-13-2.
GenBank accession numbers—CGMCC 3.28971 (ITS, PV819213; LSU, PV833756; RPB1, PV973985), XG10424-13-2 (ITS, PV819214; LSU, PV833757; RPB1, PV973986).
Notes—Based on the ITS-LSU-RPB1 phylogenetic tree, two strains of the Mucor xizangensis sp. nov. formed a fully supported independent lineage (MLBV = 100, BIPP = 1.00; Figure 1), which is closely related to M. multimorphus. Morphologically, the new species differs from M. multimorphus in sporangiophores, sporangia, sporangiospores, and chlamydospores. Specifically, the species does not form swelling on sporangiophores, while M. multimorphus occasionally develops a swelling on these structures. The sporangia of the new species are smaller than those of M. multimorphus (23.3–58.6 μm vs. 33.8–70.0 μm). In sporangiospores and chlamydospores, the new species is distinguished from M. multimorphus by smaller size and fewer shapes. More precisely, the new species produces predominantly ellipsoidal sporangiospores (3.9–8.4 × 2.4–4.9 μm) and globose chlamydospores (4.0–11.1 μm × 3.9–10.7 μm), while M. multimorphus develops fusiform and ellipsoidal sporangiospores (5.4–18.0 × 3.3–7.8 μm) and ellipsoidal or irregular chlamydospores (6.3–16.8 μm × 8.1–12.5 μm).
4. Morphological Comparison and Key to the Genus Mucor in China
Together with the four new Mucor species proposed, 28 species of this genus have been found in China. Since the detailed description of Mucor gigasporus was not available, the remaining 27 species were compared (Table S2), and a concise key to these species was provided. The characteristics used in the key include colonies, hyphae, sporangiophores, sporangia, collars, columellae, sporangiospores, chlamydospores, and zygospores.
- Zygospores known………………………………………………..……….M. homothallicusZygospores unknown…………………………………………………………………….....2
- Aborted sporangia known……………………………………………………………….....3Aborted sporangia unknown……………………………………………………………....6
- Fertile sporangia globose only………………………………………………….……..…...4Fertile sporangia globose and subglobose………………………………...……………...5
- Fertile sporangia 19.0–30.0 μm diameter……………………………M. abortisporangiumFertile sporangia 19.0–34.5 μm diameter………………………………………M. radiates
- Columellae 7.5–24.0 μm diameter…………………………………..…M. chlamydosporusColumellae 17.0–49.5 μm diameter……………………………….…….…… M. orientalis
- Collars known………………………………………………………………………………..7Collars unknown…………………………………………………………..……………….22
- Hyphae branched………………………………………………………….……..………….8Hyphae unbranched……………………………………………...…….…….………..…..19
- Sporangiospores shape > 2 kinds…………………………………………………..……...9Sporangiospores shape ≤ 2 kinds……………………………………………....………...10
- Fertile sporangia 17.0–52.5 μm diameter…………………………….……M. breviphorusFertile sporangia 33.8–70.0 μm diameter…………………………….…M. multimorphus
- Sporangiophores branched………………………………………….…………….…..….11Sporangiophores unbranched………………………………………………………...….14
- Sporangia > 65 μm diameter……………………………………………………………...12Sporangia < 65 μm diameter……………………………………………………………...13
- Sporangiospores 4.0–7.0 × 3.5–6.0 μm………………………………………...M. robustusSporangiospores 4.5–10.5 × 3.0–7.0μm…………………………….…..M. sinosaturninus
- Sporangiospores 4.0–7.0 × 3.0–5.0 μm…………………………………..M. changshaensisSporangiospores 3.5–5.5 × 2.5–3.5 μm……………………….…………...M. moniliformis
- Chlamydospores known……………………………………………………………….….15Chlamydospores unknown……………………………………………………………….18
- Sporangia > 38 μm diameter………………………………………………………...……16Sporangia < 38 μm diameter……………………………………………………...………17
- Columellaesubglobose, globose, ellipsoidal……………………………...M. globosporusColumellae hemispherical or depressed globose………………….....M. hemisphaericus
- Sporangiospores usually ellipsoid, occasionally reniform………..M. heilongjiangensisSporangiospores fusiform or ellipsoid……………………………….…....M. rhizosporus
- Collars obvious………………………………...……………………………M. amphisporusCollars not obvious………………………….…………………….…….M. fusiformisporus
- Chlamydospores known……………………………………………………………….....20Chlamydospores unknown……………………………………………………………….21
- Sporangiospores usually fusiform……………………………….……….M. polymorphusSporangiospores usually ellipsoidal……………………………………….M. xizangensis
- Columellae multiple shaped, with pyriform……………………….…………….M. tofusColumellae multiple shaped, without pyriform…………………..………M. brunneolus
