Abstract
Candidozyma auris is a multidrug-resistant, thermo- and osmotolerant yeast capable of persisting on biotic and abiotic surfaces, attributes likely linked to its cell wall composition. Here, seven putative genes encoding yapsins, aspartyl proteases GPI-anchored to the membrane or cell wall, were identified in the genomes of C. auris CJ97 and 20-1498, from clades III and IV, respectively. The C. auris YPS1 gene is orthologous to the SAP9 of C. albicans. The YPS7 gene is orthologous to YPS7 in C. glabrata and S. cerevisiae, so that they may share similar roles. An in silico analysis suggested an interaction between pepstatin and the catalytic domain of Yps1 and Yps7. Although this inhibitor, when combined with caffeine, had a subtle effect on the growth of C. auris, it induced alterations in the cell wall. CauYPS1 and CauYPS7 expression increased under nutrient starvation and NaCl, and at 42 °C. The transcriptome of the 20-1498 strain suggests that autophagy may play a role in thermal stress, probably degrading deleterious proteins or maintaining cell wall and vacuolar homeostasis. Therefore, CauYps1 and CauYps7 may play a role in the cell wall integrity of C. auris in stress conditions, and they could be a target of new antifungal or antivirulence agents.
1. Introduction
Aspartyl proteases are enzymes that have gained attention due to their involvement in the infection processes of pathogenic fungi. Yeast aspartyl proteases, or yapsins, are anchored to the cell wall through covalent bonds to β-1,6-glucans or the cell membrane via the GPI (glycosylphosphatidylinositol) group. They participate in cell wall remodeling by processing proteins located here, a process essential for the integrity of this structure during morphogenesis, and adaptation to environmental changes adhesion, biofilm development, and nutrient acquisition by degrading extracellular proteins, such as host tissue and immune defense proteins [1,2,3,4]. Since 1990, when Egel-Mitani et al. [5] described in S. cerevisiae GPI-aspartyl proteases capable of partially complementing kex2 mutants by processing the α-mating factor precursor when it is overexpressed, it has been shown that, in contrast to other aspartyl proteases with an affinity to hydrophobic residues, S. cerevisiae yapsins’s cleave dibasic residues [6]. Then, Bourbonnais et al. [7] showed that the Yps1 of S. cerevisiae also cleaves monobasic residues, unlike Kex2. Similarly, Sap9 and Sap10 of C. albicans partially complemented kex2 mutants by processing monobasic and dibasic residues of peptides. Although Sap9 and Sap10 C. albicans still retain the nomenclature of the secreted aspartyl protease (Sap), proteases without GPI modification, in silico, molecular, and biochemical studies have shown that they differ from other Sap members. For example, they exhibit enzymatic activity over a broad range of pH values, even at a neutral (physiological) pH, have their distinct cleavage preferences, and are cell-surface-associated proteases that function in cell wall integrity and the interaction with human epithelial cells and neutrophils [2].
Yapsins are synthesized as zymogens, containing an N-terminal signal peptide removed after translocation into the endoplasmic reticulum by a signal peptidase. These enzymes present a catalytic domain with two aspartic residues, or at least one, in the Asp-Thr(Ser)-Gly(DT(S)G) consensus, which is crucial for their enzymatic function, the N-terminal propeptide removed in the Golgi apparatus at the dibasic or monobasic residues by Kex2 or by yapsin peptidases [1,2,8]. The C-terminal is a hydrophobic domain for transient binding to the ER membrane and the ω site where the protein is linked to the GPI anchor. The ω site is at the consensus sequence [NSGADC]–[GASVIETKDLF]–[GASV]–X(4,19)–[FILMVAGPSTCYWN]. Like other aspartyl proteases from the A1 family, they contain four cysteines forming disulfide bridges [1,9,10]. It has been observed that yapsins, like Saps, of human pathogenic yeasts, occur as multigene families, and the expansion of these families seems to be related to their success as pathogens and in their survival [11].
Candidozyma auris (syn. Candida auris) is an opportunistic multidrug-resistant pathogen first described in 2009 by Satoh et al. [12]. Since then, it has been identified as being responsible for hospital outbreaks worldwide, causing systemic infections with high mortality. As a result, the World Health Organization issued an epidemiological alert to identify, notify, and manage cases and outbreaks caused by C. auris promptly [13]. Additionally, C. auris is thermotolerant and halotolerant, which contributes to its ability to colonize various environments. This yeast also forms pseudohyphae and aggregates under specific conditions [14,15,16,17]. Among other characteristics that distinguish C. auris from other pathogenic yeasts are its capacity to form multilayered biofilm and cause invasive infections without forming mycelium, and immune evasion, features attributed in part to the yeast cell wall [18,19,20]. C. auris isolates have been grouped into six main geographical clades through phylogenetic analyses. The strains of each clade differ by hundreds of thousands of single-nucleotide polymorphisms (SNPs). Genetic differences between clades have been linked to differences in the cell wall and membrane composition [13,21].
In contrast to C. albicans, whose genome carries 10 genes encoding Saps, C. auris has a multigene family of 14 or 15 genes encoding these proteins [20,22]. Particularly, Sap3 contributes to extracellular proteolytic activity, biofilm formation, and virulence in animal models, suggesting that its inhibition could be a promising therapeutic strategy [22]. Given the importance of secreted aspartyl proteases as virulence factors in C. auris, this study addressed the identification of GPI-anchored aspartyl proteases encoding genes of two strains of clades III and IV each and through the use of the aspartyl protease inhibitor, pepstatin A, and the expression analysis of the C. auris YPS1 and YPS7 genes, which seem to be the most probable orthologs of YPS1, YPS7, and SAP9 of S. cerevisiae and other Candida, which encode the standout GPI-proteases in cell wall homeostasis [23,24].
Understanding the molecular mechanisms by which YPS1 and YPS7 contribute to stress tolerance in Candida auris could aid in the development of therapeutic and control strategies against this yeast.
2. Materials and Methods
2.1. Strains
In this work, C. auris CJ97 [25,26] and 20-1498 [27] strains isolated from the blood of hospitalized patients from Spain and Mexico were used, grouped into clades III and IV, respectively. They were stored frozen at −70 °C and routinely grown in YPD (yeast extract 1%, peptone 2%, and dextrose 2%) at 37 °C with shaking at 100 rpm.
2.2. In Silico Analysis
The sequences of putative yapsin-coding genes and putative targets of yapsins were identified through BLAST+ 2.17.0+ analysis and hidden Markov models (HMMs) in the previously reported genomes of the two C. auris strains CJ97 and 20-1498 (GCA_034640365.1) [28], and C. auris B11220 strain from clade II (GCA_003013715.2). Sequences of the Saccharomyces cerevisiae yapsin genes (ScYPS 1, 2, 3, 6, and 7) from the genome (https://www.yeastgenome.org/, accessed on 12 January 2024) were used as a template. The predicted open reading frame and 1000 bp upstream (considered the promoter sequence) of each selected gene were analyzed using various tools. The SignalP v6.0 (10 February 2024; https://services.healthtech.dtu.dk/services/SignalP-6.0/) [29], ScanProsite (13 February 2024; https://prosite.expasy.org/scanprosite/), and NetGPI v1.1 (15 March 2024; https://services.healthtech.dtu.dk/services/NetGPI-1.1/) [30] were used to find motifs in protein sequences, while, to explain a possible differential expression of the genes, promoters were analyzed in the YEASTRACT server, looking for probable transcription factors binding sites (TFBSs) (27 June 2024; https://saves.mbi.ucla.edu/) [31]. Moreover, the percentage of similarity and identity of protein sequences was stated with MatGat v2.01 [32].
2.3. Phylogeny of C. auris Putative Yapsins
First, proteins were aligned with Clustal X [33]. The most conserved regions of GPI-anchored aspartyl peptidases of S. cerevisiae, C. glabrata (Nakaseomyces glabratus), C. albicans, and C. auris CJ97 were used to construct the phylogeny, with the IQ-TREE software v2.4.0 [34], using the best substitution model (WAG+F+R4). One thousand bootstrap replicates were applied, and the tree was visualized with Figtree v1.4.5. Accession numbers of protein sequence used were as follows: S. cerevisiae: ScYps1 (NP_013221.1), ScYps2 (NP_010428.3), ScYps3 (NP_013222.1), ScYps6 (NP_012305.3), and ScYps7 (NP_010636.1); C. albicans: CaSap9 (KAL1572689.1), and CaSap10 (XP_717243.1); C. glabrata CgYps1 (XP_449529.1), CgYps2 (XP_445750.2), CgYps3 (XP_445764.1), CgYps4 (XP_445765.1), CgYps5 (XP_445766.1), CgYps6 (XP_445767.1), CgYps7 (XP_444870.1), CgYps8 (XP_445768.1), CgYps9 (XP_445769.1), CgYps10 (XP_445770.1), and CgYps11 (XP_445771.1); and C. auris B11220: CauYps1 (XP_028889414.2), CauYps2 (XP_028891323.2), CauYps3 (XP_028888407.2), CauYps4 (XP_028892789.1), CauYps5 (XP_028891322.2), CauYps6 (XP_028890862.2), and CauYps7 (XP_028892266.2).
2.4. Prediction of the Secondary and Tertiary Structure of Putative Yapsins
The mature forms of putative yapsins CauYps1 (amino acids 52–108 and 199–566) and CauYps7 (amino acids 57–527) were used to predict the secondary and tertiary structure of C. auris Yps proteins. Prediction of the secondary and tertiary structure was carried out in ENDscript v3.0 (18 July 2024; https://endscript.ibcp.fr/ESPript/ENDscript/) [35] and the SWISS-MODEL servers, respectively (12 October 2024; https://swissmodel.expasy.org) [36], using the better models selected by the programs, as templates. Once the theoretical tertiary structures were obtained, their quality was analyzed using the SAVES server (15 October 2024; https://saves.mbi.ucla.edu/).
