National Belgian Study on Terbinafine Resistance in Trichophyton interdigitale/mentagrophytes/indotineae (2022–2023): Epidemiology and Molecular Features
Abstract
1. Introduction
2. Materials and Methods
2.1. Samples
2.2. Screening Method “Derma-Check”
2.3. EUCAST E.Def 11.0 Microdilution Plates
2.4. Whole Genome Sequencing
2.5. Statistics
3. Results
3.1. Species Identification and Phylogenetic Classification
3.2. Screening for Terbinafine Resistance Using the Agar Dilution Method
3.3. Antifungal Susceptibility Testing by Microdilution (EUCAST E.Def 11.0)
3.3.1. Terbinafine Susceptibility Profile
3.3.2. Itraconazole Susceptibility Profile
3.3.3. Voriconazole Susceptibility Profile
3.3.4. Amorolfine Susceptibility Profile
3.4. Squalene Epoxidase Substitutions and Their Association with Terbinafine Resistance
3.5. Impact of K276N on Antifungal Susceptibility
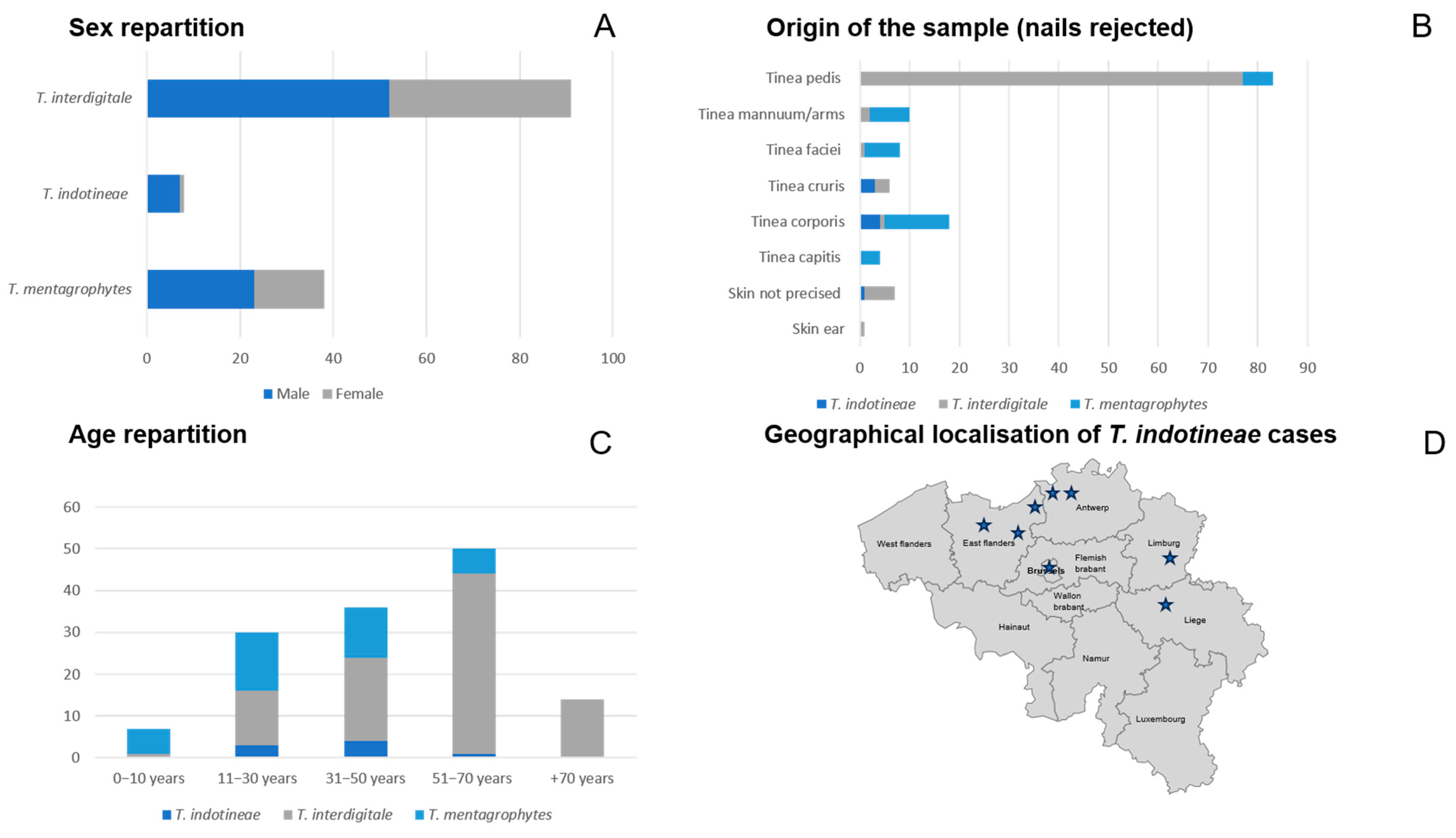
3.6. Epidemiological Characteristics of Collected Dermatophyte Strains
3.7. Type VII T. mentagrophytes: Epidemiological and Resistance Profile
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Havlickova, B.; Czaika, V.A.; Friedrich, M. Epidemiological trends in skin mycoses worldwide. Mycoses 2008, 51, 2–15. [Google Scholar] [CrossRef] [PubMed]
- Dolenc-Voljč, M. Dermatophyte infections in humans: Current trends and future prospects. In Medical Mycology: Current Trends and Future Prospects; CRC Press: Boca Raton, FL, USA, 2015. [Google Scholar]
- Thakur, R.; Kushwaha, P.; Kalsi, A.S. Tinea universalis due to Trichophyton indotineae in an adult male. Indian J. Med. Microbiol. 2023, 46, 100476. [Google Scholar] [CrossRef]
- Uhrlaß, S.; Verma, S.B.; Gräser, Y.; Rezaei-Matehkolaei, A.; Hatami, M.; Schaller, M.; Nenoff, P. Trichophyton indotineae—An Emerging Pathogen Causing Recalcitrant Dermatophytoses in India and Worldwide—A Multidimensional Perspective. J. Fungi 2022, 8, 757. [Google Scholar] [CrossRef]
- Singh, A.; Masih, A.; Khurana, A.; Singh, P.K.; Gupta, M.; Hagen, F.; Meis, J.F.; Chowdhary, A. High terbinafine resistance in Trichophyton interdigitale isolates in Delhi, India harbouring mutations in the squalene epoxidase gene. Mycoses 2018, 61, 477–484. [Google Scholar] [CrossRef]
- Singh, A.; Masih, A.; Monroy-Nieto, J.; Singh, P.K.; Bowers, J.; Travis, J.; Khurana, A.; Engelthaler, D.M.; Meis, J.F.; Chowdhary, A. A unique multidrug-resistant clonal Trichophyton population distinct from Trichophyton mentagrophytes/Trichophyton interdigitale complex causing an ongoing alarming dermatophytosis outbreak in India: Genomic insights and resistance profile. Fungal Genet. Biol. 2019, 133, 103266. [Google Scholar] [CrossRef]
- Kano, R.; Kimura, U.; Kakurai, M.; Hiruma, J.; Kamata, H.; Suga, Y.; Harada, K. Trichophyton indotineae sp. nov.: A New Highly Terbinafine-Resistant Anthropophilic Dermatophyte Species. Mycopathologia 2020, 185, 947–958. [Google Scholar] [CrossRef]