- Chlamydospores known…………………………………………………………….…….23Chlamydospores unknown……………………………………………………………….26
- Fertile sporangia > 44 μm diameter………………………………………………..…….24Fertile sporangia < 44 μm diameter…………………………..…………........................25
- Columellae elongated-conical, cylindrical……………………………..M. chuxiongensisColumellae globose to subglobose………………………………………M. donglingensis
- Columellae hemispherical or depressed globose…………………………...M. floccosusColumellae conical to cylindrical………………………………………..M. hyalinosporus
- Sporangiophores unbranched……………………………………………….…..M. lobatusSporangiophores branched……………………………………….…………..…..M. rongii
5. Discussion
The first description of the genus Mucor dates back to 1850 [21], and since then its taxonomic studies have been continuously deepened. In 2016, Spatafora et al. [2] conducted phylogenetic analyses based on genomic data, segregating the Mucor-lineage fungi from the traditional phylum Zygomycota and establishing a distinct phylum, Mucoromycota. This taxonomic revision precisely defined their phylogenetic position in the fungal kingdom.
Modern fungal taxonomy relies predominantly on molecular data as the primary criterion to establish new taxonomic groups or evaluate interspecific relationships. Classical morphological characteristics (e.g., hyphal structure and spore morphology) and physiological traits (e.g., temperature tolerance) remain essential supplements for species delimitation. In phylogenetic research, integrating multigene markers such as ITS, LSU [25,26,57], and protein-coding genes like RPB1 is crucial for resolving the evolutionary relationships in taxonomically complex lineages [28]. Most Mucor species delimitation studies commonly use ITS and LSU as genetic markers due to their high availability, while genes like RPB1 aid in fine-scale analyses. Phylogenetic inferences via Maximum Likelihood (ML) and Bayesian Inference (BI) consistently showed that the novel species occupy stable phylogenetic positions with strong statistical support.
In this study, four novel Mucor species (M. globosporus sp. nov., M. multimorphus sp. nov., M. polymorphus sp. nov., and M. xizangensis sp. nov.) from China were identified through the integration of molecular data of ITS, LSU, and RPB1, combined with phenotypic observation and physiological trait assessments. Additionally, a systematic comparison of the morphological characteristics between these four novel species and their close relatives was performed (Table 2).

Table 2.
Morphological characteristics of Mucor species involved in this study.
Mucor globosporus is closely related to M. moniliformis. In contrast to M. moniliformis, M. globosporus possesses distinct columellae and chlamydospore shapes, and rhizoids. Mucor multimorphus and M. xizangensis are sister taxa. Morphologically, M. multimorphus exhibits larger sporangia, and its sporangiospores and chlamydospores are both larger in size and more varied in shape. Additionally, M. multimorphus occasionally forms swellings on sporangiophores. Mucor polymorphus is closely related to M. multimorphus and M. xizangensis. Compared with the latter two, M. polymorphus has thinner sporangiophores, smaller columellae, and smaller sporangiospores. Moreover, the shapes of its columellae, sporangiospores, and chlamydospores differ significantly.
Physiologically, the thermal tolerance thresholds of these novel taxonomic groups were determined using a temperature gradient cultivation technique. Growth characterization revealed significant differences in the maximum growth temperatures among the four new Mucor species: M. globosporus 32 °C, M. multimorphus 30 °C, M. polymorphus 31 °C, and M. xizangensis 31 °C. These temperature parameters are consistent with the mesophilic physiological characteristics of most Mucor species, further supporting the taxonomic affiliation of these novel species groups with the Mucor genus.
Over the past five years, at least 45 new species of the genus Mucor have been discovered and described from diverse habitats such as soil, insects, plants, fungi, Mao-tofu, and animal dung (http://www.indexfungorum.org/, accessed on 30 June 2025), suggesting that this taxon retains extensive distribution potential in numerous understudied habitats. The four new species described herein further update the globally recognized species count of the genus to 137.