2.5. Molecular Docking of C. auris Putative Yps1 and Yps7
The 3D models of predicted mature and CauYps7 from C. auris, obtained from the SWISS-MODEL server, were used for molecular docking analysis with the specific and reversible inhibitor of aspartic proteases, pepstatin A, retrieved from the PDB (PRD_000557). This was modified using the Avogadro 2 software to prepare it for interaction [37]. The docking process was performed in ChimeraX [38] with the aid of AutoDock Vina [39].
To test the potential activity of yapsins on a probable molecular target, a molecular docking analysis was conducted using a peptide (MALWMRLLPLLALLALWGPDPAAAFVNQHLCGSHLVEALYLVCGERGFFYTPKTRREAEDLQVGQVELGGGPGAGSLQPLALEGSLQKRGIVEQCCTSICSLYQLENYCN) of the proinsulin hormone (AAW83741.1). The docking was then performed using the H-Dock server (28 October 2024; http://hdock.phys.hust.edu.cn/) [40], which predicts possible molecular interactions between two proteins.
2.6. Evaluation of C. auris Growth Under Different Conditions
The C. auris strains CJ97 and 20-1498 were pre-cultured for 24 h in YPD medium at 37 °C with shaking at 100 rpm. A 96-well plate was inoculated with 100 µL of yeast at 0.05 of OD600 containing YPD medium with different compounds at a final concentration as indicated: 1.5 M NaCl, 12 mM caffeine, 10 mM H2O2, 0.05% SDS all from Sigma-Aldrich (St. Louis, MO, USA), in the presence and absence of 25 μM pepstatin A (ChemCruz, Dallas, TX, USA). YPD without any compounds was considered the growth control, and YPD without compounds and inoculum was used as the sterility control. Plates were incubated for 20 h, at 37 °C, with shaking, in a Multiskan FC 5111900 (Thermo Fisher Scientific, Waltham, MA, USA) and OD620 measured. Three independent replicates were performed, and the standard deviation and significance were determined by two-way ANOVA with SigmaPlot 15.1 software.
2.7. Expression of YPS1 and YPS7 Genes by RT-qPCR
Starting from a 48 h pre-culture in a YPD medium, an early stationary phase culture in YPD was obtained by inoculating yeast at an OD600 of 0.05 after 15 h of incubation at 37 °C and 100 rpm. These non-proliferating cultures were harvested at 2400 g for 5 min, washed twice, and inoculated into an equal volume of YNB (USBiological, Swampscott, MA, USA) with 2% dextrose (Sigma-Aldrich, St. Louis, MO, USA) as a carbon source and 0.5% (NH4)2SO4 (Gibco, Waltham, MA, USA) or BSA (Sigma-Aldrich, St. Louis, MO, USA) as a nitrogen source (YNB+C+N); YNB with only carbon (YNB+C-N) or nitrogen (YNB-C+N), and neither carbon nor nitrogen (YNB-C-N), also were tested. The cultures were incubated at 37 °C with shaking at 100 rpm. To evaluate the effect on gene expression at 42 °C and NaCl, cells were transferred to YPD without/with 1.5 M NaCl, incubated at 37 °C, or to YPD alone and incubated at 42 °C. Two independent experiments, each performed in triplicate, were conducted for each condition.
Cells were harvested after 3 and 6 h to extract total RNA using the Schmitt et al. method [41], based on hot phenol extraction. Cells from the desired condition were resuspended in AE buffer (50 mM sodium acetate, 10 mM EDTA) and disrupted with 10% SDS and pH 5 phenol. Samples were placed in a water bath at 65 °C for 5 min and immediately ultra-frozen for 10 min. After centrifugation at 12,879× g for 10 min, the aqueous phase was recovered, followed by two washes with chloroform-isoamyl alcohol. Then, absolute and 70% ethanol with DEPC-treated water were added to precipitate and wash RNA. Finally, it was resuspended in nuclease-free water, and its quantity and quality were assessed.
All the RT-qPCR reagents were from Thermo Fisher Scientific (Waltham, MA; USA) SYBR Select Master Mix. cDNA synthesis was performed using 2 μg of DNA-free RNA, treated with DNase I, and the RevertAid reverse transcriptase, along with the oligo (dT)18 primer, according to the manufacturer’s instructions. SYBR Select Master was used according to the supplier’s instructions, using the primers YPS1 Fw: CTCGAACGAGGAGGACATCG, Rev: TGCTTACAGTCACCGAG; and YPS7 Fw: CCTTAAAAACTTCAGAGCAGTAGG, Rev GACCACACCAACAAACGCTC. The reaction was subjected to initial denaturation of 95 °C for 5 min, with subsequent forty-five cycles at 95 °C for 30 s, annealing at 59 °C for 30 s, and extension at 72 °C for 1 min 15 s, in a thermal cycler Corbett Research RG-6000 (Corbett Robotic Inc., San Francisco, CA, USA). The data were analyzed with the 2-ΔΔct methodology [42]. The ACT1 gene (CJI96_0003605) [43] was used as the normalizing gene, and the control conditions were YNB+C+N at 37 °C and YPD at 37 °C. Reactions were performed in triplicate in two biological experiments.
2.8. Transcriptomic Analysis of C. auris 20-1498 Under Heat Stress
The C. auris strain 20-1498, belonging to clade IV, was cultured in YPD medium until the early stationary phase. Cells were harvested by centrifugation (2400× g, 5 min, 4 °C) and washed twice with minimal YNB medium without supplements. They were then incubated for six hours in complete YNB medium (YNB+C+N) at 37 °C and 42 °C.
Total RNA was extracted using the GeneJET RNA Purification Kit (Thermo Scientific; Waltham, MA, USA) following the manufacturer’s instructions. RNA quality was assessed by RNA integrity number (RIN). Library preparation, based on poly-A enrichment, and sequencing on an Illumina NovaSeq X Plus platform were performed by NOVOGENE (Sacramento, CA, USA). Three biological replicates were sequenced per condition.
Raw reads were processed with Trimmomatic v0.39. The quality of filtered reads was evaluated using FastQC v0.12.1. The reference genome of C. auris clade IV B11243 (GCA_003014415.1) was downloaded from FungiDB (https://fungidb.org/fungidb/app/, accessed on 12 January 2024). Clean reads were mapped with STAR v2.7.11b, and read counts per gene were obtained using FeatureCounts v2.1.1. Differential expression analysis was performed in R v4.2.2 using DESeq2 v1.44.0. Genes with log2FoldChange ≥ 0.585 or ≤−0.585 and padj <0.05 were considered differentially expressed.
Results were processed in Python v3.13.2. Three-dimensional principal component analysis was performed with scikit-learn v1.2.2 and visualized using plotly v5.15.0. The sample distance matrix was generated by calculating Euclidean distances and represented as a heatmap with seaborn v0.12.0. The volcano plot was constructed from the differential expression analysis using matplotlib v3.7.1. Differentially expressed genes were annotated in FungiDB (https://fungidb.org/fungidb/app/, accessed on 12 January 2024) and assigned putative functions by identifying orthologs in CandidaDB (http://www.candidagenome.org, accessed on 12 January 2024) using the pBLAST tool.
Based on functional annotation, Gene Ontology terms associated with biological processes were retrieved, and over-represented terms were visualized in enrichment plots. Finally, differentially expressed orthologous genes were depicted in a lollipop plot using matplotlib v3.7.1.
Processed RNA-Seq data, including gene-level count data and differential expression analyses, are available in the NCBI Gene Expression Omnibus (GEO) under accession number GSE302916. The corresponding raw sequencing reads have been deposited in the NCBI Sequence Read Archive (SRA) under BioProject accession PRJNA1291775.
2.9. Effect of Pepstatin A on the Microscopic Morphology of C. auris
Scanning electron microscopy (SEM): Yeast cells were inoculated at 0.05 OD600 in YPD medium without or with different concentrations of pepstatin A (12.5, 25, and 50 μM), 12 mM caffeine, or 0.01% methanol, from an inoculum of 24 h in YPD. Cells were treated for 22 h, harvested, and washed with Sörensen’s PBS. They were then fixed overnight at room temperature by adding 2.5% glutaraldehyde. Cells were washed three times with Sörensen’s PBS and sequentially dehydrated with increasing ethanol concentrations, from 50% to 100%. Then, samples were dried for 3 h at 35 °C/1350 psi using a critical point dryer K850 (Quorum Technologies, Lewes, UK) and observed using a scanning electron microscope Quanta FEG 250 (FEI Company, Eindhoven, The Netherlands).
Atomic force microscopy (AFM): Cells treated or not with 50 mM of pepstatin were harvested and washed three times with sterile distilled water. Cells resuspended in an equal volume of water as a medium were placed on a slide, air-dried, and fixed for 1 h with formaldehyde vapors. Afterwards, slides were observed (TT-AFM., Workshop, SC, USA).
3. Results
3.1. C. auris Presents a Family of YPS Multigene Encoding Putative Yapsins
Genes encoding putative GPI-anchored aspartyl proteases (yapsins) were identified by Blast and HMMs in the genome of three strains of C. auris: B11220 (reference genome), CJ97, and 20-1498 from clades II, III, and IV, respectively. We found 15 putative genes encoding aspartyl proteases based on information retrieved from the bioinformatic tools. Seven of these sequences presented the omega site of GPI anchorage (ω). Both CauYps2 and CauYps5 amino acid sequences do not show the typical catalytic aspartic residue of this type of proteases (Figure S1). The putative yapsins ranged in size from 372 to 703 amino acids. C. auris yapsins from the three clades showed identities greater than 95.6% and virtually identical motifs, except for CauYps5. CauYps5 from B11220 (clade II) was shorter at the N-terminus than the yapsins from strains CJ97 and 20-1498, and they shared identities of approximately 50% and 60%, respectively (Figure S1).