- Tang, C.; Kong, X.; Ahmed, S.A.; Thakur, R.; Chowdhary, A.; Nenoff, P.; Uhrlass, S.; Verma, S.B.; Meis, J.F.; Kandemir, H.; et al. Taxonomy of the Trichophyton mentagrophytes/T. interdigitale Species Complex Harboring the Highly Virulent, Multiresistant Genotype T. indotineae. Mycopathologia 2021, 186, 315–326. [Google Scholar] [CrossRef]
- Ebert, A.; Monod, M.; Salamin, K.; Burmester, A.; Uhrlaß, S.; Wiegand, C.; Hipler, U.C.; Krüger, C.; Koch, D.; Wittig, F.; et al. Alarming India-wide phenomenon of antifungal resistance in dermatophytes: A multicentre study. Mycoses 2020, 63, 717–728. [Google Scholar] [CrossRef] [PubMed]
- Kumar, P.; Ramachandran, S.; Das, S.; Bhattacharya, S.N.; Taneja, B. Insights into Changing Dermatophyte Spectrum in India Through Analysis of Cumulative 161,245 Cases Between 1939 and 2021. Mycopathologia 2023, 188, 183–202. [Google Scholar] [CrossRef]
- De, A.; Das, S.; Saha, R.; Sharma, N.; Khemka, M.; Singh, S.; Reja, A.H.H.; Kumar, P. The current Indian epidemic of dermatophytosis: A study on causative agents and sensitivity patterns. Indian J. Dermatol. 2020, 65, 118–122. [Google Scholar] [CrossRef] [PubMed]
- Khurana, A.; Masih, A.; Chowdhary, A.; Sardana, K.; Borker, S.; Gupta, A.; Gautam, R.K.; Sharma, P.K.; Jain, D. Correlation of invitro susceptibility based on MICs and SQLE mutations with clinical response to terbinafine in patients with tinea corporis/cruris. Antimicrob Agents Chemother 2018, 62, e01038-18. [Google Scholar] [CrossRef]
- Hsieh, A.; Quenan, S.; Riat, A.; Toutous-Trellu, L.; Fontao, L. A new mutation in the SQLE gene of Trichophyton mentagrophytes associated to terbinafine resistance in a couple with disseminated tinea corporis. J. Mycol. Med. 2019, 29, 352–355. [Google Scholar] [CrossRef]
- Sacheli, R.; Harag, S.; Dehavay, F.; Evrard, S.; Rousseaux, D.; Adjetey, A.; Seidel, L.; Laffineur, K.; Lagrou, K.; Hayette, M.P. Belgian national survey on tinea capitis: Epidemiological considerations and highlight of terbinafine-resistant T. mentagrophytes with a mutation on SQLE gene. J. Fungi 2020, 6, 195. [Google Scholar] [CrossRef]
- Osborne, C.S.; Leitner, I.; Favre, B.; Ryder, N.S. Amino acid substitution in Trichophyton rubrum squalene epoxidase associated with resistance to terbinafine. Antimicrob. Agents Chemother. 2005, 49, 2840–2844. [Google Scholar] [CrossRef]
- Taghipour, S.; Shamsizadeh, F.; Pchelin, I.M.; Rezaei-Matehhkolaei, A.; Mahmoudabadi, A.Z.; Valadan, R.; Ansari, S.; Katiraee, F.; Pakshir, K.; Zomorodian, K.; et al. Emergence of terbinafine resistant trichophyton mentagrophytes in iran, harboring mutations in the squalene epoxidase (Sqle) gene. Infect. Drug Resist. 2020, 13, 845–850. [Google Scholar] [CrossRef]
- Rudramurthy, S.M.; Shankarnarayan, S.A.; Dogra, S.; Shaw, D.; Mushtaq, K.; Paul, R.A.; Narang, T.; Chakrabarti, A. Mutation in the squalene epoxidase gene of trichophyton interdigitale and trichophyton rubrum associated with allylamine resistance. Antimicrob. Agents Chemother. 2018, 62, e02522-17. [Google Scholar] [CrossRef]