As a region of high fungal diversity, China contributes substantially to global Mucor diversity. Based on existing records, the inclusion of these four new species brings the total number of recognized Mucor species in China to 28 (https://www.catalogueoflife.org/, accessed on 30 June 2025), accounting for approximately 20% of the globally recognized Mucor species pool. This high regional richness is presumably attributed to the diverse climate zones and ecosystems in China. Taxonomic sampling for fungi in China has primarily focused on provinces with distinct ecosystems such as Yunnan, Sichuan, Guangdong, and Fujian, where intensive surveys have also facilitated Mucor discoveries. Notably, Yunnan leads in the description of new fungal species, including Mucor species. Beyond Mucor, China’s fungal diversity has also helped refine global fungal diversity estimates by filling gaps in East Asian mycobiota data, improving global distribution models, and expanding knowledge of extremophilic soil fungi.
Despite progress in characterizing Mucor’s species diversity and global distribution, its ecological functions have not been systematically analyzed, and Mucor groups in some extreme habitats remain underinvestigated. Future research should therefore prioritize under-sampled regions. Based on the above, this study not only provides new materials for Mucor taxonomy but also further highlights the necessity of continuously exploring their diversity and ecological functions.
Supplementary Materials
The following supporting information can be downloaded at https://www.mdpi.com/article/10.3390/jof11090682/s1: Supplementary Table S1: GenBank accession numbers of Mucor and Backusella strains in this study; Supplementary File S1: the combined ITS-LSU-RPB1 sequence matrix used in this study; Supplementary Table S2: morphological description table; References [58,59,60] are cited in Supplementary Table S2.
Author Contributions
Z.-Y.D. was responsible for DNA sequencing, photo editing, and paper drafting; X.-Y.J. and W.-X.L. were responsible for data analyses; F.L. collected soil samples; S.W. and H.Z. were responsible for manuscript revision, conceiving and revising the paper; X.-Y.L. took charge of naming the new species, conceiving and revising the paper, and providing funding. All authors have read and agreed to the published version of the manuscript.
Funding
This study was supported by the National Natural Science Foundation of China (Nos. 32170012, 32470004, and 32300011), Ji’nan City’s ‘New University 20 Policies’ Initiative for Innovative Research Teams Project (202228028), Innovative Agricultural Application Technology Project of Jinan City (CX202210), and Key Technological Innovation Program of Shandong Province, China (2022CXGC020710).
Institutional Review Board Statement
Not applicable for studies involving humans or animals.
Informed Consent Statement
Not applicable.
Data Availability Statement
The original contributions presented in this study are included in the article/supplementary material. Further inquiries can be directed to the corresponding author.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Nicolás, F.E.; Murcia, L.; Navarro, E.; Navarro-Mendoza, M.I.; Pérez-Arques, C.; Garre, V. Mucorales Species and Macrophages. J. Fungi 2020, 6, 94. [Google Scholar] [CrossRef]