Additionally, to the two catalytic aspartic residues and the ω site, the N-terminal signal peptide and the typical KR sites, cleavage by Kex2 or autoprocessing at the N-terminal propeptide and the C-terminal, before the ω site, N-glycosylation sites, and low pI, all characteristics of the yapsins, were also predicted. The sequences of the C. auris CJ97 yapsins were usually more conserved with their cognate sequences in B11220 (Figures S1 and S2); thereby, the sequences of the putative yapsins of the last two strains were used for the in silico analysis.
3.2. Yps1 and Yps7 of C. auris are Orthologous to the Yapsins of C. albicans, C. glabrata (Nakaesomyces glabratus), and S. cerevisiae
To examine the phylogenetic relationship between the putative yapsins of C. auris and those of other yeast species, an alignment of the most conserved regions of 25 sequences was performed. This analysis showed that C. auris Yps1 is related to C. albicans Sap9 and Sap10 (Figure 1A). CauYps1 showed a probable internal loop that could be removed similarly to ScYps1 by the cleavage of the monobasic or dibasic residues into its region (Figure 1B and Figure S2). On the other hand, C. auris Yps7 is associated with the clade of C. glabrata and S. cerevisiae Yps7. However, C. glabrata and S. cerevisiae did not show well-conserved aspartic and disulfide bridges (Figure 1D). Moreover, C. auris Yps3, Yps4, and Yps6 were less related to yapsin proteins, while C. auris Yps2 and Yps5 showed no relation to the yapsins included in the analysis (Figure 1A). The CauYPS1–CauYPS7 genes are distributed across different chromosomes in the C. auris genome (Figure 1C). Although CauYPS2 and CauYPS5 are on the same chromosome, there is no phylogenetic relationship between them (Figure 1 and Figure S1).
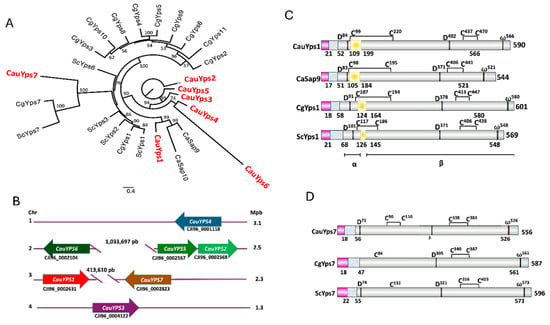
Figure 1.
Characteristics of C. auris predicted yapsins. (A) Phylogenetic analysis of the yapsins multigene family of yeasts. The maximum likelihood reconstruction was performed using amino acid sequences of S. cerevisiae (Sc), C. albicans (Ca), C. glabrata (Cg), and C. auris (Cau) in the IQ-TREE software with 1000 bootstrap replicates. Figtree services were used for visualization and edition, and bootstrap support with a confidence level >50% was included in branches. Protein accession numbers were indicated in Section 2. The scale bar shows the number of substitutions per position. (B) Schematic representation of C. auris YPS gene loci. (C) Schematic representation of the domains or motifs of predicted CauYps1 and its homologs of C. albicans (Sap9), C. glabrata (CgYps1), and S. cerevisiae (ScYps1). This yapsin undergoes self-processing in an internal loop (yellow rectangle) and remains split into two subunits (α and β) through a disulfide bridge. The numbers below indicate the numbers where processing takes place. Active aspartic residues (D), omega site (ω), and cysteines of the disulfide bridges (C) are indicated: pink rectangle: signal peptide, grey–blue rectangle: propeptide. (D) Motifs of CauYps7 predicted and its homologs yapsin of C. glabrata (CgYps7) and S. cerevisiae (ScYps7). CgYps7 and ScYps7 did not show one of the cysteines (C) involved in the disulfide bridges, and CgYPS7 did not show the first catalytic aspartic residue (D).
3.3. The C. auris Proteins Yps1 and Yps7 Present a Secondary and Tertiary Structure of Aspartyl Proteases, Capable of Binding to the Specific Inhibitor Pepstatin A and Interacting with Other Proteins
The secondary and tertiary structures of mature CauYps1 and CauYps7 were predicted, as mentioned in Section 2. The most conserved regions of the yapsins showed a secondary structure similar to the templates C. tropicalis Sap and C. albicans Sap5 (Figure S3). In the case of the 3D structure, the templates selected were C. tropicalis Sap and C. albicans Sap1, which showed an identity and similarity of 33.5% and 52.7% with Yps1, and 18.2% and 33.1% with Yps7, respectively. The quality of the predicted tertiary structures of CauYps1 and CauYps7, suggested that they are suitable models. The percentage of residues in most favored regions in the Ramachandran plot, the overall quality factor (from ERRAT), and the percentage of residues showing an averaged 3D/1D score ≥ 0.1 (from VERIFY3D) correspond to 81.9%, 77.86%, and 90.07% for CauYps1 and 88.7%, 81.68%, and 50.85% for CauYps7.
Both protein structures of CauYps1 and CauYps7 exhibited a bilobed arrangement, forming a catalytic pocket. Each lobe contains one of the two catalytic aspartic residues, both of which are oriented toward the pocket. The molecular docking showed that pepstatin A (a hexapeptide inhibitor of aspartyl protease) fits into the pocket of the predicted structures. The binding energy of pepstatin A for Yps1 was −6.5 kJ/mol, while, for Yps7, it was −6.9 kJ/mol. Furthermore, the catalytic aspartic residues bind to the statine amino acid of pepstatin A and some amino acids of the proinsulin peptide (Figure 2).
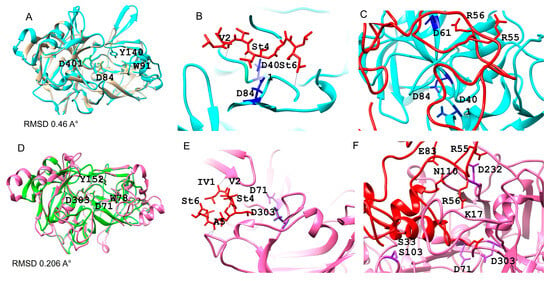
Figure 2.
The predicted tertiary structure of the C. auris Yps1 and Yps7 and their interaction with the inhibitor pepstatin A and a synthetic substrate. (A,D) Overlap of the mature CauYps1 (cyan) and the C. tropicalis Sap (1J71) (brown), and CauYps7 (pink) and the C. albicans Sap1 (2QZW). Molecular docking of the CauYps1 (B) and CauYps7 (E) with pepstatin A (red), and (C,F) with a derived peptide of proinsulin (red), respectively. The catalytic residues Asp84 and Asp401 of CauYps1 interact with statines 4 and 6 of the inhibitor and Asp61 with Arg56 of the substrate. In contrast, the residue Asp232 of CauYps7 interacts with the statine 4 of the inhibitor and Arg55, Glu83, Gly84, and Asn110 of the substrate. Amino acid numbers are given based on the complete protein or peptide sequences.
In addition to proinsulin-derived peptide, sequences of homologous proteins to the targets of Yps1 of S. cerevisiae and Sap9 of C. albicans were identified in C. auris. The GPI-anchored cell wall putative proteins Utr2, Gas1, and Msb2, the first two involved in cell wall homeostasis, and the latter acts as a stress sensor, showed conserved monobasic (K or R) and dibasic (KR or RR) cleavage sites. Furthermore, the cell wall CauPir1 protein could be processed by Yps1 at the propeptide in a similar fashion to ScPir1; however, CauPir1 lacks the processing sites present in the internal repeats of ScPir1, whose function is related to cell wall stability through the linkage of the repeats to glucans. Additionally, the putative Matα precursor exhibited well-conserved KR sites, which were processed by Kex2 and, in its absence, by Yps1 (Figure S4).
3.4. Growth of C. auris CJ97 and 20-1498 in the Presence of Different Stressor Compounds
C. auris yeast were cultured for 20 h at 37 °C in the presence of H2O2, caffeine, NaCl, and SDS without or with pepstatin A to determine whether aspartyl protease inhibition affected yeast growth. The results showed that H2O2, NaCl, and SDS inhibit the growth of C. auris. When pepstatin A was also present in media containing H2O2 and caffeine, it exhibited a subtle but sustained effect on the growth of the yeasts compared to the controls (without pepstatin A) (Figure 3). In contrast, C. auris strains grown in media with pepstatin plus SDS and NaCl did not exhibit changes in growth compared to the controls (Figure S3).
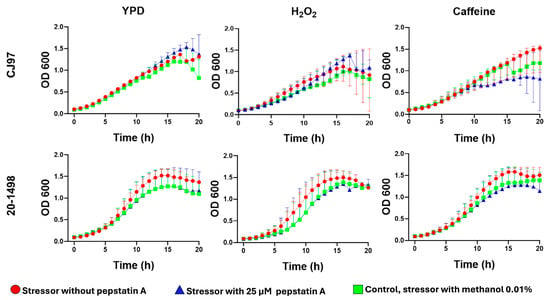
Figure 3.
Impact of pepstatin A on the growth of C. auris strains CJ97 (clade III) and 20-1498 (clade IV). Yeast cultures were grown in YPD medium alone or supplemented with 10 mM H2O2 or 12 mM caffeine, either without pepstatin A or with 25 μM pepstatin A. Each stressor plus 0.01% methanol, the solvent used for pepstatin A, served as a control in addition to YPD. Experiments were conducted in triplicate, and error bars represent standard deviation.