- Yamada, T.; Maeda, M.; Alshahni, M.M.; Tanaka, R.; Yaguchi, T.; Bontems, O.; Salamin, K.; Fratti, M.; Monod, M. Terbinafine resistance of Trichophyton clinical isolates caused by specific point mutations in the squalene epoxidase gene. Antimicrob. Agents Chemother. 2017, 61, e00115-17. [Google Scholar] [CrossRef] [PubMed]
- Burmester, A.; Hipler, U.C.; Uhrlaß, S.; Nenoff, P.; Singal, A.; Verma, S.B.; Elsner, P.; Wiegand, C. Indian Trichophyton mentagrophytes squalene epoxidase erg1 double mutants show high proportion of combined fluconazole and terbinafine resistance. Mycoses 2020, 63, 1175–1180. [Google Scholar] [CrossRef]
- Kong, X.; Tang, C.; Singh, A.; Ahmed, S.A.; Al-Hatmi, A.M.S.; Chowdhary, A.; Nenoff, P.; Gräser, Y.; Hainsworth, S.; Zhan, P.; et al. Antifungal susceptibility and mutations in the squalene epoxidase gene in dermatophytes of the trichophyton mentagrophytes species complex. Antimicrob. Agents Chemother. 2021, 65, e00056-21. [Google Scholar] [CrossRef] [PubMed]
- Burmester, A.; Hipler, U.C.; Hensche, R.; Elsner, P.; Wiegand, C. Point mutations in the squalene epoxidase gene of Indian ITS genotype VIII T. mentagrophytes identified after DNA isolation from infected scales. Med. Mycol. Case Rep. 2019, 26, 23–24. [Google Scholar] [CrossRef] [PubMed]
- Łagowski, D.; Gnat, S.; Nowakiewicz, A.; Osińska, M.; Dyląg, M. Intrinsic resistance to terbinafine among human and animal isolates of Trichophyton mentagrophytes related to amino acid substitution in the squalene epoxidase. Infection 2020, 48, 889–897. [Google Scholar] [CrossRef]
- Mirhendi, H.; Aboutalebian, S.; Jahanshiri, Z.; Rouhi, F.; Shidfar, M.R.; Chadeganipour, A.S.; Shadzi, S.; Kharazi, M.; Erami, M.; Rizi, M.H. Increasing and Alarming Prevalence of Trichophyton indotineae as the Primary Causal Agent of Skin Dermatophytosis in Iran. Mycoses 2025, 68, e70013. [Google Scholar] [CrossRef] [PubMed]
- Spivack, S.; Gold, J.A.W.; Lockhart, S.R.; Anand, P.; Quilter, L.A.S.; Smith, D.J.; Bowen, B.; Gould, J.M.; Eltokhy, A.; Gamal, A.; et al. Potential Sexual Transmission of Antifungal- Resistant Trichophyton indotineae. Emerg. Infect. Dis. 2024, 30, 807. [Google Scholar] [CrossRef]
- Jabet, A.; Chiarabini, T.; Hennequin, C.; Bouscarat, F.; Valin, N.; Cruchet, R.; Boo, N.; Chanal, J.; Schuttler, C.; Kirstetter, M.; et al. Autochthonous transmission of Trichophyton indotineae through sexual contact, France, 2024. Eurosurveillance 2025, 30, 2500416. [Google Scholar] [CrossRef] [PubMed Central]
- Moreno-Sabater, A.; Normand, A.C.; Bidaud, A.L.; Cremer, G.; Foulet, F.; Brun, S.; Bonnal, C.; Aït-Ammar, N.; Jabet, A.; Ayachi, A.; et al. Terbinafine Resistance in Dermatophytes: A French Multicenter Prospective Study. J. Fungi 2022, 8, 220. [Google Scholar] [CrossRef] [PubMed]
