Insights into Persian Gulf Beach Sand Mycobiomes: Promises and Challenges in Fungal Diversity
Abstract
1. Introduction
2. Materials and Methods
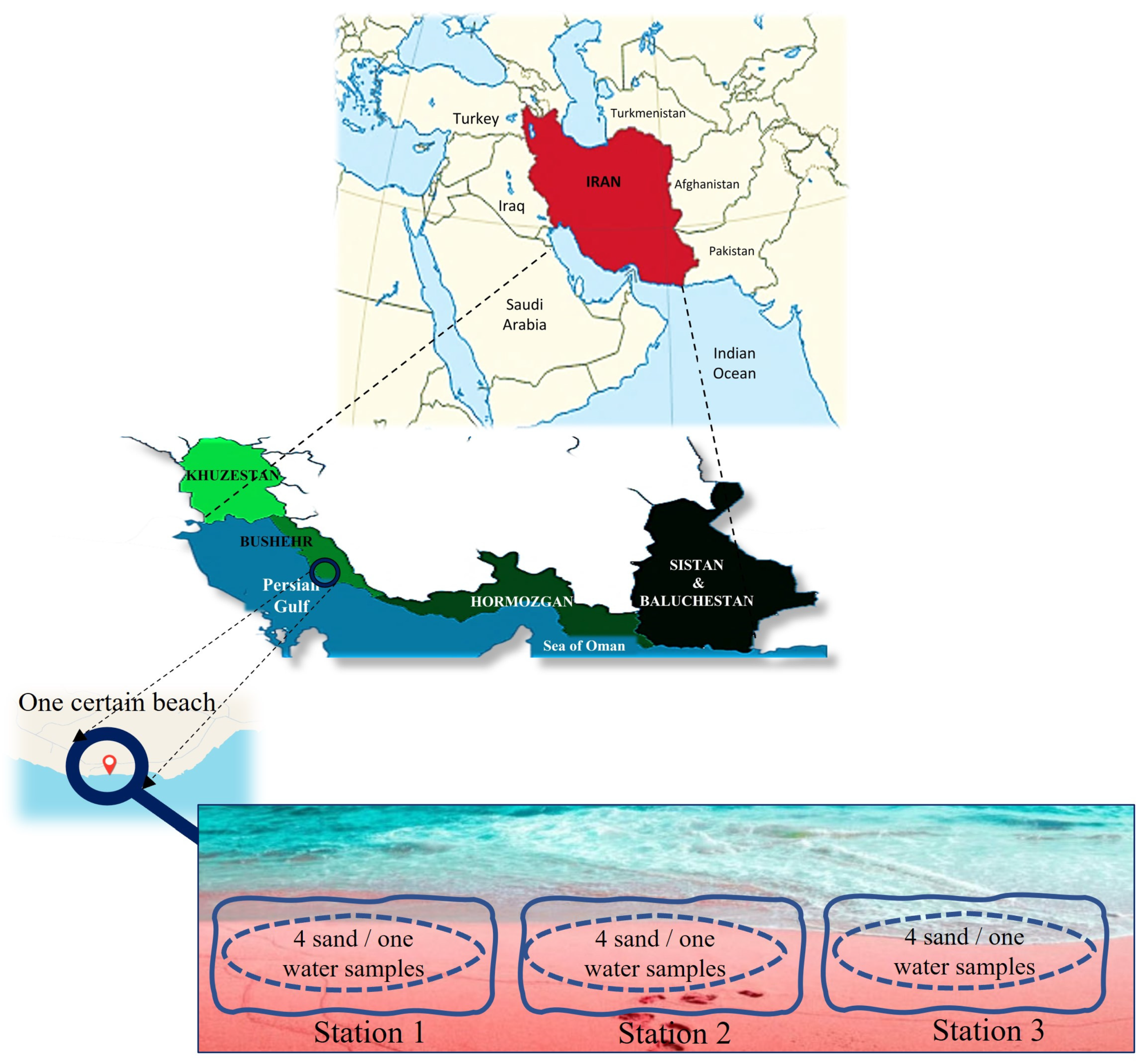
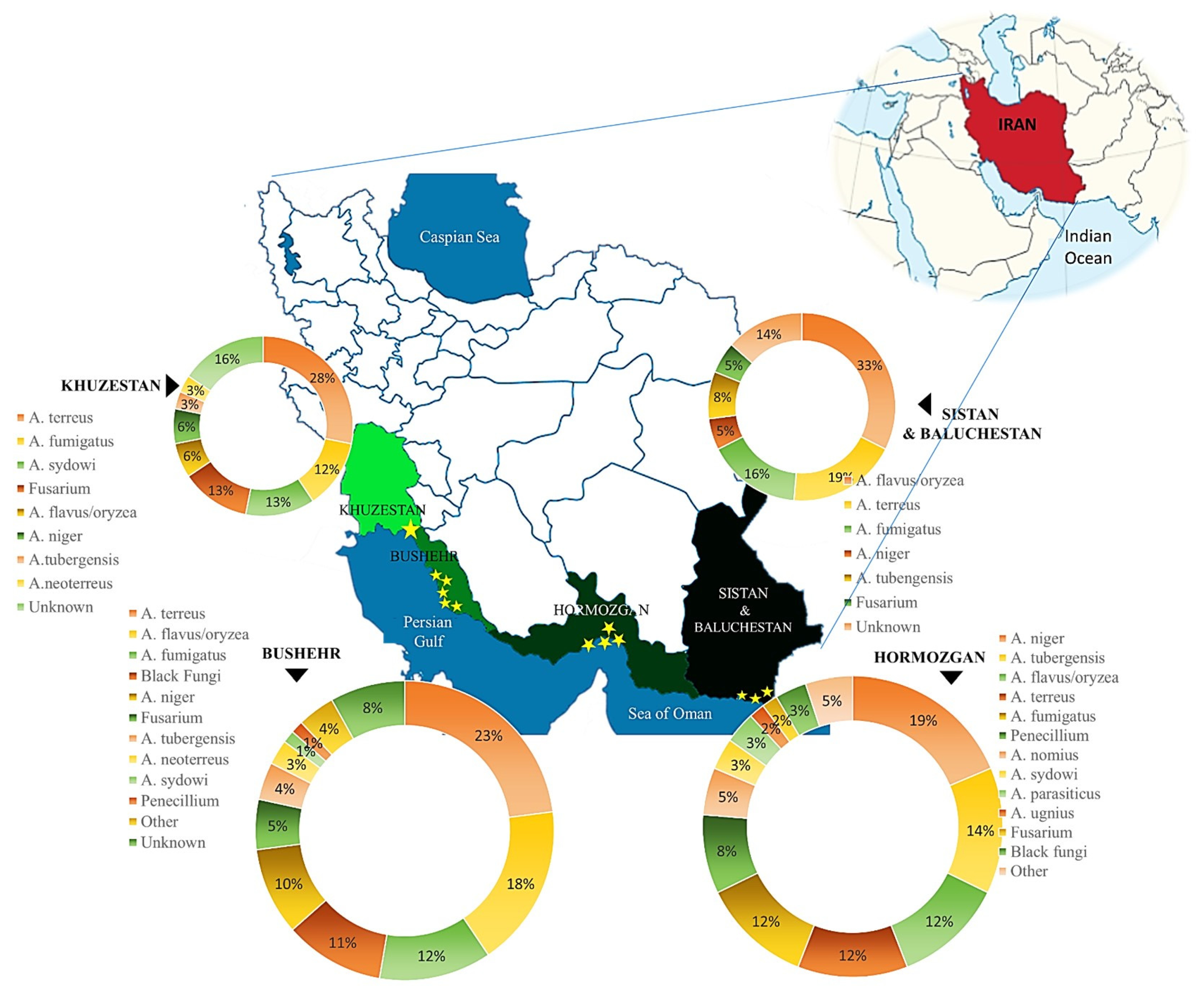
2.1. Sites and Sampling
2.2. Sample Preparation, Fungal Load, and Phenotypic Identification
DNA Extraction, Amplification, and Sequencing
2.3. Antifungal Susceptibility Test Assay for Selected Isolates
2.4. Statistical Analyses
3. Results
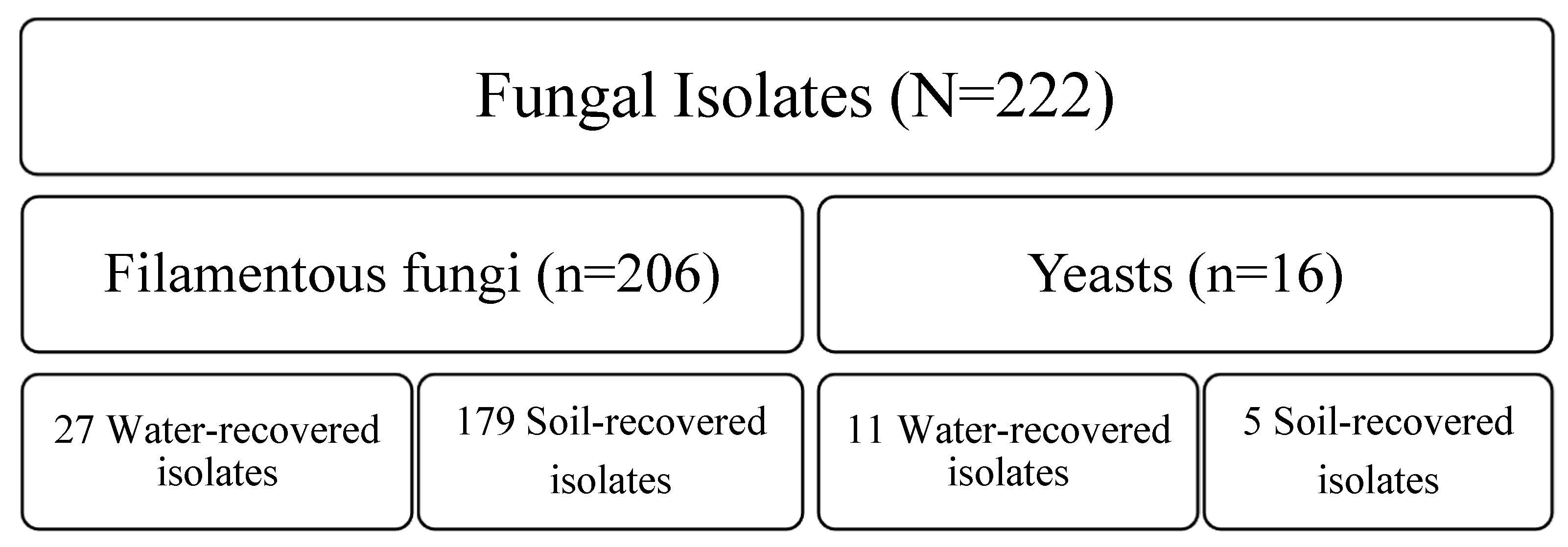
3.1. Fungal Community Flora in Sand and Seawater
3.2. Fungal Load (CFUs) in Sand and Seawater
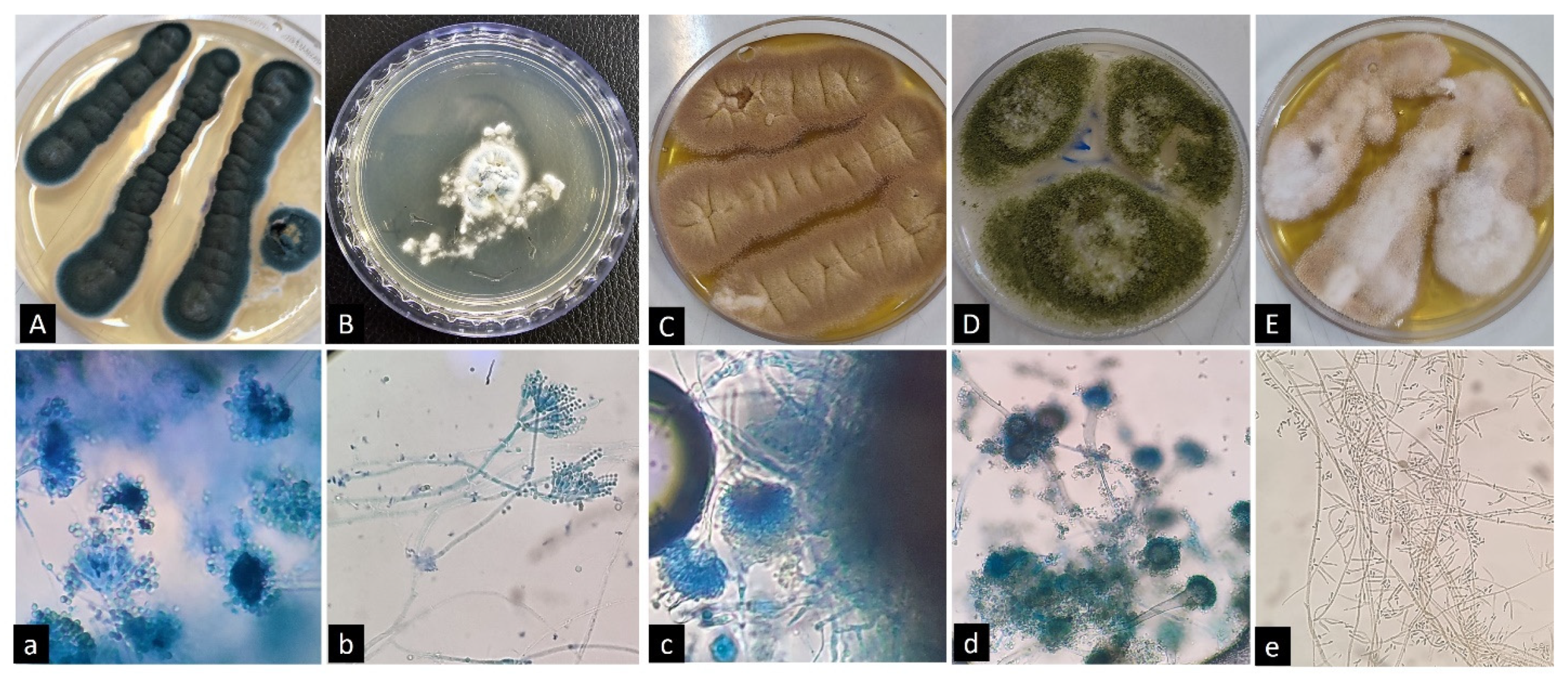
3.3. Molecular Identification of Fungal Isolates
3.4. Antifungal Susceptibility Test Assay:
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| CLSI | Clinical & Laboratory Standards Institute |
| DOAJ | Voriconazole |
| TLA | Itraconazole |
| LD | Posaconazole |
| PG | Persian Gulf |
References
- Pessoa, M.F.; Lidon, F.C. Impact of human activities on coastal vegetation-A review. Emir. J. Food Agric. (EJFA) 2013, 25, 926–944. [Google Scholar]
- Solo-Gabriele, H.M.; Harwood, V.J.; Kay, D.; Fujioka, R.S.; Sadowsky, M.J.; Whitman, R.L.; Wither, A.; Caniça, M.; Da Fonseca, R.C.; Duarte, A. Beach sand and the potential for infectious disease transmission: Observations and recommendations. J. Mar. Biol. Assoc. United Kingd. 2016, 96, 101–120. [Google Scholar] [CrossRef]