3.5. Differential Expression of YPS1 and YPS7 Genes Under Different Stress Conditions
Yeasts of C. auris from clades III and IV were grown in YPD to the early stationary phase and then subjected to different nutritional conditions, 1.5 mM NaCl, and at 42 °C. Gene expression was conducted 3 and 6 h after each stimulus. The results were grouped in a heatmap according to the expression levels of genes (Figure 4). It was observed that the two genes of the CJ97 and 20-1498 strains were differentially expressed. CauYPS1 of both strains was overexpressed mainly after 6 h without a carbon source compared to the complete medium. The CauYPS7 of 20-1498 (clade IV) showed a higher expression than the CauYPS7 of the CJ97 strain (clade III) after 3 and 6 h of carbon and/or nitrogen starvation. Instead of ammonium sulfate as a nitrogen source, BSA induced the CauYPS7 of 20-1498 after 3 h, while, after 6 h, the CauYPS1 and CauYPS7 of both strains were repressed.
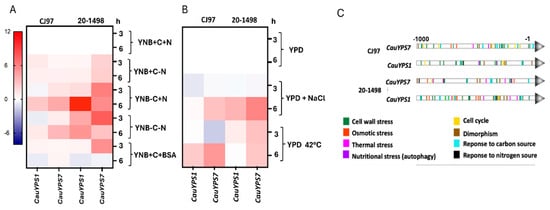
Figure 4.
Differential expression of CauYPS1 and CauYPS7 genes of the strains CJ97 (clade III) and 20-1498 (clade IV). Heatmap representing the log2 of the gene expression change determined by RT-qPCR. (A) RNA extracted from yeast after 3 and 6 h of treatment in YNB medium supplemented with +C = 2% dextrose (as carbon source), +N = 0.5% ammonium sulfate, or +BSA = 0.5% BSA (as nitrogen source). Negative signal indicates the absence of the corresponding stimulus. (B) RNA extracted from yeast after 3 and 6 h of treatment at 37 °C in YPD medium without or with 1.5 M NaCl (-NaCl and +NaCl, respectively) or YPD at 42 °C. Two independent experiments were carried out in triplicate. (C) Predicted TFBSs into the 1000 base pairs upstream of the CauYPS1 and CauYPS7 genes using the YEASTRACT database.
In general, the CauYPS1 and YPS7 genes of both strains were overexpressed after 6 h at 42 °C and with NaCl compared to YPD at 37 °C, and the expression of CauYPS7 of the 20-1498 strain was usually higher than that of CJ97. On the other hand, CauYPS1 and YPS7 showed repression after treating yeasts of the CJ97 and 20-1498 with NaCl for 3 h (Figure 4A,B).
The expression levels of the C. auris YPS1 and YPS7 genes agreed with their theoretical TFBSs found 1000 bp upstream of the encoding regions. These sites could bind transcription factors linked to, for example, the response to the cell wall, osmotic and thermal stress, and nutritional stress. Moreover, CauYPS1 and CauYPS7 showed differences in the disposition of the TFBSs along the analyzed upstream regions between the strains (Figure 4C).
3.6. Transcriptional Response of C. auris Strain 20-1498 Under Heat Stress
Given the overexpression of CauYPS1 and CauYPS7 detected by RT-qPCR under heat stress conditions, we performed a transcriptomic analysis of the first Mexican isolate of C. auris (strain 20-1498) to identify genes involved in the heat stress response. The total RNA was extracted from stationary-phase cultures grown in nutrient-rich medium (YNB supplemented with carbon and nitrogen sources) and incubated for six hours at either 37 °C (control) or 42 °C, using three biological replicates per condition.
A sample distance matrix and three-dimensional principal component analysis (PCA 3D) revealed a clear separation of samples according to incubation temperature, indicating a high reproducibility among replicates and distinct transcriptional profiles between conditions (Figure S5).
Applying cutoffs of log2FoldChange ≥ 0.585 or ≤−0.585 and an adjusted p-value (padj) < 0.05, we identified 1341 differentially expressed genes (DEGs) out of 5538 mapped genes, with 630 upregulated and 711 downregulated under heat stress (Figure 5A). A functional enrichment analysis of these DEGs indicated the activation of pathways related to transmembrane transport, redox processes, proteolysis, and phosphorylation, whereas genes associated with cellular component synthesis, DNA repair and replication, and vesicular transport were downregulated under these conditions (Figure 5B,C).
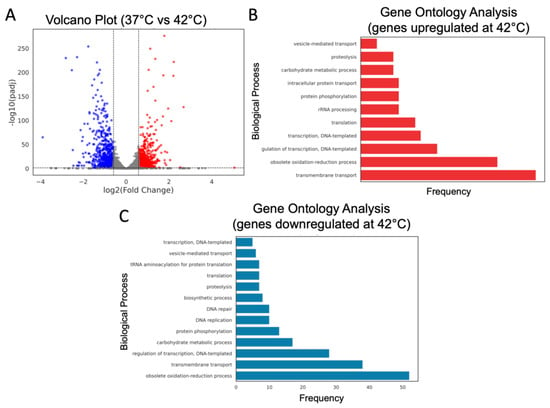
Figure 5.
Differentially expressed genes and biological process ontology in C. auris under heat stress. (A) Volcano plot of differentially expressed genes. A threshold of log2FoldChange ≥ 0.585 or ≤−0.585 along with an adjusted p-value (padj) < 0.05 was applied. Downregulated genes are shown in blue, upregulated genes in red, and genes with no significant change in gray. (B,C) Functional enrichment analysis of GO terms for upregulated (B) and downregulated (C) genes at 42 °C.
Additionally, orthologous genes from C. albicans were identified within the set of differentially expressed genes in C. auris through a pBLAST analysis focused on specific biological pathways of interest. These included sterol, phosphatidylinositol, and cell wall biosynthesis, as well as stress response genes associated with the cell wall integrity pathway, heat-stress-induced autophagy, the HOG (High Osmolarity Glycerol) pathway, the calcineurin pathway, and other genes linked to thermal stress (Figure 6, Table S1).
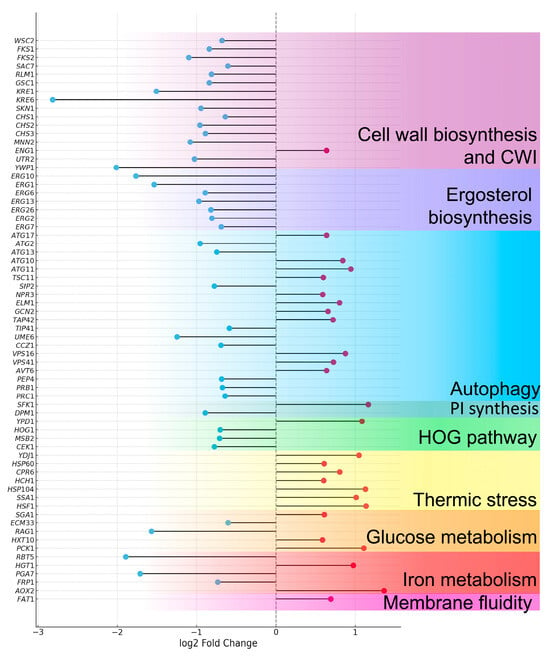
Figure 6.
Lollipop plot of differentially expressed genes in C. auris under heat stress conditions. Strain 20-1498 (clade IV) was cultured in YNB+C+N medium at 37 °C and 42 °C. A threshold of log2FoldChange ≥ 0.585 or ≤−0.585 with an adjusted p-value (padj) < 0.05 was applied. Genes are arranged in order of their functional relevance. Red circles indicate upregulated genes, while blue circles correspond to downregulated genes.
3.7. Effect of Pepstatin A on the Microscopic Morphology of C. auris 20-1498
C. auris cells were cultured in YPD medium with 12.5, 25, and 50 μM of pepstatin A, and microscopic morphology was observed using scanning electron microscopy (SEM). Cells without the inhibitor plus 0.01% methanol or not were used as a control. In the control conditions, oval-shaped yeasts of approximately 2–2.5 × 3–4 μm were observed with some budding and smooth surfaces. At 12.5 μM, yeasts with a rough appearance were observed at a percentage of 3.5%. This effect was dose-dependent, as yeasts with this morphology increased to 4.5% at 25 μM and 6.5% at 50 μM of pepstatin A. At the highest concentration, concave-looking cells were also observed in 60%.
A synergistic effect was observed when cells were grown in YPD with 25 μM pepstatin A and 12 mM caffeine. Nearly 90% of the cells showed evident damage to the cell wall, with a wrinkled appearance and presumptive holes, in contrast to the yeasts grown in the medium with only caffeine or pepstatin (Figure 7A).
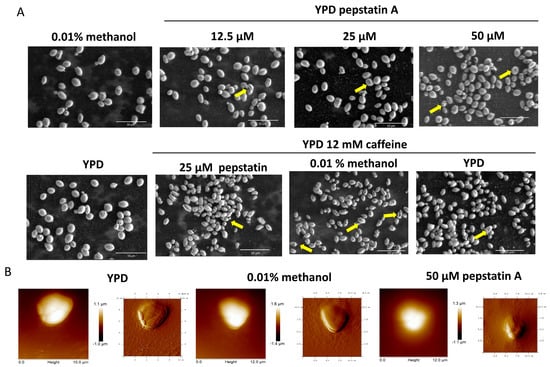
Figure 7.
Effect of pepstatin A on the microscopic morphology of C. auris 20-1498. (A) SEM micrographs showing cells with rough and concave surfaces (arrows) after being grown for 22 h in YPD supplemented with pepstatin A and/or caffeine at the indicated concentration. YPD and YPD with methanol are the control conditions. Images of the fixed cells were acquired using an SEM microscope Quanta FEG 250 (FEI Company, Eindhoven, The Netherlands). (B) Height (left) and amplitude (right) images showing the yeast surface of cells grown as mentioned before and treated for AFM observations as indicated in Section 2. Images were acquired with an atomic force microscope.