- Jabet, A.; Brun, S.; Normand, A.C.; Imbert, S.; Akhoundi, M.; Dannaoui, E.; Audiffred, L.; Chasset, F.; Izri, A.; Laroche, L.; et al. Extensive Dermatophytosis Caused by Terbinafi ne-Resistant Trichophyton indotineae, France. Emerg. Infect. Dis. 2022, 28, 229. [Google Scholar] [CrossRef]
- Dellière, S.; Joannard, B.; Benderdouche, M.; Mingui, A.; Gits-Muselli, M.; Hamane, S.; Alanio, A.; Petit, A.; Gabison, G.; Bagot, M.; et al. Emergence of Diffi cult-to-Treat Tinea Corporis Caused by Trichophyton mentagrophytes Complex Isolates, Paris, France. Emerg. Infect. Dis. 2022, 28, 224–228. [Google Scholar] [CrossRef] [PubMed]
- Klinger, M.; Theiler, M.; Bosshard, P.P. Epidemiological and clinical aspects of Trichophyton mentagrophytes/Trichophyton interdigitale infections in the Zurich area: A retrospective study using genotyping. J. Eur. Acad. Dermatol. Venereol. 2021, 35, 1017–1025. [Google Scholar] [CrossRef]
- Russo, G.; Toutous Trellu, L.; Fontao, L.; Ninet, B. Towards an Early Clinical and Biological Resistance Detection in Dermatophytosis: About 2 Cases of Trichophyton indotineae. J. Fungi 2023, 9, 733. [Google Scholar] [CrossRef]
- Siopi, M.; Efstathiou, I.; Theodoropoulos, K.; Pournaras, S.; Meletiadis, J. Molecular epidemiology and antifungal susceptibility of trichophyton isolates in greece: Emergence of terbinafine-resistant trichophyton mentagrophytes type viii locally and globally. J. Fungi 2021, 7, 419. [Google Scholar] [CrossRef]
- Astvad, K.M.T.; Hare, R.K.; Jørgensen, K.M.; Saunte, D.M.L.; Thomsen, P.K.; Arendrup, M.C. Increasing Terbinafine Resistance in Danish Trichophyton Isolates 2019–2020. J. Fungi 2022, 8, 150. [Google Scholar] [CrossRef] [PubMed]
- Crotti, S.; Cruciani, D.; Spina, S.; Piscioneri, V.; Natalini, Y.; Pezzotti, G.; Sabbatucci, M.; Papini, M. A Terbinafine Sensitive Trichophyton indotineae Strain in Italy: The First Clinical Case of tinea corporis and onychomycosis. J. Fungi 2023, 9, 865. [Google Scholar] [CrossRef] [PubMed]
- Abdolrasouli, A.; Borman, A.M.; Johnson, E.M.; Hay, R.J.; Arias, M. Terbinafine-resistant Trichophyton indotineae causing extensive dermatophytosis in a returning traveller, London, UK. Clin. Exp. Dermatol. 2024, 49, 635–637. [Google Scholar] [CrossRef]
- Gawaz, A.; Nenoff, P.; Uhrlaß, S.; Schaller, M. Treatment of a terbinafine-resistant trichophyton mentagrophytes type VIII. Hautarzt 2021, 72, 900–904. [Google Scholar] [CrossRef]
- Süß, A.; Uhrlaß, S.; Ludes, A.; Verma, S.B.; Monod, M.; Krüger, C.; Nenoff, P. Extensive tinea corporis due to a terbinafine-resistant Trichophyton mentagrophytes isolate of the Indian genotype in a young infant from Bahrain in Germany. Hautarzt 2019, 70, 888–896. [Google Scholar] [CrossRef]