- Brandão, J.; Gangneux, J.-P.; Arikan-Akdagli, S.; Barac, A.; Bostanaru, A.C.; Brito, S.; Bull, M.; Çerikçioğlu, N.; Chapman, B.; Efstratiou, M. Mycosands: Fungal diversity and abundance in beach sand and recreational waters—Relevance to human health. Sci. Total Environ. 2021, 781, 146598. [Google Scholar] [CrossRef]
- Byappanahalli, M.; Nevers, M.; Korajkic, A.; Staley, Z.; Harwood, V. Enterococci in the environment. Microbiol. Mol. Biol. Rev. 2012, 76, 685–706. [Google Scholar] [CrossRef]
- Jang, J.; Hur, H.G.; Sadowsky, M.J.; Byappanahalli, M.; Yan, T.; Ishii, S. Environmental Escherichia coli: Ecology and public health implications—A review. J. Appl. Microbiol. 2017, 123, 570–581. [Google Scholar] [CrossRef]
- Pinto, K.C.; Hachich, E.M.; Sato, M.I.; Di Bari, M.; Coelho, M.C.; Matté, M.H.; Lamparelli, C.C.; Razzolini, M.T. Microbiological quality assessment of sand and water from three selected beaches of South Coast, São Paulo State, Brazil. Water Sci. Technol. A J. Int. Assoc. Water Pollut. Res. 2012, 66, 2475–2482. [Google Scholar] [CrossRef]
- Novak Babič, M.; Gunde-Cimerman, N.; Breskvar, M.; Džeroski, S.; Brandão, J. Occurrence, diversity and anti-fungal resistance of fungi in sand of an urban beach in Slovenia—Environmental monitoring with possible health risk implications. J. Fungi 2022, 8, 860. [Google Scholar] [CrossRef]
- Gangneux, J.-P.; Brandao, J.; Segal, E. Knowledge and regulation on fungal contamination of sand and water: Progress report and perspectives. Med. Mycol. 2024, 62, myad137. [Google Scholar] [CrossRef]