The effect of pepstatin A on the cell surface topography of C. auris was also evaluated by atomic force microscopy (AFM). The roughness of cells with pepstatin A 50 μM increased in contrast to cell growth in control conditions (Figure 7B). The AFM analysis suggests a possible decrease in the size of C. auris yeasts when treated with pepstatin A. The length of the yeast would be affected, but not the height. The length varies from 4.2–4.7 μm of yeast in the control condition to 3.7–3.9 μm of yeast treated with pepstatin (Figure 7B).
4. Discussion
C. auris has emerged as a multidrug-resistant pathogen associated with intra-hospital outbreaks that cause systemic infections with high mortality, becoming a serious public health problem worldwide. This yeast is distinguished from other human pathogenic species by its thermo- and osmotolerance, characteristics linked to its ability to survive and persist in various harsh natural and healthcare facility environments, as well as on the human skin. Combined with the effects of climate change and possibly the excessive use of antifungals, these traits could have contributed to the recent and simultaneous emergence of this yeast as a pathogen in several regions of the world [15,16,17].
Among other characteristics that distinguish C. auris from other pathogenic yeasts are its capacity to form a multilayered biofilm, to cause invasive infections without forming mycelium, and immune evasion, features contributing to virulence and attributed in part to the yeast cell wall [19]. The use of this structure as a helpful target to control fungal infections has been widely discussed, and, because of their contribution to cell wall integrity, GPI-anchored cell wall proteins have been studied for this purpose. The APX001A is a novel agent that inhibits fungal Gwt1 protein, an enzyme involved in the synthesis of the glycosylphosphatidylinositol moiety of GPI-anchored proteins, and the treatment of C. auris with this compound impairs the maturation and localization of GPI proteins, thereby compromising cell viability [19,44].
Fungal yapsins are extracellular aspartyl proteases GPI-anchored to the yeast cell wall or membrane. These enzymes play a role in nutrient acquisition, dimorphism, and cell wall integrity, accounting for the virulence in pathogenic fungi [23,45]. Here, we identified seven genes encoding putative yapsins in the genome of C. auris B11220 (clade II), which were distributed in four chromosomes. In contrast to the multigene family of C. glabrata yapsin, the genes of C. auris did not exhibit a tandem arrangement, suggesting recent duplication [3]. The putative yapsins of C. auris range in size from 372 to 696 amino acids, suggesting potential functional variability among them, typical of this multigene family [3,23]. The seven yapsin-encoding sequences were also identified in the genomes of the CJ97 (clade III) and 20-1498 (clade IV) C. auris strains, which were practically identical between strains, except for putative CauYps5, which is shorter in B11220 than in CJ97 and 20-1498 (from 697 in the aforementioned strain to 703 amino acids in the two latter ones). Most of these amino acid sequences exhibited typical characteristics of yapsin-type aspartyl proteases [9,46]: the two conserved aspartic residues within the catalytic domain, except for Yps2 and Yps5, the cysteines responsible for disulfide bridge formation, the signal peptide, propeptides with dibasic or basic sites processed by Kex2 or autoprocessed, and the omega site characteristic of GPI-anchored proteins [2,5,46,47]. The predicted CauYps2 and CauYps5 do not present the aspartic catalytic residues in the consensus sequence, in contrast to yapsins of S. cerevisiae and C. glabrata; the exception is the CgYps7 [3,48]. In addition, the phylogenetic analysis showed that CauYps2 and CauYps5 are not related to the canonical yapsins of S. cerevisiae, C. glabrata, and C. albicans. Particularly, putative CauYps1 clustered with the Sap9 and Sap10 of C. albicans and the CauYps7 to S. cerevisiae and C. glabrata Yps7 proteins. It suggests that YPS1 and YPS7 of C. auris are homologous to the YPS1, SAP9, and YPS7 of S. cerevisiae and C. albicans, in agreement with the situation previously described by Alvarado et al. [20].
In addition, CauYps1 and CauYps7 also showed similar domains to those described for the Yps1, Sap9, and Yps7 of other yeasts. Notably, C. auris Yps1 showed an internal loop like that of the Sap9 and Yps1 of C. albicans and S. cerevisiae, proteins where this region undergoes autoprocessing, generating two subunits linked through a disulfide bridge [2]. Dibasic sites upstream of the ω site seem to contribute to the shedding activity of the Yps1 of S. cerevisiae, acting on itself and under other GPI-anchored proteins such as Gas1, Utr2, and Msb2 [2,49]. Moreover, the conserved regions of Yps1 and Yps7 with SAPs of C. albicans showed moderate identity percentages. Nevertheless, they are considered adequate for generating predictive models in structural biology, especially for medically relevant proteins such as aspartic proteases [50]. The secondary and tertiary structures like the A1 family of aspartyl protease showed two non-identical lobes, each one containing one of the two catalytic aspartic residues, and the flap, a loop part of the S1 subsite [10], and their catalytic pockets interact through their aspartic catalytic residues with pepstatin A and other residues with proinsulin-delivered peptide containing mono or dibasic sites. The above observations suggest that C. auris yapsins are potential targets of this inhibitor peptide, such as the yapsin of S. cerevisiae, Aspergillus oryzae, and C. albicans, besides the affinity of these yapsins for substrates rich in basic residues [4,48,51].
Since the theoretical analysis points to the probable inhibition of at least CauYps1 and CauYps7 by pepstatin A, the effect of this inhibitor on the growth and the superficial structures of C. auris of the clades III and IV was evaluated to provide insights into the role of yapsins. Pepstatin A is a known inhibitor of aspartic proteases and has helped investigate the fungal protease function during morphogenesis, and growth in stress conditions, and in superficial structure morphology [52,53]. In the study conditions, the growth of C. auris in media containing pepstatin A was not affected, despite other aspartic proteases, such as the previously described SAPs or the vacuolar PrA, being inhibited by pepstatin [22,53]. Furthermore, pepstatin was tested in concert with H2O2, caffeine, NaCl, or SDS, stress conditions, where yapsins of other yeasts seem to be involved in maintaining the cell wall homeostasis [3,23]. Although all the compounds inhibited the growth of both the clade III and IV C. auris strains, in a similar behavior as observed before [54], together with pepstatin, only peroxide and caffeine influenced the growth of the yeast, suggesting that aspartic proteases may be involved in C. auris’s handling of stress associated with the cell wall. Thus, C. auris 20-1498 of the clade IV was subjected to SEM and AFM; while the former has been widely used to observe the superficial structures of yeast, the latter has been used more recently for this purpose with other yeasts, showing its utility [55]. Under control conditions, yeast cells exhibited a typical oval morphology with budding evidence. However, as the concentration of the inhibitor increased, progressive changes in cell morphology were observed, from a rough appearance to cells with a concave shape at higher concentrations, which increased further when caffeine was added. All of this supports the idea that cell wall integrity depends partially on the activity of aspartic proteases, as has been shown in P. brasiliensis and C. glabrata. In the latter, using a strain carrying an inactive Yps1D91A, it was evidenced that active yapsins are required for cell wall homeostasis [47,52]. Although, now, it is not possible to assign this result to a specific aspartyl protease, similar behavior on the cell wall was observed in C. glabrata by SEM, as a consequence of the deletion of their 11 yapsins [56], and at least the Sap9 of C. auris, here called CauYps1, seem to be involved in the biofilm formation through its regulation by the HOG pathway [57].
To relate the function of C. auris yapsins to the stress response of the yeast, the expression of the genes YPS1 and YPS7 was evaluated during nutrient scarcity, high temperature, and the presence of NaCl, which also activate the response of the cell through the cell wall integrity response [58]. The analysis showed that, while the lack of carbon sources induces the expression of YPS1 of both strains CJ97 (clade III) and 20-1498 (Clade IV), the lack of nitrogen source or the presence of BSA induced mainly that of the YPS7 gene of the 20-1498 strain. These results agree with previous reports on the regulation of gene expression encoding yapsins 1 and 7 of C. glabrata in response to dextrose and nitrogen depletion [24,59]. Moreover, S. cerevisiae YPS7 is induced by alternate carbon sources such as mucin, a polysaccharide encountered in the host cell [60]. Osmotic stress conditions (NaCl) and thermal stress (42 °C) also regulate the expression of CauYPS1 and CauYPS7, as observed before in C. glabrata. In the late case, authors suggested that different clinical strains showed a differential expression of CgYPS1 and CgYPS7, which may be a consequence of the differences in promoter gene sequences [59]. The prediction of TFBSs 1000 bp upstream of the coding sequences of the YPS1 and YPS7 genes of both strains displayed dissimilar arrangements. However, these promoters may be regulated by the CWI, HOG, and calcineurin pathways implicated in membrane and cell wall stability [61]. The relatively mild change in expression, mainly of CauYPS1 under these conditions, could be a result of the suggested efficient homeostatic mechanisms that allow C. auris to maintain its physiology without the need for a massive transcriptional response, which may be related to post-translational regulation or epigenetic mechanisms [61].
Given the predominantly CauYPS1 and CauYPS7 behavior, and considering the thermotolerance of C. auris, we assessed the transcriptomic profile of C. auris 20-1498 after 6 h at 42 °C compared to 37 °C. This analysis showed that genes such as HSF1 and HSP90, as well as others that influence the response to heat stress, are overexpressed, as previously described in other strains of C. auris and other pathogenic yeasts [62]. Along with heat shock proteins, autophagy helps cells to maintain protein homeostasis after stress stimuli [63]. In this case, some autophagy genes are overexpressed, which could suggest a role in the degradation of misfolded proteins. Moreover, genes involved in iron homeostasis, and glucose and pyruvate metabolism are upregulated, as has been reported for this yeast [62]. Moreover, genes encoding proteins that regulate membrane fluidity showed an increased expression, as observed in other fungi [64]. On the other hand, the HOG1 gene was downregulated at 42 °C in contrast to 37 °C, in agreement with the results recently reported by Xiao et al. [62]. Furthermore, genes involved in the synthesis of chitin and β-glucans are downregulated, as well as cell surface proteins that are involved in the remodeling of the C. albicans cell wall at 42 °C [65]. It is worth noting that, in contrast to C. albicans, C. auris exhibits a lower chitin content, which is likely related to its cell wall flexibility, allowing the yeast to adapt to osmotic stress [21].