- Cañete-Gibas, C.F.; Mele, J.; Patterson, H.P.; Sanders, C.J.; Ferrer, D.; Garcia, V.; Fan, H.; David, M.; Wiederhold, N.P.; Hanson, K.E. Terbinafine-Resistant Dermatophytes and the Presence of Trichophyton indotineae in North America. J. Clin. Microbiol. 2023, 61, e0056223. [Google Scholar] [CrossRef] [PubMed]
- Caplan, A.S.; Chaturvedi, S.; Zhu, Y.; Todd, G.C.; Yin, L.; Lopez, A.; Travis, L.; Smith, D.J.; Chiller, T.; Lockhart, S.R.; et al. Notes from the Field: First Reported U.S. Cases of Tinea Caused by Trichophyton indotineae—New York City, December 2021–March 2023. MMWR Morb. Mortal. Wkly. Rep. 2023, 72, 536–537. [Google Scholar] [CrossRef]
- de Almeida, J.N.; Santos, A.R.D.; de S Trindade, M.R.; Gold, J.A.W.; Razo, F.P.M.; Gonçalves, S.S.; Dorlass, E.G.; de Mello Ruiz, R.; Pasternak, J.; Mangueira, C.L.P.; et al. Trichophyton indotineae Infection, São Paulo, Brazil, 2024. Emerg. Infect. Dis. 2025, 31, 1049–1051. [Google Scholar] [PubMed]
- Jia, S.; Long, X.; Hu, W.; Zhu, J.; Jiang, Y.; Ahmed, S.; de Hoog, G.S.; Liu, W.; Jiang, Y. The epidemic of the multiresistant dermatophyte Trichophyton indotineae has reached China. Front. Immunol. 2023, 13, 1113065. [Google Scholar] [CrossRef]
- Ngo, T.M.C.; Ton Nu, P.A.; Le, C.C.; Ha, T.N.T.; Do, T.B.T.; Tran Thi, G. First detection of Trichophyton indotineae causing tinea corporis in Central Vietnam. Med. Mycol. Case Rep. 2022, 36, 37–41. [Google Scholar] [CrossRef]
- Posso-De Los Rios, C.J.; Tadros, E.; Summerbell, R.C.; Scott, J.A. Terbinafine Resistant Trichophyton indotineae Isolated in Patients With Superficial Dermatophyte Infection in Canadian Patients. J. Cutan. Med. Surg. 2022, 26, 371–376. [Google Scholar] [CrossRef]
- Nenoff, P.; Verma, S.B.; Ebert, A.; Süß, A.; Fischer, E.; Auerswald, E.; Dessoi, S.; Hofmann, W.; Schmidt, S.; Neubert, K.; et al. Spread of terbinafine-resistant trichophyton mentagrophytes type VIII (India) in Germany–“the tip of the iceberg?”. J. Fungi 2020, 6, 207. [Google Scholar] [CrossRef] [PubMed]
- Dashti, Y.; Alobaid, K.; Al-Rashidi, S.; Dashti, M.; AbdulMoneim, M.H.; Al-Enezi, M.; Abou-Chakra, N.; Jørgensen, K.M. Autochthonous case of Trichophyton indotineae in Kuwait. J. Med. Mycol. 2023, 33, 101432. [Google Scholar] [CrossRef]
- Ferreira, C.B.; Lisboa, C. A systematic review on the emergence of terbinafine-resistant Trichophyton indotineae in Europe: Time to act? J. Eur. Acad. Dermatol. Venereol. 2025, 39, 364–376. [Google Scholar] [CrossRef] [PubMed]
- Arendrup, M.C.; Kahlmeter, G.; Guinea, J.; Meletiadis, J. How to: Perform antifungal susceptibility testing of microconidia-forming dermatophytes following the new reference EUCAST method E.Def 11.0, exemplified by Trichophyton. Clin. Microbiol. Infect. 2021, 27, 55–60. [Google Scholar] [CrossRef] [PubMed]
- Andrews, S.; Krueger, F.; Segonds-Pichon, A.; Biggins, L.; Krueger, C.; Wingett, S. FastQC: A Quality Control Tool for High Throughput Sequence Data. 2010. Available online: https://www.bioinformatics.babraham.ac.uk/projects/fastqc/ (accessed on 12 January 2025).