- Prigitano, A.; Trovato, L.; Esposto, M.C.; Brandão, J.; Cogliati, M.; Gatta, G.D.; Grancini, A.; Migliorisi, G.; Oliveri, S.; Romanò, L. Fungal diversity in lake and sea beaches of Italy: Relevance to human health. Sci. Total Environ. 2023, 859, 160417. [Google Scholar] [CrossRef]
- Banchi, E.; Manna, V.; Muggia, L.; Celussi, M. Marine fungal diversity and dynamics in the Gulf of Trieste (Northern Adriatic Sea). Microb. Ecol. 2024, 87, 78. [Google Scholar] [CrossRef]
- Moazeni, M.; Hedayati, M.T.; Haghani, I.; Abastabar, M.; Jahantigh, A.S.; Kheshteh, M.; Nabili, M.; Brandão, J. Caspian Sea Mycosands: The variety and abundance of medically important fungi in beach sand and water. Int. J. Environ. Res. Public Health 2022, 20, 459. [Google Scholar] [CrossRef]
- Pak, A.; Farajzadeh, M. Iran's integrated coastal management plan: Persian Gulf, Oman Sea, and southern Caspian Sea coastlines. Ocean Coast. Manag. 2007, 50, 754–773. [Google Scholar] [CrossRef]
- Bordbar, M.H.; Nasrolahi, A.; Lorenz, M.; Moghaddam, S.; Burchard, H. The Persian Gulf and Oman Sea: Climate variability and trends inferred from satellite observations. Estuar. Coast. Shelf Sci. 2024, 296, 108588. [Google Scholar] [CrossRef]
- Noori, R.; Tian, F.; Berndtsson, R.; Abbasi, M.R.; Naseh, M.V.; Modabberi, A.; Soltani, A.; Kløve, B. Recent and future trends in sea surface temperature across the Persian Gulf and Gulf of Oman. PLoS ONE 2019, 14, e0212790. [Google Scholar] [CrossRef]