- Spatafora, J.W.; Chang, Y.; Benny, G.L.; Lazarus, K.; Smith, M.E.; Berbee, M.L.; Bonito, G.; Corradi, N.; Grigoriev, I.; Gryganskyi, A.; et al. A phylum-level phylogenetic classification of zygomycete fungi based on genome-scale data. Mycologia 2016, 108, 1028–1046. [Google Scholar] [CrossRef]
- Pitt, J.I.; Hocking, A.D. Fungi and Food Spoilage; Springer: New York, NY, USA, 2009; pp. 151–157. [Google Scholar]
- Walther, G.; Pawłowska, J.; Alastruey-Izquierdo, A.; Wrzosek, M.; Rodriguez-Tudela, J.L.; Dolatabadi, S.; Chakrabarti, A.; de Hoog, G.S. DNA barcoding in Mucorales: An inventory of biodiversity. Persoonia 2013, 30, 11–47. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, T.T.T.; Lee, H.B. Mucor cheongyangensis, a new species isolated from the surface of Lycorma delicatula in Korea. Phytotaxa 2020, 446, 33–42. [Google Scholar] [CrossRef]
- Boonmee, S.; Wanasinghe, D.N.; Calabon, M.S.; Huanraluek, N.; Chandrasiri, S.K.U.; Jones, G.E.B.; Rossi, W.; Leonardi, M.; Singh, S.K.; Rana, S.; et al. Fungal diversity notes 1387–1511: Taxonomic and phylogenetic contributions on genera and species of fungal taxa. Fungal Divers. 2021, 111, 1–335. [Google Scholar] [CrossRef]
- Lima, D.X.; De Azevedo Santiago, A.L.C.M.; De Souza-Motta, C.M. Diversity of Mucorales in natural and degraded semi-arid soils. Braz. J. Bot. 2016, 39, 1127–1133. [Google Scholar] [CrossRef]
- de Souza, C.A.F.; Lima, D.X.; Gurgel, L.M.S.; de Azevedo Santiago, A.L.C.M. Coprophilous Mucorales (ex Zygomycota) from three areas in the semi-arid of Pernambuco, Brazil. Braz. J. Microbiol. 2017, 48, 79–86. [Google Scholar] [CrossRef][Green Version]
- Ribes, J.A.; Vanover-Sams, C.L.; Baker, D.J. Zygomycetes in Human Disease. Clin. Microbiol. Rev. 2000, 13, 236–301. [Google Scholar] [CrossRef] [PubMed]
- Chandra, S.; Woodgyer, A. Primary cutaneous zygomycosis due to Mucor circinelloides. Australas. J. Dermatol. 2002, 43, 39–42. [Google Scholar] [CrossRef]
- Alvarez, E.; Cano, J.; Stchigel, A.M.; Sutton, D.A.; Fothergill, A.W.; Salas, V.; Rinaldi, M.G.; Guarro, J. Two new species of Mucor from clinical samples. Med. Mycol. 2011, 49, 62–72. [Google Scholar] [CrossRef]
- Nout, M.J.R.; Aidoo, K.E. Hofrichter, M., Ed.; Asian Fungal Fermented Food. In Industrial Applications; Springer: Berlin/Heidelberg, Germany, 2010; Volume 10, pp. 29–58. [Google Scholar] [CrossRef]
- Hermet, A.; Méheust, D.; Mounier, J.; Barbier, G.; Jany, J.L. Molecular systematics in the genus Mucor with special regards to species encountered in cheese. Fungal Biol. 2012, 116, 692–705. [Google Scholar] [CrossRef]
- Alves, M.H.; de Campos-Takaki, G.M.; Okada, K.; Ferreira-Pessoa, I.H.; Milanez, A.I. Detection of extracellular protease in Mucor species. Rev. Iberoam. Micol. 2005, 22, 114–117. [Google Scholar] [CrossRef]
- Roopesh, K.; Ramachandran, S.; Nampoothiri, K.M.; Szakacs, G.; Pandey, A. Comparison of phytase production on wheat bran and oilcakes in solid-state fermentation by Mucor racemosus. Bioresour. Technol. 2006, 97, 506–511. [Google Scholar] [CrossRef] [PubMed]
- Yazdi, M.T.; Zarrini, G.; Mohit, E.; Faramarzi, M.A.; Setayesh, N.; Sedighi, N.; Mohseni, F.A. Mucor hiemalis: A new source for uricase production. World J. Microbiol. Biotechnol. 2006, 22, 325–330. [Google Scholar] [CrossRef]
- de Carvalho, A.K.F.; Faria, E.; Rivaldi, J.D.; Andrade, G.S.S.; Oliveira, P.C.; de Castro, H.F. Performance of whole-cells lipase derived from Mucor circinelloides as a catalyst in the ethanolysis of non-edible vegetable oils under batch and continuous run conditions. Ind. Crops Prod. 2015, 67, 287–294. [Google Scholar] [CrossRef]