- Ewels, P.; Magnusson, M.; Lundin, S.; Käller, M. MultiQC: Summarize analysis results for multiple tools and samples in a single report. Bioinformatics 2016, 32, 3047–3048. [Google Scholar] [CrossRef]
- Chen, S.; Zhou, Y.; Chen, Y.; Gu, J. Fastp: An ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 2018, 4, i884–i890. [Google Scholar] [CrossRef]
- Prjibelski, A.; Antipov, D.; Meleshko, D.; Lapidus, A.; Korobeynikov, A. Using SPAdes De Novo Assembler. Curr. Protoc. Bioinform. 2020, 70, e102. [Google Scholar] [CrossRef]
- Bankevich, A.; Nurk, S.; Antipov, D.; Gurevich, A.A.; Dvorkin, M.; Kulikov, A.S.; Lesin, V.M.; Nikolenko, S.I.; Pham, S.; Prjibelski, A.D.; et al. SPAdes: A new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 2012, 19, 455–477. [Google Scholar] [CrossRef]
- Bertels, F.; Silander, O.K.; Pachkov, M.; Rainey, P.B.; van Nimwegen, E. Automated Reconstruction of Whole-Genome Phylogenies from Short-Sequence Reads. Mol. Biol. Evol. 2014, 31, 1077–1088. [Google Scholar] [CrossRef]
- Kozlov, A.M.; Darriba, D.; Flouri, T.; Morel, B.; Stamatakis, A. RAxML-NG: A fast, scalable and user-friendly tool for maximum likelihood phylogenetic inference. Bioinformatics 2019, 35, 4453–4455. [Google Scholar] [CrossRef]
- Taghipour, S.; Pchelin, I.M.; Zarei Mahmoudabadi, A.; Ansari, S.; Katiraee, F.; Rafiei, A.; Shokohi, T.; Abastabar, M.; Taraskina, A.E.; Kermani, F.; et al. Trichophyton mentagrophytes and T interdigitale genotypes are associated with particular geographic areas and clinical manifestations. Mycoses 2019, 62, 1084–1091. [Google Scholar] [CrossRef] [PubMed]
- Tóth, Z.; Ványai, B.; Kovács, R.; Jakab, Á.; Szegedi, A.; Balázs, B.; Majoros, L. First Report of Trichophyton indotineae Infection in Hungary. J. Fungi 2025, 11, 248. [Google Scholar] [CrossRef] [PubMed]
- De Paepe, R.; Normand, A.C.; Uhrlaß, S.; Nenoff, P.; Piarroux, R.; Packeu, A. Resistance Profile, Terbinafine Resistance Screening and MALDI-TOF MS Identification of the Emerging Pathogen Trichophyton indotineae. Mycopathologia 2024, 189, 29. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Bhuiyan, M.S.I.; Verma, S.B.; Illigner, G.M.; Uhrlaß, S.; Klonowski, E.; Burmester, A.; Noor, T.; Nenoff, P. Trichophyton mentagrophytes ITS Genotype VIII/Trichophyton indotineae Infection and Antifungal Resistance in Bangladesh. J. Fungi 2024, 10, 768. [Google Scholar] [CrossRef] [PubMed Central]
- Gupta, A.K.; Polla Ravi, S.; Wang, T.; Cooper, E.A.; Lincoln, S.A.; Foreman, H.C.; Bakotic, W.L. Antifungal Resistance, Susceptibility Testing and Treatment of Recalcitrant Dermatophytosis Caused by Trichophyton indotineae: A North American Perspective on Management. Am. J. Clin. Dermatol. 2023, 24, 927–938. [Google Scholar] [CrossRef] [PubMed]
- Kolarczyková, D.; Lysková, P.; Švarcová, M.; Kuklová, I.; Dobiáš, R.; Mallátová, N.; Kolařík, M.; Hubka, V. Terbinafine resistance in Trichophyton mentagrophytes and Trichophyton rubrum in the Czech Republic: A prospective multicentric study. Mycoses 2024, 67, e13708. [Google Scholar] [CrossRef]
- Monod, M.; Feuermann, M.; Salamin, K.; Fratti, M.; Makino, M.; Alshahni, M.M.; Makimura, K.; Yamada, T. Trichophyton rubrum azole resistance mediated by a new ABC transporter, TruMDR3. Antimicrob. Agents Chemother. 2019, 63, e00863-19. [Google Scholar] [CrossRef]
- Yamada, T.; Yaguchi, T.; Maeda, M.; Alshahni, M.M.; Salamin, K.; Guenova, E.; Feuermann, M.; Monod, M. Gene Amplification of CYP51B: A New Mechanism of Resistance to Azole Compounds in Trichophyton indotineae. Antimicrob. Agents Chemother. 2022, 66, e0005922. [Google Scholar] [CrossRef]