- Sabino, R.; Veríssimo, C.; Cunha, M.A.; Wergikoski, B.; Ferreira, F.C.; Rodrigues, R.; Parada, H.; Falcão, L.; Rosado, L.; Pinheiro, C. Pathogenic fungi: An unacknowledged risk at coastal resorts? New insights on microbiological sand quality in Portugal. Mar. Pollut. Bull. 2011, 62, 1506–1511. [Google Scholar] [CrossRef]
- Petersen, C.; Sørensen, T.; Westphal, K.R.; Fechete, L.I.; Sondergaard, T.E.; Sørensen, J.L.; Nielsen, K.L. High molecular weight DNA extraction methods lead to high quality filamentous ascomycete fungal genome assemblies using Oxford Nanopore sequencing. Microb. Genom. 2022, 8, 000816. [Google Scholar] [CrossRef]
- Karimizadeh Esfahani, M.; Eslampoor, A.; Dolatabadi, S.; Najafzadeh, M.J.; Houbraken, J. First case of fungal keratitis due to Aspergillus minisclerotigenes in Iran. Curr. Med. Mycol. 2019, 5, 45–48. [Google Scholar] [CrossRef]
- Haghani, I.; Hedayati, M.T.; Shokohi, T.; Kermani, F.; Ghazanfari, M.; Javidnia, J.; Khojasteh, S.; Roohi, B.; Badali, H.; Fathi, M.; et al. Onychomycosis due to Fusarium species in different continents, literature review on diagnosis and treatment. Mycoses 2024, 67, e13652. [Google Scholar] [CrossRef]
- Nabili, M.; Shokohi, T.; Moazeni, M.; Khodavaisy, S.; Aliyali, M.; Badiee, P.; Zarrinfar, H.; Hagen, F.; Badali, H. High prevalence of clinical and environmental triazole-resistant Aspergillus fumigatus in Iran: Is it a challenging issue? J. Med. Microbiol. 2016, 65, 468–475. [Google Scholar] [CrossRef]
- CLSI Standard M38; Reference Method for Broth Dilution Antifungal Susceptibility Testing of Filamentous fungi, 3rd ed. Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2017.