- de Souza, P.M.; de Assis Bittencourt, M.L.; Caprara, C.C.; de Freitas, M.; de Almeida, R.P.C.; Silveira, D.; Fonseca, Y.M.; Filho, E.X.F.; Junior, A.P.; Magalhães, P.O. A biotechnology perspective of fungal proteases. Braz. J. Microbiol. 2015, 46, 337–346. [Google Scholar] [CrossRef]
- Voigt, K.; Wolf, T.; Ochsenreiter, K.; Nagy, G.; Kaerger, K.; Shelest, E.; Papp, T. Hoffmeister, D., Ed.; 15 Genetic and metabolic aspects of primary and secondary metabolism of the Zygomycetes. In Biochemistry and Molecular Biology, 3rd ed.; Springer: Berlin, Germany, 2016; Volume III, pp. 361–385. [Google Scholar] [CrossRef]
- Morin-Sardin, S.; Nodet, P.; Coton, E.; Jany, J.L. Mucor: A Janus-faced fungal genus with human health impact and industrial applications. Fungal Biol. Rev. 2017, 33, 12–32. [Google Scholar] [CrossRef]
- Fresenius, G. Beiträge Zur Mykologie; Bei Heinrich Ludwig Brönner: Frankfurt, Germany, 1850; pp. 1–38. [Google Scholar]
- Benny, G.L.; Smith, M.E.; Kirk, P.M.; Tretter, E.D.; White, M.M. Li, D.W., Ed.; Challenges and Future Perspectives in the Systematics of Kickxellomycotina, Mortierellomycotina, Mucoromycotina, and Zoopagomycotina. In Biology of Microfungi (Fungal Biology); Springer: Cham, Switzerland, 2016. [Google Scholar] [CrossRef]
- Hurdeal, V.G.; Gentekaki, E.; Hyde, K.D.; Nguyen, T.T.T.; Lee, H.B. Novel Mucor species (Mucoromycetes, Mucoraceae) from northern Thailand. MycoKeys 2021, 84, 57–78. [Google Scholar] [CrossRef]
- Nguyen, T.T.T.; Jeon, Y.J.; Mun, H.Y.; Goh, J.; Chung, N.; Lee, H.B. Isolation and Characterization of Four Unrecorded Mucor Species in Korea. Mycobiology 2019, 48, 29–36. [Google Scholar] [CrossRef] [PubMed]
- Lima, D.X.; Barreto, R.W.; Lee, H.B.; Cordeiro, T.R.L.; de Souza, C.A.F.; de Oliveira, R.J.V.; Santiago, A.L.C.M. Novel Mucoralean Fungus from a Repugnant Substrate: Mucor merdophylus sp. nov., isolated from dog excrement. Curr. Microbiol. 2020, 77, 2642–2649. [Google Scholar] [CrossRef] [PubMed]
- Lima, C.L.F.; Lima, D.X.; de Souza, C.A.F.; de Oliveira, R.J.V.; Cavalcanti, I.B.; Gurgel, L.M.S.; Santiago, A. Description of Mucor pernambucoensis (Mucorales, mucoromycota), a new species isolated from the Brazilian upland rainforest. Phytotaxa 2018, 350, 274. [Google Scholar] [CrossRef]
- Zhao, H.; Nie, Y.; Zong, T.; Wang, K.; Lv, M.; Cui, Y.; Tohtirjap, A.; Chen, J.; Zhao, C.; Wu, F.; et al. Species diversity, updated classification and divergence times of the phylum Mucoromycota. Fungal Divers. 2023, 123, 49–157. [Google Scholar] [CrossRef]
- Walther, G.; Wagner, L.; Kurzai, O. Updates on the Taxonomy of Mucorales with an Emphasis on Clinically Important Taxa. J. Fungi 2019, 5, 106. [Google Scholar] [CrossRef]
- Ferroni, G.D.; Kaminski, J.S. Psychrophiles, psychrotrophs, and mesophiles in an environment which experiences seasonal temperature fluctuations. Can. J. Microbiol. 1980, 26, 1184–1190. [Google Scholar] [CrossRef]
- Sandle, T.; Skinner, K. Study of psychrophilic and psychrotolerant micro-organisms isolated in cold rooms used for pharmaceutical processing. J. Appl. Microbiol. 2013, 114, 1166–1174. [Google Scholar] [CrossRef]
- Linnaeus, C. Species Plantarum; Laurentius Salvius: Stockholm, Sweden, 1753; 1200p. [Google Scholar]