The role of yapsins in cell wall homeostasis could be due to their ability to release cell wall proteins, since these proteins contain similar dibasic or monobasic residues recognized by this protease and by Kex2, as previously suggested [48]. For example, GPI-anchored the proteins Gas1 (Phr family), Utr2 (Chr family), and Msb2, which are a β-1,3-glucan transferase and chitin transferase, respectively, both of which are involved in conveying these moieties to the growing β-1,3-glucans, allowing the cross-linking of the cell wall. On the other hand, Msb2, a transmembrane protein that regulates the MAPK pathway in response mainly to nutrient scarcity (glucose) and osmotic stress, is activated after the removal of the extracellular HMD domain by Yps1 action on the cleavage domain (CD) [49,66,67,68]. Furthermore, proteins as the soluble cell wall protein Pir1 of Saccharomyces and C. albicans are cleaved by Yps1 and Sap9 and Sap10, respectively, between binding repeats, probably altering their binding toβ-1,3-glucans [4,49]. On the other hand, the α-mating factor (Matα), a natural target of Kex2, is processed by S. cerevisiae Yps1 in Δkex2 strains [5,46,47,68]. In contrast, in the double mutant Δerg6Δpep4, YPs1 processes the carboxypeptidase Y (CpY) to the mature form CPY, suggesting a role in vacuole homeostasis, similar to the yapsins of C. glabrata [56,68,69]. Other proteins released by yapsins are the adhesins Epa1 and Als1 of C. glabrata and C. albicans, respectively. All of this, together with the suggested capacity of yapsins to degrade host proteins, could be relevant in their role as virulence factors [3,4] (Figure 8).
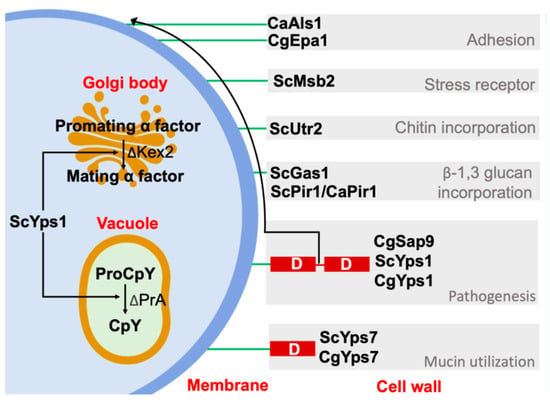
Figure 8.
Functions of Yps1 and Yps7 of yeast S. cerevisiae (Sc), C. albicans (Ca), and C. glabrata (Cg). The yapsins appear to be redundant to carboxypeptidase Kex2; both cleave monobasic or dibasic residues. Furthermore, a sheddase activity of yapsins on cell wall proteins and itself could be related to cell wall homeostasis, and other functions shown in the figure. GPI-anchor: green line. [3,4,5,43,56,66,67,68]. Figure generated by the authors.
Thus, the C. auris genome holds five genes encoding well-conserved putative extracellular yapsins that may be anchored to the membrane or cell wall. Two are less related to aspartyl proteases. Nevertheless, some aspartic proteases with an alanine residue instead of the catalytic aspartic acid have been described [10]. Using pepstatin A, an inhibitor of aspartyl proteases, the growth of both strains of C. auris was slightly and differentially affected, mainly in the presence of compounds that induce the cell wall integrity pathway, such as H2O2 and caffeine, the latter through the TOR pathway [70,71]. Moreover, after caffeine with pepstatin A, the cell wall became rough with depressions. These results suggest a role of aspartic proteases in maintaining cell wall integrity during stress conditions. Other well-known conditions that induce the reorganization of the cell wall of Candida spp. included nutrient, temperature, and osmotic stress [58], conditions that differentially regulate the CauYPS1 and CauYPS7 expression between the tested strains, showing a specific clade transcriptome response. Jenull et al. [61] suggest that the transcriptional profile of C. auris associates with phenotypic traits of the strains of each clade. Moreover, the transcriptomic profile of the strain 20-1498 (clade IV) showed an increased expression of genes related to chaperone function, glucose and iron homeostasis, and membrane fluidity, as previously shown [62]. In addition, autophagy genes (ATGs) are also overexpressed, suggesting a role in the removal of damaged proteins. Thus, as in other fungi, the TOR pathway could be a crosstalk signaling of stress conditions to regulate the cell wall remodeling and the vacuole function [70], where yapsin Yps1 and CauYps7 might play a role, through the processing of proteins to mature or release them from the cell wall or membrane.
In conclusion, the results of pepstatin A inhibition on cell wall morphology, growth, and YPS1 and YPS7 gene expression suggest that the yapsins CauYps1 and CauYps7 may play a role in C. auris cell wall integrity under stress conditions. However, further efforts are needed in order to obtain corresponding mutants lacking the corresponding enzymes and to evaluate the role of the other yapsins. C. auris yapsins could be a therapeutic target for the design of new antifungal or antivirulence agents.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/jof11080573/s1, Figure S1: Characteristics of predicted yapsins of three C. auris strains of clades II, III, and IV; Figure S2: Predicted secondary structure and motifs of yeast yapsins; Figure S3: Effect of pepstatin A on the growth of C. auris in the presence of different compounds; Figure S4: Predicted protein targets of Yps1 of C. auris; Figure S5: Global transcriptomic analysis of C. auris. Strain 20-1498, belonging to clade IV, was cultured in YNB+C+N medium at 37 °C and 42 °C for 6 h [68,70]; Table S1: Accession numbers and data generated from the transcriptomic analysis of C. auris at 42 °C.
Author Contributions
Conceptualization, L.V.-T., M.J.-M., and C.H.-R.; writing—original draft preparation, A.V.-M., L.V.-T., and M.J.-M.; writing—review and editing, A.V.-M., D.C.-F., L.V.-T., M.J.-M., C.H.-R., J.P.L.-A., and E.V.-G.; bioinformatic analysis, A.V.-M. and D.C.-F.; effect of pepstatin and cell wall stressor on the growth an microscopic morphology, A.V.-M.; total RNA extraction and relative expression analysis, A.V.-M. and E.R.-C.; project administration, L.V.-T., M.J.-M., and C.H.-R.; funding acquisition, L.V.-T. and M.J.-M. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by SIP20251179, SIP20251308, SIP20240946, and SIP20242205 (Instituto Politécnico Nacional, Mexico), CBF-2025-I-2871, Bordallo-Landa-Guillén Foundation (grant 2025-07).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The original contributions presented in this study are included in the article/Supplementary Materials. Further inquiries can be directed to the corresponding authors.