- CLSI Supplement M60; Performanse Standard for Antifungal Susceptibility Testing of Yeasts, 1st ed. Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2017.
- CLSI Supplement M59; Epidemiological Cutoff Values for Antifungal Susceptibility Testing, 2nd ed. Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2018.
- Almeida, B.D.; Viegas, C. Fungal Contamination of Swimming Pools and Fitness Centers. In Encyclopedia of Mycology; Zaragoza, Ó., Casadevall, A., Eds.; Elsevier: Oxford, UK, 2021; pp. 84–90. [Google Scholar]
- Straus, D.C.; Danny, C.J.; Wong, W.C.; Jumper, C.A. Studies on the Role of Fungi in Sick Building Syndrome. Arch. Environ. Health: Int. J. 2003, 58, 475–478. [Google Scholar] [CrossRef]
- Sharifi, A.; Baubekova, A.; Patro, E.R.; Klöve, B.; Haghighi, A.T. The combined effects of anthropogenic and climate change on river flow alterations in the Southern Caspian Sea Iran. Heliyon 2024, 10, e31960. [Google Scholar] [CrossRef]
- Weiskerger, C.J.; Brandão, J.; Ahmed, W.; Aslan, A.; Avolio, L.; Badgley, B.D.; Boehm, A.B.; Edge, T.A.; Fleisher, J.M.; Heaney, C.D. Impacts of a changing earth on microbial dynamics and human health risks in the continuum between beach water and sand. Water Res. 2019, 162, 456–470. [Google Scholar] [CrossRef]
- Cogliati, M.; Arikan-Akdagli, S.; Barac, A.; Bostanaru, A.; Brito, S.; Çerikçioğlu, N.; Efstratiou, M.; Ergin, Ç.; Esposto, M.; Frenkel, M. Environmental and bioclimatic factors influencing yeasts and molds distribution along European shores. Sci. Total Environ. 2023, 859, 160132. [Google Scholar] [CrossRef]
- Casadevall, A. Global warming could drive the emergence of new fungal pathogens. Nat. Microbiol. 2023, 8, 2217–2219. [Google Scholar] [CrossRef]
- Lass-Flörl, C.; Dietl, A.-M.; Kontoyiannis, D.P.; Brock, M. Aspergillus terreus Species Complex. Clin. Microbiol. Rev. 2021, 34, e00311–e00320. [Google Scholar] [CrossRef]
- Hachem, R.Y.; Dagher, H.; Chaftari, A.-M.; Jiang, Y.; Haddad, A.; Wehbe, S.; Shrestha, J.; Sherchan, R.; Lamie, P.; Makhoul, J.; et al. Decreased Frequency and Improved Outcomes in Invasive Aspergillosis Caused by Aspergillus terreus After the Introduction of Anti-Mold Azole Agents: A 30-Year Study at a Tertiary Cancer Center. J. Fungi 2025, 11, 119. [Google Scholar] [CrossRef]
- Vahedi-Shahandashti, R.; Dietl, A.-M.; Binder, U.; Nagl, M.; Würzner, R.; Lass-Flörl, C. Aspergillus terreus and the Interplay with Amphotericin B: From Resistance to Tolerance? Antimicrob. Agents Chemother. 2022, 66, e02274-21. [Google Scholar] [CrossRef]
- Najafzadeh, M.J.; Dolatabadi, S.; Zarrinfar, H.; Houbraken, J. Molecular Diversity of Aspergilli in Two Iranian Hospitals. Mycopathologia 2021, 186, 519–533. [Google Scholar] [CrossRef]
- Borjian Boroujeni, Z.; Shamsaei, S.; Yarahmadi, M.; Getso, M.I.; Salimi Khorashad, A.; Haghighi, L.; Raissi, V.; Zareei, M.; Saleh Mohammadzade, A.; Moqarabzadeh, V.; et al. Distribution of invasive fungal infections: Molecular epidemiology, etiology, clinical conditions, diagnosis and risk factors: A 3-year experience with 490 patients under intensive care. Microb. Pathog. 2021, 152, 104616. [Google Scholar] [CrossRef]