- Wijayawardene, N.N.; Pawłowska, J.; Letcher, P.M.; Kirk, P.M.; Humber, R.A.; Schüßler, A.; Wrzosek, M.; Muszewska, A.; Okrasinka, A.; Istel, L.; et al. Notes for genera: Basal clades of Fungi (including Aphelidiomycota, Basidiobolomycota, Blastocladiomycota, Calcarisporiellomycota, Caulochytriomycota, Chytridiomycota, Entomophthoromycota, Glomeromycota, Kickxellomycota, Monoblepharomycota, Mortierellomycota, Mucoromycota, Neocallimastigomycota, Olpidiomycota, Rozellomycota and Zoopagomycota). Fungal Divers. 2018, 92, 43–129. [Google Scholar] [CrossRef]
- Nguyen, T.T.T.; Lee, H.B. Discovery of Three New Mucor Species Associated with Cricket Insects in Korea. J. Fungi 2022, 8, 601. [Google Scholar] [CrossRef]
- Li, N.; Bowling, J.; de Hoog, S.; Aneke, C.I.; Youn, J.; Shahegh, S.; Cuellar-Rodriguez, J.; Kanakry, C.G.; Rodriguez Pena, M.; Ahmed, S.A.; et al. Mucor germinans, a novel dimorphic species resembling Paracoccidioides in a clinical sample: Questions on ecological strategy. mBio 2024, 15, e00144-24. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, T.T.T.; de Santiago, A.L.C.M.A.; Hallsworth, J.E.; Cordeiro, T.R.L.; Voigt, K.; Kirk, P.M.; Crous, P.W.; Júnior, M.A.M.; Elsztein, C.; Lee, H.B. New Mucorales from opposite ends of the world. Stud. Mycol. 2024, 109, 273–321. [Google Scholar] [CrossRef]
- Zhao, H.; Nie, Y.; Huang, B.; Liu, X.Y. Unveiling species diversity within early-diverging fungi from China I: Three new species of Backusella (Backusellaceae, Mucoromycota). MycoKeys 2024, 109, 285–304. [Google Scholar] [CrossRef]
- Tao, M.F.; Ding, Z.Y.; Wang, Y.X.; Zhang, Z.X.; Zhao, H.; Meng, Z.; Liu, X.Y. Unveiling species diversity within early-diverging fungi from China II: Three new species of Absidia (Cunninghamellaceae, Mucoromycota) from Hainan Province. MycoKeys 2024, 110, 255–272. [Google Scholar] [CrossRef]
- Wang, Y.X.; Zhao, H.; Jiang, Y.; Liu, X.Y.; Tao, M.F.; Liu, X.Y. Unveiling species diversity within early-diverging fungi from China III: Six new species and a new record of Gongronella. (Cunninghamellaceae, Mucoromycota). MycoKeys 2024, 110, 287–317. [Google Scholar] [CrossRef]
- Ding, Z.Y.; Ji, X.Y.; Tao, M.F.; Jiang, Y.; Liu, W.X.; Wang, Y.X.; Meng, Z.; Liu, X.Y. Unveiling species diversity within early-diverging fungi from China IV: Four new species of Absidia (Cunninghamellaceae, Mucoromycota). MycoKeys 2025, 119, 29–46. [Google Scholar] [CrossRef] [PubMed]
- Ji, X.Y.; Ding, Z.Y.; Nie, Y.; Zhao, H.; Wang, S.; Huang, B.; Liu, X.Y. Unveiling species diversity: Within early-diverging fungi from China V: Five new species of Absidia (Cunninghamellaceae, Mucoromycota). MycoKeys 2025, 117, 267–288. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.X.; Ding, Z.Y.; Ji, X.Y.; Meng, Z.; Liu, X.Y. Unveiling species diversity within early-diverging fungi from China VI: Four Absidia sp. nov. (Mucorales) in Guizhou and Hainan. Microorganisms 2025, 13, 1315. [Google Scholar] [CrossRef] [PubMed]
- Ding, Z.Y.; Tao, M.F.; Ji, X.Y.; Jiang, Y.; Wang, Y.X.; Liu, W.X.; Wang, S.; Liu, X.Y. Unveiling species diversity within early-diverging fungi from China VII: Seven new species of Cunninghamella (Mucoromycota). J. Fungi 2025, 11, 417. [Google Scholar] [CrossRef]
- Ji, X.Y.; Jiang, Y.; Li, F.; Ding, Z.Y.; Meng, Z.; Liu, X.Y. Unveiling species diversity within early-diverging fungi from China VIII: Four new species in Mortierellaceae (Mortierellomycota). Microorganisms 2025, 13, 1330. [Google Scholar] [CrossRef]