Acknowledgments
We thank Mayahuel Ortega Avilés and Juan Vicente Méndez Méndez for their help in processing the samples for SEM and AFM and for image acquisition, at the Centro de Nanociencias y Micro y Nanotecnologías, IPN, Mexico.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Hube, B.; Naglik, J. Candida albicans proteinases: Resolving the mystery of a gene family. Microbiology 2001, 147, 1997–2005. [Google Scholar] [CrossRef]
- Albrecht, A.; Felk, A.; Pichova, I.; Naglik, J.R.; Schaller, M.; de Groot, P.; MacCallum, D.; Odds, F.C.; Schäfer, W.; Michel, M.; et al. Glycosylphosphatidylinositol-anchored proteases of Candida albicans target proteins necessary for both cellular processes and host-pathogen interactions. J. Biol. Chem. 2006, 281, 688–694. [Google Scholar] [CrossRef]
- Kaur, R.; Ma, B.; Cormack, B.P. A family of glycosylphosphatidylinositol-linked aspartyl proteases is required for virulence of Candida glabrata. Proc. Natl. Acad. Sci. USA 2007, 104, 7628–7633. [Google Scholar] [CrossRef]
- Schild, L.; Heyken, A.; de Groot, P.W.; Hiller, E.; Mock, M.; de Koster, C.; Horn, U.; Rupp, S.; Hube, B. Proteolytic cleavage of covalently linked cell wall proteins by Candida albicans Sap9 and Sap10. Eukaryot. Cell 2011, 10, 98–109. [Google Scholar] [CrossRef]
- Egel-Mitani, M.; Flygenring, H.P.; Hansen, M.T. A novel aspartyl protease allowing KEX2-independent MF alpha propheromone processing in yeast. Yeast 1990, 6, 127–137. [Google Scholar] [CrossRef] [PubMed]
- Olsen, V.; Loh, Y.P. In vivo processing of nonanchored Yapsin 1 (Yap3p). Arch. Biochem. Biophys. 2000, 375, 315–321. [Google Scholar] [CrossRef] [PubMed]
- Bourbonnais, Y.; Ash, J.; Daigle, M.; Thomas, D.Y. Isolation and characterization of S. cerevisiae mutants defective in somatostatin expression: Cloning and functional role of a yeast gene encoding an aspartyl protease in precursor processing at monobasic cleavage sites. EMBO J. 1993, 12, 285–294. [Google Scholar] [CrossRef]
- Gagnon-Arsenault, I.; Tremblay, J.; Bourbonnais, Y. Fungal yapsins and cell wall: A unique family of aspartic peptidases for a distinctive cellular function. FEMS Yeast Res. 2006, 6, 966–978. [Google Scholar] [CrossRef] [PubMed]
- de Groot, P.W.J.; Brandt, B.W. ProFASTA: A pipeline web server for fungal protein scanning with integration of cell surface prediction software. Fungal Genet. Biol. 2012, 49, 173–179. [Google Scholar] [CrossRef]
- Rawlings, N.D.; Salvesen, G. Handbook of Proteolytic Enzymes; Academic Press: San Diego, CA, USA, 2013. [Google Scholar]
- Parra-Ortega, B.; Cruz-Torres, H.; Villa-Tanaca, L.; Hernandez-Rodriguez, C. Phylogeny and evolution of the aspartyl protease family from clinically relevant Candida species. Mem. Inst. Oswaldo Cruz. 2009, 104, 505–512. [Google Scholar] [CrossRef]
- Satoh, K.; Makimura, K.; Hasumi, Y.; Nishiyama, Y.; Uchida, K.; Yamaguchi, H. Candida auris sp. nov., a novel ascomycetous yeast isolated from the external ear canal of an inpatient in a Japanese hospital. Microbiol. Immunol. 2009, 53, 41–44. [Google Scholar] [CrossRef] [PubMed]
- Santana, D.J.; Zhao, G.; O’Meara, T.R. The many faces of Candida auris: Phenotypic and strain variation in an emerging pathogen. PLoS Pathog. 2024, 20, e1012011. [Google Scholar] [CrossRef]
- Yue, H.; Bing, J.; Zheng, Q.; Zhang, Y.; Hu, T.; Du, H.; Wang, H.; Huang, G. Filamentation in Candida auris, an emerging fungal pathogen of humans: Passage through the mammalian body induces a heritable phenotypic switch. Emerg. Microbes Infect. 2018, 7, 188. [Google Scholar] [CrossRef]
- Casadevall, A.; Kontoyiannis, D.P.; Robert, V. On the emergence of Candida auris: Climate change, azoles, swamps, and birds. mBio 2019, 10, e01397-19. [Google Scholar] [CrossRef]
- Arora, P.; Singh, P.; Wang, Y.; Yadav, A.; Pawar, K.; Singh, A.; Padmavati, G.; Xu, J.; Chowdhary, A. Environmental isolation of Candida auris from the coastal wetlands of Andaman Islands, India. mBio 2021, 12, e03181-20. [Google Scholar] [CrossRef]
- Lockhart, S.R.; Chowdhary, A.; Gold, J.A. The rapid emergence of antifungal-resistant human-pathogenic fungi. Nat. Rev. Microbiol. 2023, 21, 818–832. [Google Scholar] [CrossRef]
- Horton, M.V.; Johnson, C.J.; Zarnowski, R.; Andes, B.D.; Schoen, T.J.; Kernien, J.F.; Lowman, D.; Kruppa, M.D.; Ma, Z.; Williams, D.L.; et al. Candida auris cell wall mannosylation contributes to neutrophil evasion through pathways divergent from Candida albicans and Candida glabrata. mSphere 2021, 6, e0040621. [Google Scholar] [CrossRef] [PubMed]
- Dakalbab, S.; Hamdy, R.; Holigová, P.; Abuzaid, E.J.; Abu-Qiyas, A.; Lashine, Y.; Mohammad, M.G.; Soliman, S.S. Uniqueness of Candida auris cell wall in morphogenesis, virulence, resistance, and immune evasion. Microbiol. Res. 2024, 286, 127797. [Google Scholar] [CrossRef] [PubMed]
- Alvarado, M.; Gómez-Navajas, J.A.; Blázquez-Muñoz, M.T.; Gómez-Molero, E.; Fernández-Sánchez, S.; Eraso, E.; Munro, C.A.; Valentín, E.; Mateo, E.; de Groot, P.W.J. The good, the bad, and the hazardous: Comparative genomic analysis unveils cell wall features in the pathogen Candidozyma auris typical for both baker’s yeast and Candida. FEMS Yeast Res. 2024, 24, foae039. [Google Scholar] [CrossRef]
- Pezzotti, G.; Kobara, M.; Nakaya, T.; Imamura, H.; Fujii, T.; Miyamoto, N.; Adachi, T.; Yamamoto, T.; Kanamura, N.; Ohgitani, E.; et al. Raman metabolomics of Candida auris clades: Profiling and barcode identification. Int. J. Mol. Sci. 2022, 23, 11736. [Google Scholar] [CrossRef]
- Kim, J.S.; Lee, K.T.; Bahn, Y.S. Secreted aspartyl protease 3 regulated by the Ras/cAMP/PKA pathway promotes the virulence of Candida auris. Front. Cell. Infect. Microbiol. 2023, 13, 1257897. [Google Scholar] [CrossRef]
- Krysan, D.J.; Ting, E.L.; Abeijon, C.; Kroos, L.; Fuller, R.S. Yapsins are a family of aspartyl proteases required for cell wall integrity in Saccharomyces cerevisiae. Eukaryot. Cell 2005, 4, 1364–1374. [Google Scholar] [CrossRef]
- Askari, F.; Rasheed, M.; Kaur, R. The yapsin family of aspartyl proteases regulate glucose homeostasis in Candida glabrata. J. Biol. Chem. 2022, 298, 101593. [Google Scholar] [CrossRef] [PubMed]
- Ruiz Gaitán, A.C.; Moret, A.; López Hontangas, J.L.; Molina, J.M.; Aleixandre López, A.I.; Cabezas, A.H.; Mollar Maseres, J.; Arcas, R.C.; Gómez Ruiz, M.D.; Chiveli, M.Á.; et al. Nosocomial fungemia by Candida auris: First four reported cases in continental Europe. Rev. Iberoam. Micol. 2017, 34, 23–27. [Google Scholar] [CrossRef] [PubMed]
- Caballero, U.; Eraso, E.; Quindós, G.; Jauregizar, N. In vitro interaction and killing-kinetics of amphotericin B combined with anidulafungin or caspofungin against Candida auris. Pharmaceutics 2021, 13, 1333. [Google Scholar] [CrossRef]
- Ayala-Gaytán, J.J.; Montoya, A.M.; Martínez-Resendez, M.F.; Guajardo-Lara, C.E.; Treviño-Rangel, R.D.J.; Salazar-Cavazos, L.; Llaca-Díaz, J.M.; González, G.M. First case of Candida auris isolated from the bloodstream of a Mexican patient with serious gastrointestinal complications from severe endometriosis. Infection 2021, 49, 523–525. [Google Scholar] [CrossRef]
- Casimiro-Ramos, A.; Bautista-Crescencio, C.; Vidal-Montiel, A.; González, G.M.; Hernández-García, J.A.; Hernández-Rodríguez, C.; Villa-Tanaca, L. Comparative genomics of the first resistant Candida auris strain isolated in Mexico: Phylogenomic and pan-genomic analysis and mutations associated with antifungal resistance. J. Fungi 2024, 10, 392. [Google Scholar] [CrossRef]
- Teufel, F.; Almagro Armenteros, J.J.; Johansen, A.R.; Gíslason, M.H.; Pihl, S.I.; Tsirigos, K.D.; Winther, O.; Brunak, S.; von Heijne, G.; Nielsen, H. SignalP 6.0 predicts all five types of signal peptides using protein language models. Nat. Biotechnol. 2022, 40, 1023–1025. [Google Scholar] [CrossRef]
- Gíslason, M.H.; Nielsen, H.; Almagro-Armenteros, J.J.; Johansen, A. Prediction of GPI-Anchored proteins with pointer neural networks. Curr. Res. Biotechnol. 2021, 3, 6–13. [Google Scholar] [CrossRef]
- Monteiro, P.T.; Oliveira, J.; Pais, P.; Antunes, M.; Palma, M.; Cavalheiro, M.; Teixeira, M.C. YEASTRACT+: A portal for cross-species comparative genomics of transcription regulation in yeasts. Nucleic Acids Res. 2020, 48, D642–D649. [Google Scholar] [CrossRef] [PubMed]
- Campanella, J.J.; Bitincka, L.; Smalley, J. MatGAT: An application that generates similarity/identity matrices using protein or DNA sequences. BMC Bioinform. 2003, 4, 29. [Google Scholar] [CrossRef]
- Larkin, M.A.; Blackshields, G.; Brown, N.P.; Chenna, R.; McGettigan, P.A.; McWilliam, H.; Valentin, F.; Wallace, I.M.; Wilm, A.; Lopez, R.; et al. Clustal W and Clustal X version 2.0. Bioinformatics 2007, 23, 2947–2948. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, L.T.; Schmidt, H.A.; von Haeseler, A.; Minh, B.Q. IQ-TREE: A fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 2015, 32, 268–274. [Google Scholar] [CrossRef]