- Beni, A.N.; Marriner, N.; Sharifi, A.; Azizpour, J.; Kabiri, K.; Djamali, M.; Kirman, A. Climate change: A driver of future conflicts in the Persian Gulf Region? Heliyon 2021, 7, e06288. [Google Scholar] [CrossRef]
- Abolghasemi, S.; Hakamifard, A.; Sharifynia, S.; Pourabdollah Toutkaboni, M.; Azhdari Tehrani, H. Fatal invasive pulmonary aspergillosis in an immunocompetent patient with COVID-19 due to Aspergillus terreus: A case study. Clin. Case Rep. 2021, 9, 2414–2418. [Google Scholar] [CrossRef]
- Yoshii, N.; Yamada, K.; Niki, M.; Imoto, W.; Yamairi, K.; Shibata, W.; Namikawa, H.; Sakatoku, K.; Sato, K.; Nakai, T.; et al. Invasive pulmonary aspergillosis caused by Aspergillus terreus diagnosed using virtual bronchoscopic navigation and endobronchial ultrasonography with guide sheath and successfully treated with liposomal amphotericin B. Infection 2021, 49, 1049–1054. [Google Scholar] [CrossRef]
- Badali, H.; Shokohi, T.; Khodavaisy, S.; Moazeni, M.; Farhadi, M.; Nabili, M. Molecular typing of clinical and environmental Aspergillus fumigatus isolates from Iran using microsatellites. Curr. Med. Mycol. 2021, 7, 25. [Google Scholar] [CrossRef]
- Tashiro, M.; Nakano, Y.; Shirahige, T.; Kakiuchi, S.; Fujita, A.; Tanaka, T.; Takazono, T.; Izumikawa, K. Comprehensive Review of Environmental Surveillance for Azole-Resistant Aspergillus fumigatus: A Practical Roadmap for Hospital Clinicians and Infection Control Teams. J. Fungi 2025, 11, 96. [Google Scholar] [CrossRef]
- Hosseini, P.; Keniya, M.V.; Sagatova, A.A.; Toepfer, S.; Müller, C.; Tyndall, J.D.A.; Klinger, A.; Fleischer, E.; Monk, B.C. The Molecular Basis of the Intrinsic and Acquired Resistance to Azole Antifungals in Aspergillus fumigatus. J. Fungi 2024, 10, 820. [Google Scholar] [CrossRef]
- WHO. Fungal Priority Pathogens List to Guide Research, Development and Public Health Action; WHO: Geneva, Switzerland, 2022. [Google Scholar]
- Casadevall, A. Expanding the pathogenic potential concept to incorporate fulminancy, time, and virulence factors. Msphere 2022, 7, e01021. [Google Scholar] [CrossRef]
- WHO. Guidelines for Safe Recreational Water Environments: Coastal and Fresh Waters; WHO: Geneva, Switzerland, 2003; Volume 1. [Google Scholar]
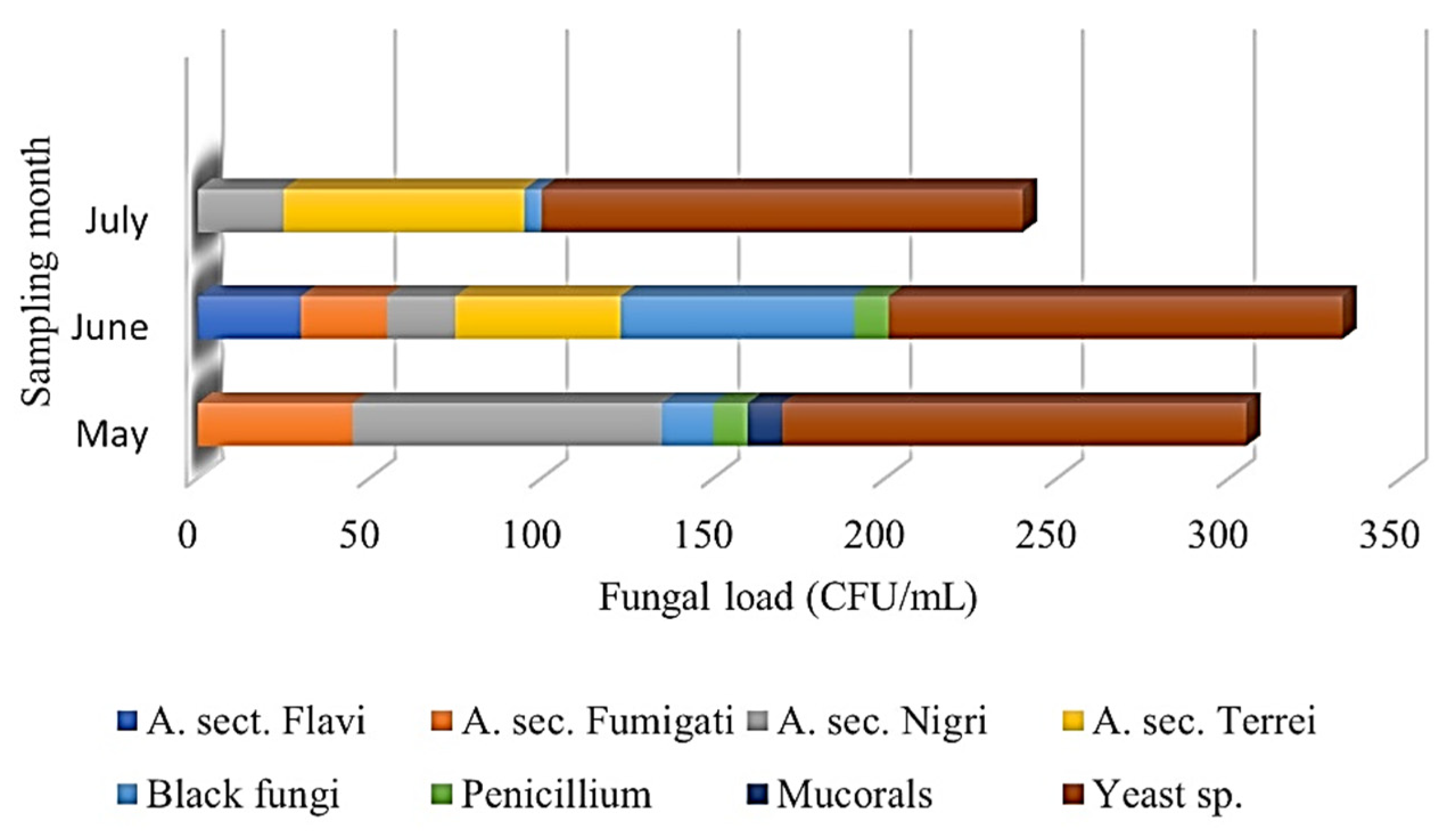
| PROVINCE | Sample Type | Fungal load (CFUs) (30 °C) | First Round (May) | ||||||||||||||||||
| Temperature (°C) | Humidity (%) | A. Section Flavi | A. section Fumigati | A. section Nigri | A. section Terrei | Black fungi | Penicillium sp. | Mucorales | Yeast sp. | Other | |||||||||||
| Khuzestan | Sand | 10 | |||||||||||||||||||
| 29 | 55 | Water | 30 | ||||||||||||||||||
| Bushehr | Sand | 25 | 75 | 5 | 5 | 55 | |||||||||||||||
| 28 | 51 | Water | 5 | 25 | |||||||||||||||||
| Hormozgan | Sand | 10 | 15 | 40 | |||||||||||||||||