- Li, W.X.; Wei, Y.H.; Zou, Y.; Liu, P.; Li, Z.; Gontcharov, A.A.; Stephenson, S.L.; Wang, Q.; Zhang, S.H.; Li, Y. Dictyostelid cellular slime molds from the Russian Far East. Protist 2020, 171, 125756. [Google Scholar] [CrossRef]
- Zou, Y.; Hou, J.G.; Guo, S.N.; Li, C.T.; Li, Z.; Stephenson, S.L.; Pavlov, I.N.; Liu, P.; Li, Y. Diversity of dictyostelid cellular slime molds, including two species new to science, in forest soils of Changbai Mountain, China. Microbiol. Spectr. 2022, 10, 02402–02422. [Google Scholar] [CrossRef]
- Corry, J.E.L. Rose bengal chloramphenicol (RBC) agar. In Progress in Industrial Microbiology; Elsevier: Amsterdam, The Netherlands, 1995; pp. 431–433. [Google Scholar]
- Zheng, R.Y.; Chen, G.Q.; Huang, H.; Liu, X.Y. A monograph of Rhizopus. Sydowia 2007, 59, 273–372. [Google Scholar]
- Zheng, R.Y.; Liu, X.Y.; Li, R.Y. More Rhizomucor causing human mucormycosis from China: R. chlamydosporus sp. nov. Sydowia 2009, 61, 135–147. [Google Scholar]
- Zong, T.K.; Zhao, H.; Liu, X.Y.; Ren, L.Y.; Zhao, C.L.; Liu, X.Y. Taxonomy and phylogeny of the Absidia (Cunninghamellaceae, Mucorales) introducing nine new species and two new combinations from China. Res. Sq. 2021. [Google Scholar] [CrossRef]
- Guo, L.D.; Hyde, K.D.; Liew, E.C.Y. Identification of endophytic fungi from Livistona chinensis based on morphology and rDNA sequences. New Phytol. 2000, 147, 617–630. [Google Scholar] [CrossRef] [PubMed]
- White, T.J.; Bruns, T.D.; Lee, S.B.; Taylor, J.W.; Innis, M.A.; Gelfand, D.H.; Sninsky, J. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In PCR Protocols: A Guide to Methods and Applications; Academic Press: New York, NY, USA, 1990; pp. 315–322. [Google Scholar]
- Vilgalys, R.; Hester, M. Rapid genetic identification and mapping of enzymatically amplified ribosomal DNA from several Cryptococcus species. J. Bacteriol. 1990, 172, 4238–4246. [Google Scholar] [CrossRef]
- Stiller, J.W.; Hall, B.D. The origin of red algae: Implications for plastid evolution. Proc. Natl. Acad. Sci. USA 1997, 94, 4520–4525. [Google Scholar] [CrossRef]
- Nylander, J. MrModeltest V2. program distributed by the author. Bioinformatics 2004, 24, 581–583. [Google Scholar] [CrossRef]
- Stamatakis, A. RAxML Version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 2014, 30, 1312–1313. [Google Scholar] [CrossRef] [PubMed]
- Huelsenbeck, J.P.; Ronquist, F. MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics 2001, 17, 754–755. [Google Scholar] [CrossRef] [PubMed]
- Madden, A.A.; Stchigel, A.M.; Guarro, J.; Sutton, D.; Starks, P.T. Mucor nidicola sp. nov., a fungal species isolated from an invasive paper wasp nest. Int. J. Syst. Evol. Microbiol. 2011, 62, 1710–1714. [Google Scholar] [CrossRef]
- Chai, C.Y.; Liu, W.J.; Cheng, H.; Hui, F.L. Mucor chuxiongensis sp. nov., a novel fungal species isolated from rotten wood. Int. J. Syst. Evol. Microbiol. 2019, 69, 1881–1889. [Google Scholar] [CrossRef]
- Bai, F.R.; Yao, S.; Cai, C.S.; Zhang, T.C.; Wang, Y.; Liu, W.; Ma, B.W.; Rong, C.Y.; Cheng, C. Mucor rongii sp. nov., a new cold-tolerant species from China. Curr. Microbiol. 2021, 78, 2464–2469. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.N.; Zheng, R.Y. Mucor tofus, a new species isolated from Mao-tofu (a fermented soybean food) in China. Phytotaxa 2022, 567, 233–244. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).