- Berstecher, N.; Burmester, A.; Gregersen, D.M.; Tittelbach, J.; Wiegand, C. Trichophyton indotineae Erg1Ala448Thr Strain Expressed Constitutively High Levels of Sterol 14-α Demethylase Erg11B mRNA, While Transporter MDR3 and Erg11A mRNA Expression Was Induced After Addition of Short Chain Azoles. J. Fungi 2024, 10, 731. [Google Scholar] [CrossRef]
- Yamada, T.; Maeda, M.; Nagai, H.; Salamin, K.; Chang, Y.T.; Guenova, E.; Feuermann, M.; Monod, M.; Groll, A.H. Two different types of tandem sequences mediate the overexpression of TinCYP51B in azole-resistant Trichophyton indotineae. Antimicrob. Agents Chemother. 2023, 67, e0093323. [Google Scholar] [CrossRef]
- Nenoff, P.; Wendrock-Shiga, G.; Mechtel, D.; Schubert, K.; Jarsumbeck, R.; Lusmöller, E.; Stadler, R.; Ginter-Hanselmayer, G.; Tietz, H.-J.; Pfüller, R.; et al. Trichophyton mentagrophytes ITS Genotype VII from Thailand. In Dermatophytes and Dermatophytoses; Springer International Publishing: Berlin/Heidelberg, Germany, 2021; pp. 231–256. [Google Scholar]
- Wendrock-Shiga, G.; Mechtel, D.; Uhrlaß, S.; Koch, D.; Krüger, C.; Nenoff, P. Tinea barbae profunda due to Trichophyton mentagrophytes after journey to Thailand: Case report and review. Hautarzt 2017, 68, 639–648. [Google Scholar] [CrossRef] [PubMed]
- Nenoff, P.; Uhrlaß, S.; Wendrock-Shiga, G.; Mechtel, D.; Schubert, K.; Jarsumbeck, R.; Lusmoeller, E.; Krueger, C.; Stadler, R. Trichophyton mentagrophytes genotype Thailand Type 1 as causative pathogen of abscessing dermatophytoses-tinea genitalis and barbae-in Germany. Med. Mycol. 2018, 56 (Suppl. S2), S24. [Google Scholar]
- Jabet, A.; Dellière, S.; Seang, S.; Chermak, A.; Schneider, L.; Chiarabini, T.; Teboul, A.; Hickman, G.; Bozonnat, A.; Brin, C.; et al. Sexually Transmitted Trichophyton mentagrophytes Genotype VII Infection among Men Who Have Sex with Men. Emerg. Infect. Dis. 2023, 29, 1411–1414. [Google Scholar] [CrossRef]
- Jabet, A.; Bérot, V.; Chiarabini, T.; Dellière, S.; Bosshard, P.P.; Siguier, M.; Tubiana, R.; Favier, M.; Canestri, A.; Makhloufi, S.; et al. Trichophyton mentagrophytes ITS genotype VII infections among men who have sex with men in France: An ongoing phenomenon. J. Eur. Acad. Dermatol. Venereol. 2024, 39, 407–415. [Google Scholar] [CrossRef] [PubMed]
- Müller, V.L.; Kreuter, A.; Uhrlaß, S.; Nenoff, P. Trichophyton mentagrophytes genotype VII increasingly causes anogenital infections. Dermatologie 2024, 75, 48–54. [Google Scholar] [CrossRef]

| Terbinafine (Ecoff: 0.125 µg/mL) | ||||
| Species | Range Terbinafine MIC (µg/mL) | Resistance Rate | Geometric Mean S/WT Strains (µg/mL) | Geometric Mean R/Non-WT Strains (µg/mL) |
| T. indotineae | 0.03–8 | 7/8 (87.7%) | 0.03 | 4.41 |
| T. interdigitale | 0.008–0.125 | 0/91 (0%) | 0.016 | NA |
| T. mentagrophytes | 0.008–0.5 | 6/38 (15.8%) | 0.047 | 0.35 |
| Itraconazole (Ecoff: 0.25 µg/mL) | ||||
| Species | Range itraconazole MIC (µg/mL) | Resistance rate | Geometric mean S/WT strains (µg/mL) | Geometric mean R/non-WT strains (µg/mL) |
| T. indotineae | 0.008–4 | 1/8 (12.5%) | 0.054 | 4 |
| T. interdigitale | 0.008–0.25 | 0/91 (0%) | 0.035 | NA |
| T. mentagrophytes | 0.03–1 | 2/38 (5.2%) | 0.116 | 1 |
| Voriconazole (Ecoff: 1 µg/mL) | ||||
| Species | Range voriconazole MIC (µg/mL) | Resistance rate | Geometric mean S/WT strains (µg/mL) | Geometric mean R/non-WT strains (µg/mL) |