| 27 | 69 | Water | 5 | 15 | 5 | 65 | 40 | ||||||||||||||
| Sistan-Baluchestan | Sand | 5 | 5 | 20 | 20 | ||||||||||||||||
| 28 | 52 | Water | |||||||||||||||||||
| PROVINCE | Sample type | Fungal load (CFUs) (30 °C) | Second Round (June) | ||||||||||||||||||
| Temperature (°C) | Humidity (%) | A. Section Flavi | A. section Fumigati | A. section Nigri | A. section Terrei | Black fungi | Penicillium sp. | Mucorales | Yeast sp. | Other | |||||||||||
| Khuzestan | Sand | 10 | 5 | 5 | |||||||||||||||||
| 37 | 62 | Water | 5 | 10 | 5 | 25 | 5 | ||||||||||||||
| Bushehr | Sand | 10 | 5 | 10 | 33 | 68 | 5 | 47 | 122 | ||||||||||||
| 35 | 60 | Water | 5 | 5 | |||||||||||||||||
| Hormozgan | Sand | 5 | 25 | ||||||||||||||||||
| 38 | 68 | Water | 5 | ||||||||||||||||||
| Sistan-Baluchestan | Sand | 5 | 5 | 15 | 60 | ||||||||||||||||
| 39 | 66 | Water | 5 | 45 | 5 | ||||||||||||||||
| PROVINCE | Sample type | Fungal load (CFUs) (30 °C) | Third Round (July) | ||||||||||||||||||
| Temperature (°C) | Humidity (%) | A. Section Flavi | A. section Fumigati | A. section Nigri | A. section Terrei | Black fungi | Penicillium sp. | Mucorales | Yeast sp. | Other | |||||||||||
| Khuzestan | Sand | 10 | 115 | ||||||||||||||||||
| 41 | 59 | Water | 25 | ||||||||||||||||||
| Bushehr | Sand | 10 | 55 | 10 | 70 | ||||||||||||||||
| 40 | 57 | Water | |||||||||||||||||||
| Hormozgan | Sand | 15 | 15 | 5 | |||||||||||||||||
| 42 | 71 | Water | 125 | ||||||||||||||||||
| Sistan-Baluchestan | Sand | 5 | 15 | ||||||||||||||||||
| 41 | 61 | Water | |||||||||||||||||||
| Province | Fungal Genus | Fungal Species | Number of Isolates |
|---|---|---|---|
| Khuzestan | Aspergillus | flavus/oryzae | 2 |
| terreus | 9 | ||
| fumigatus | 4 | ||
| niger | 2 | ||
| tubingensis | 1 | ||
| neoterreus | 1 | ||
| sydowii | 4 | ||
| Fusarium | sarocladium | 1 | |
| incarnatum-equiseti species complex | 3 | ||
| Yeasts | C. parapsilosis | 2 | |
| Rhodotorula | 2 | ||
| Unknown | 5 | ||
| Bushehr | Penicillium | citrinum | 1 |
| Aspergillus | flavus/oryzae | 13 | |
| terreus | 17 | ||
| fumigatus | 9 | ||
| niger | 7 | ||
| tubingensis | 3 | ||
| neoterreus | 2 | ||
| sydowii | 1 | ||
| Fusarium | solani | 2 | |
| sarocladium | 1 | ||
| incarnatum-equiseti species complex | 1 | ||
| Black fungi | Alternaria alternata | 3 | |
| Other | Lecanicillium psalliotae | 1 | |
| Apiospora marii | 1 | ||
| Scopolariopsis brevicaulis | 1 | ||
| Unknown | 11 | ||
| Hormozgan | Penicillium | citrinum | 3 |
| chrysogenom | 2 | ||
| Aspergillus | flavus/oryzae | 7 | |
| terreus | 7 | ||
| fumigatus | 7 | ||
| niger | 11 | ||
| tubingensis | 8 | ||
| unguis | 1 | ||
| sydowii | 2 | ||
| nomius | 3 | ||
| parasiticus | 2 | ||
| Fusarium | sarocladium | 1 | |
| Black fungi | Stachybotrys chartarum | 1 | |
| Epicoccum | 1 | ||
| Other | Trichoderma asperellum | 1 | |
| Simplicillium species | 2 | ||
| Unknown | 4 | ||
| Sistan and Baluchestan | Aspergillus | flavus/oryzae | 12 |