- Robert, X.; Gouet, P. Deciphering key features in protein structures with the new ENDscript server. Nucleic Acids Res. 2014, 42, W320–W324. [Google Scholar] [CrossRef]
- Waterhouse, A.; Bertoni, M.; Bienert, S.; Studer, G.; Tauriello, G.; Gumienny, R.; Heer, F.T.; de Beer, T.A.P.; Rempfer, C.; Bordoli, L.; et al. SWISS-MODEL: Homology modelling of protein structures and complexes. Nucleic Acids Res. 2018, 46, W296–W303. [Google Scholar] [CrossRef]
- Hanwell, M.D.; Curtis, D.E.; Lonie, D.C.; Vandermeersch, T.; Zurek, E.; Hutchison, G.R. Avogadro: An advanced semantic chemical editor, visualization, and analysis platform. J. Cheminform. 2012, 4, 17. [Google Scholar] [CrossRef]
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Couch, G.S.; Greenblatt, D.M.; Meng, E.C.; Ferrin, T.E. UCSF Chimera—A visualization system for exploratory research and analysis. J. Comput. Chem. 2004, 25, 1605–1612. [Google Scholar] [CrossRef]
- Eberhardt, J.; Santos-Martins, D.; Tillack, A.F.; Forli, S. AutoDock Vina 1.2.0: New docking methods, expanded force field, and python bindings. J. Chem. Inf. Model. 2021, 61, 3891–3898. [Google Scholar] [CrossRef] [PubMed]
- Yan, Y.; Tao, H.; He, J.; Huang, S.-Y. The HDOCK server for integrated protein-protein docking. Nat. Protoc. 2020, 15, 1829–1852. [Google Scholar] [CrossRef]
- Schmitt, M.E.; Brown, T.A.; Trumpower, B.L. A rapid and simple method for preparation of RNA from Saccharomyces cerevisiae. Nucleic Acids Res. 1990, 18, 3091–3092. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Carolus, H.; Pierson, S.; Muñoz, J.F.; Subotić, A.; Cruz, R.B.; Cuomo, C.A.; Van Dijck, P. Genome-wide analysis of experimentally evolved Candida auris reveals multiple novel mechanisms of multidrug resistance. mBio 2021, 12, e03333-20. [Google Scholar] [CrossRef]
- Hager, C.L.; Larkin, E.L.; Long, L.; Zohra Abidi, F.; Shaw, K.J.; Ghannoum, M.A. In vitro and in vivo evaluation of the antifungal activity of APX001A/APX001 against Candida auris. Antimicrob. Agents Chemother. 2018, 62, e02319-17. [Google Scholar] [CrossRef]
- Bras, G.; Satala, D.; Juszczak, M.; Kulig, K.; Wronowska, E.; Bednarek, A.; Karkowska-Kuleta, J. Secreted aspartic proteinases: Key factors in Candida infections and host-pathogen interactions. Int. J. Mol. Sci. 2024, 25, 4775. [Google Scholar] [CrossRef]
- Gagnon-Arsenault, I.; Parisé, L.; Tremblay, J.; Bourbonnais, Y. Activation mechanism, functional role and shedding of glycosylphosphatidylinositol-anchored Yps1p at the Saccharomyces cerevisiae cell surface. Mol. Microbiol. 2008, 69, 982–993. [Google Scholar] [CrossRef]
- Rasheed, M.; Battu, A.; Kaur, R. Aspartyl proteases in Candida glabrata are required for suppression of the host innate immune response. J. Biol. Chem. 2018, 293, 6410–6433. [Google Scholar] [CrossRef] [PubMed]
- Olsen, V.; Cawley, N.X.; Brandt, J.; Egel-Mitani, M.; Loh, Y.P. Identification and characterization of Saccharomyces cerevisiae yapsin 3, a new member of the yapsin family of aspartic proteases encoded by the YPS3 gene. Biochem. J. 1999, 339, 407–411. [Google Scholar] [CrossRef] [PubMed]
- Dubé, A.K.; Bélanger, M.; Gagnon-Arsenault, I.; Bourbonnais, Y. N-terminal entrance loop of yeast Yps1 and O-glycosylation of substrates are determinant factors controlling the shedding activity of this GPI-anchored endopeptidase. BMC Microbiol. 2015, 15, 50. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y. Progress and challenges in protein structure prediction. Curr. Opin. Struct. Biol. 2008, 18, 342–348. [Google Scholar] [CrossRef]
- Shimizu, N.; Katagiri, T.; Matsumoto, A.; Matsuda, Y.; Arai, H.; Sasaki, N.; Abe, K.; Katase, T.; Ishida, H.; Kusumoto, K.I.; et al. Oryzapsins, the orthologs of yeast yapsin in Aspergillus oryzae, affect ergosterol synthesis. Appl. Microbiol. Biotechnol. 2021, 105, 8481–8494. [Google Scholar] [CrossRef]
- Silva, R.D.S.; Segura, W.D.; Oliveira, R.S.; Xander, P.; Batista, W.L. Characterization of aspartic proteases from Paracoccidioides brasiliensis and their role in fungal thermo-dimorphism. J. Fungi 2023, 9, 375. [Google Scholar] [CrossRef]
- Clark-Flores, D.; Vidal-Montiel, A.; Mondragón-Flores, R.; Valentín-Gómez, E.; Hernández-Rodríguez, C.; Juárez-Montiel, M.; Villa-Tanaca, L. Vacuolar proteases of Candida auris from clades III and IV and their relationship with autophagy. J. Fungi 2025, 11, 388. [Google Scholar] [CrossRef]
- Day, A.M.; McNiff, M.M.; da Silva Dantas, A.; Gow, N.A.; Quinn, J. Hog1 regulates stress tolerance and virulence in the emerging fungal pathogen Candida auris. mSphere 2018, 3, e00506-18. [Google Scholar] [CrossRef]
- Gibbs, E.; Hsu, J.; Barth, K.; Goss, J.W. Characterization of the nanomechanical properties of the fission yeast (Schizosaccharomyces pombe) cell surface by atomic force microscopy. Yeast 2021, 38, 480–492. [Google Scholar] [CrossRef]
- Bairwa, G.; Rasheed, M.; Taigwal, R.; Sahoo, R.; Kaur, R. GPI (glycosylphosphatidylinositol)-linked aspartyl proteases regulate vacuole homoeostasis in Candida glabrata. Biochem. J. 2014, 458, 323–334. [Google Scholar] [CrossRef]
- Shivarathri, R.; Chauhan, M.; Datta, A.; Das, D.; Karuli, A.; Aptekmann, A.; Chauhan, N. The Candida auris Hog1 MAP kinase is essential for the colonization of murine skin and intradermal persistence. mBio 2024, 15, e02748-24. [Google Scholar] [CrossRef]
- Banda-Flores, I.A.; Torres-Tirado, D.; Mora-Montes, H.M.; Pérez-Flores, G.; Pérez-García, L.A. Resilience in resistance: The role of cell wall integrity in multidrug-resistant Candida. J. Fungi 2025, 11, 271. [Google Scholar] [CrossRef] [PubMed]
- Cortés-Acosta, E.; Ibarra, J.A.; Ramírez-Saad, H.; Vargas-Mendoza, C.F.; Villa-Tanaca, L.; Hernández-Rodríguez, C. Polymorphism in the regulatory regions of genes CgYPS1 and CgYPS7 encoding yapsins in Candida glabrata is associated with changes in expression levels. FEMS Yeast Res. 2017, 17, fox077. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Mercurio, K.; Singh, D.; Walden, E.; Baetz, K. Global analysis of Saccharomyces cerevisiae growth in mucin. G3 2021, 11, jkab294. [Google Scholar] [CrossRef] [PubMed]
- Jenull, S.; Tscherner, M.; Kashko, N.; Shivarathri, R.; Stoiber, A.; Chauhan, M.; Petryshyn, A.; Chauhan, N.; Kuchler, K. Transcriptome signatures predict phenotypic variations of Candida auris. Front. Cell. Infect. Microbiol. 2021, 11, 662563. [Google Scholar] [CrossRef]
- Xiao, W.; Zhou, H.; Huang, J.; Xin, C.; Zhang, J.; Wen, H.; Song, Z. Comparative analyses of the biological characteristics, fluconazole resistance, and heat adaptation mechanisms of Candida auris and members of the Candida haemulonii complex. Appl. Environ. Microbiol. 2025, 91, e02406-24. [Google Scholar] [CrossRef]
- Zhang, T.; Lin, Y.; Zhang, Z.; Wang, Z.; Zeng, F.; Wang, Q. Roles and applications of autophagy in guarding against environmental stress and DNA damage in Saccharomyces cerevisiae. FEBS J. 2025, 1–26. [Google Scholar] [CrossRef] [PubMed]
- Xiao, W.; Zhang, J.; Huang, J.; Xin, C.; Li, M.J.; Song, Z. Response and regulatory mechanisms of heat resistance in pathogenic fungi. Appl. Microbiol. Biotechnol. 2022, 106, 5415–5431. [Google Scholar] [CrossRef] [PubMed]
- Ikezaki, S.; Cho, T.; Nagao, J.I.; Tasaki, S.; Yamaguchi, M.; Arita-Morioka, K.I.; Yasumatsu, K.; Chibana, H.; Ikebe, T.; Tanaka, Y. Mild heat stress affects on the cell wall structure in Candida albicans biofilm. Med. Mycol. J. 2019, 60, 29–37. [Google Scholar] [CrossRef]
- Vadaie, N.; Dionne, H.; Akajagbor, D.S.; Nickerson, S.R.; Krysan, D.J.; Cullen, P.J. Cleavage of the signaling mucin Msb2 by the aspartyl protease Yps1 is required for MAPK activation in yeast. J. Cell Biol. 2008, 181, 1073–1081. [Google Scholar] [CrossRef]
- Miller, K.A.; DiDone, L.; Krysan, D.J. Extracellular secretion of overexpressed glycosylphosphatidylinositol-linked cell wall protein Utr2/Crh2p as a novel protein quality control mechanism in Saccharomyces cerevisiae. Eukaryot. Cell 2010, 9, 1669–1679. [Google Scholar] [CrossRef]
- Julius, D.; Schekman, R.; Thorner, J. Glycosylation and processing of prepro-α-factor through the yeast secretory pathway. Cell 1984, 36, 309–318. [Google Scholar] [CrossRef]
- Sievi, E.; Suntio, T.; Makarow, M. Proteolytic function of GPI-anchored plasma membrane protease Yps1p in the yeast vacuole and Golgi. Traffic 2001, 2, 896–907. [Google Scholar] [CrossRef] [PubMed]
- Jiménez-Gutiérrez, E.; Alegría-Carrasco, E.; Sellers-Moya, Á.; Molina, M.; Martín, H. Not just the wall: The other ways to turn the yeast CWI pathway on. Int. Microbiol. 2020, 23, 107–119. [Google Scholar] [CrossRef]
- Ribeiro, R.A.; Bourbon-Melo, N.; Sá-Correia, I. The cell wall and the response and tolerance to stresses of biotechnological relevance in yeasts. Front. Microbiol. 2022, 13, 953479. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).