| T. indotineae | 0.06–1 | 0/8 (0%) | 0.5 | NA |
| T. interdigitale | 0.03–1 | 0/91 (0%) | 0.179 | NA |
| T. mentagrophytes | 0.125–2 | 2/38 (5.2%) | 0.53 | 2 |
| Amorolfine (Ecoff: 0.5 µg/mL) | ||||
| Species | Range amorolfine MIC (µg/mL) | Resistance rate | Geometric mean S/WT strains (µg/mL) | Geometric mean R/non-WT strains (µg/mL) |
| T. indotineae | 0.008–0.5 | 0/8 (0%) | 0.056 | NA |
| T. interdigitale | 0.016–1 | 3/91 (3.3%) | 0.12 | 1 |
| T. mentagrophytes | 0.125–1 | 4/38 (10.5%) | 0.37 | 1 |
| DermaCheck Status 0.05/0.1/0.2 µg/mL TERB | SQLE Substitution | Number of Isolates (Species) | Susceptibility Profile | MIC TERB (µg/mL) |
|---|---|---|---|---|
| Growth (0.05/0.1/0.2 µg/mL) [4] | F397L | 4 (T. indotineae) | R | 2–8 |
| Growth (0.05/0.1/0.2 µg/mL) [2] | L393F | 2 (T. indotineae) | R | 8 |
| Growth (0.05/0.1 µg/mL) | L393S | 1 (T. indotineae) | R | 2 |
| No growth [1] | A448T | 1 (T. indotineae) | S | 0.03 |
| No growth [27] | K276N | 29 (T. mentagrophytes) | WT, (n = 14) ≥ECOFF (n = 15) | <0.125 (n = 14) 0.125–0.5 (n = 15) |
| SQLE Substitution | TERB MIC Range (µg/mL) | TERB GM (µg/mL) | p Value | ITRA MIC Range (µg/mL) | ITRA GM (µg/mL) | p Value | VOR MIC Range (µg/mL) | VOR GM (µg/mL) | p Value | AMOR MIC Range µg/mL | AMOR GM µg/mL | p Value |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| K276N [27] | 0.008–0.5 | 0.085 | 0.002 | 0.008–0.5 | 0.15 | 0.03 | 0.125–2 | 0.65 | 0.003 | 0/125–1 | 0.43 | 0.017 |
| WT [9] | 0.008–0.03 | 0.026 | 0.03–0.25 | 0.076 | 0.125–0.5 | 0.29 | 0.125–0.5 | 0.25 |
| Localization of the Lesion | SEX | Age | MIC TERB (µg/mL) | MIC ITRA (µg/mL) | MIC VOR (µg/mL) | MIC AMOR (µg/mL) | SQLE Substitution |
|---|---|---|---|---|---|---|---|
| Face | M | 34 | 0.03 | 0.06 | 0.25 | 0.125 | WT |
| Scalp and arm | M | 28 | 0.03 | 1 | 1 | 0.5 | K276N |
| Nose | M | 33 | 0.06 | 0.125 | 1 | 0.5 | K276N |
| Buttocks | M | 30 | 0.125 | 0.25 | 1 | 0.5 | K276N |
| Trunck + arm | M | 40 | 0.125 | 0.125 | 2 | 1 | K276N |
| Thigh | F | 59 | 0.125 | 0.125 | 1 | 0.5 | K276N |
| Trunck | M | 29 | 0.125 | 0.125 | 1 | 0.25 | K276N |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sacheli, R.; Egrek, S.; El Moussaoui, K.; Kurt, B.; Machowski, E.; Harag, S.; Hayette, M.-P. National Belgian Study on Terbinafine Resistance in Trichophyton interdigitale/mentagrophytes/indotineae (2022–2023): Epidemiology and Molecular Features. J. Fungi 2025, 11, 676. https://doi.org/10.3390/jof11090676
Sacheli R, Egrek S, El Moussaoui K, Kurt B, Machowski E, Harag S, Hayette M-P. National Belgian Study on Terbinafine Resistance in Trichophyton interdigitale/mentagrophytes/indotineae (2022–2023): Epidemiology and Molecular Features. Journal of Fungi. 2025; 11(9):676. https://doi.org/10.3390/jof11090676
Chicago/Turabian StyleSacheli, Rosalie, Sabrina Egrek, Khalid El Moussaoui, Bahoz Kurt, Emilie Machowski, Saadia Harag, and Marie-Pierre Hayette. 2025. "National Belgian Study on Terbinafine Resistance in Trichophyton interdigitale/mentagrophytes/indotineae (2022–2023): Epidemiology and Molecular Features" Journal of Fungi 11, no. 9: 676. https://doi.org/10.3390/jof11090676
APA StyleSacheli, R., Egrek, S., El Moussaoui, K., Kurt, B., Machowski, E., Harag, S., & Hayette, M.-P. (2025). National Belgian Study on Terbinafine Resistance in Trichophyton interdigitale/mentagrophytes/indotineae (2022–2023): Epidemiology and Molecular Features. Journal of Fungi, 11(9), 676. https://doi.org/10.3390/jof11090676