| terreus | 7 | ||
| fumigatus | 6 | ||
| niger | 2 | ||
| tubingensis | 3 | ||
| Fusarium | sarocladium | 1 | |
| incarnatum-equiseti species complex | 1 | ||
| Yeast | C. parapsilosis | 4 | |
| N. glabratus | 3 | ||
| Rhodotorula | 5 | ||
| Unknown | 5 | ||
| Total | Aspergillus | 153 | |
| Penicillium | 6 | ||
| Fusarium | 11 | ||
| Black fungi | 5 | ||
| Yeasts | 16 | ||
| Unknown | 31 |
| Isolates | Fungal Species | MIC (µg/mL) | ||
|---|---|---|---|---|
| *ITZ | £VRZ | ¥PSZ | ||
| Penicillium | P. citrinum (4) | ≥32 | ≥32 | 0.063 |
| ≥32 | ≥32 | 0.032 | ||
| ≥32 | 8 | 0.25 | ||
| 0.25 | 0.25 | 0.032 | ||
| Other Penicillium sp. (2) | ≥32 | 0.5 | 0.25 | |
| ≥32 | 2 | 0.25 | ||
| Fusarium | F. solani (1) | ≥32 | 1 | ≥32 |
| F. sarocladium (2) | 0.063 | 0.125 | 0.032 | |
| ≥32 | 0.25 | ≥32 | ||
| F. incarnatum-equiseti species complex (3) | 0.032 | 0.032 | 0.032 | |
| ≥32 | 0.25 | ≥32 | ||
| ≥32 | 1 | 1 | ||
| Scopulariopsis brevicaulis (1) | 0.032 | 0.125 | 0.032 | |
| Trichoderma asperellum (1) | ≥32 | 1 | 1 | |
| Simplicillium sp. (2) | ≥32 | 0.125 | 0.032 | |
| ≥32 | 0.25 | 0.032 | ||
| Chrysogenum (2) | 0.063 | 0.125 | 0.063 | |
| 0.032 | 0.25 | 0.032 | ||
| Black fungi | Epicoccum (1) | ≥32 | 0.25 | 0.032 |
| Other black fungi (3) | ≥32 | 0.125 | 0.032 | |
| 2 | 1 | 0.032 | ||
| 0.25 | 0.063 | 0.032 | ||
| Aspergillus Species (n) | Number of Isolates | Antifungal Agent | MIC (µg/mL) | MIC Range | MIC50 | MIC90 | GM | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Drug Susceptible | Drug Resistant | 32 | 16 | 8 | 4 | 2 | 1 | 0.5 | 0.25 | 0.125 | 0.062 | 0.031 | ||||||
| Aspergillus fumigatus (11) | 10 | 1 | *VRZ | 1 | 2 | 2 | 4 | 2 | 0.031-1 | 0.5 | 1 | 0.6299 | ||||||
| 9 | 2 | £ITZ | 2 | 2 | 1 | 1 | 1 | 4 | 0.031->32 | 0.031 | 32 | 0.6382 | ||||||
| 10 | 1 | ¥PSZ | 1 | 1 | 1 | 4 | 1 | 3 | 0.031->32 | 0.031 | 0.032 | 0.032 | ||||||
| Aspergillus flavus/oryzae (21) | 21 | - | VRZ | 1 | 8 | 11 | 1 | 0.062-1 | 0.25 | 1 | 0.3968 | |||||||
| 20 | 1 | ITZ | 1 | 1 | 5 | 9 | 5 | 0.031-32 | 0.031 | 0.25 | 0.032 | |||||||
| 20 | 1 | PSZ | 1 | 2 | 3 | 15 | 0.031-32 | 0.031 | 0.25 | 0.0505 | ||||||||
| Aspergillus terreus (27) | 27 | - | VRZ | 3 | 9 | 7 | 8 | 0.125-1 | 0.5 | 1 | 0.6040 | |||||||
| 22 | 5 | ITZ | 5 | 2 | 4 | 9 | 7 | 0.031-32 | 0.5 | 1 | 0.3314 | |||||||
| 25 | 2 | PSZ | 2 | 5 | 3 | 2 | 15 | 0.031-32 | 0.5 | 0.5 | 0.1473 | |||||||
| Aspergillus tubingensis (15) | 14 | 1 | VRZ | 1 | 6 | 1 | 4 | 3 | 0.25-1 | 0.5 | 0.5 | 0.5358 | ||||||
| - | 15 | ITZ | 15 | 32 | 32 | 32 | 32 | |||||||||||
| 14 | 1 | PSZ | 1 | 10 | 4 | 0.031-32 | 0.5 | 0.5 | 0.251 | |||||||||
| Aspergillus niger (5) | 5 | - | VRZ | 2 | 2 | 1 | 0.25-1 | 0.75 | 1 | 0.7071 | ||||||||
| 4 | 1 | ITZ | 1 | 1 | 1 | 1 | 1 | 0.031-32 | 0.063 | 25 | 0.4010 | |||||||
| 5 | - | PSZ | 1 | 4 | 0.031-1 | 0.032 | 1 | 0.075 | ||||||||||
| Aspergillus sydowii (3) | 3 | - | VRZ | 2 | 1 | - | - | - | - | |||||||||
| 2 | 1 | ITZ | 1 | 1 | 1 | - | - | - | - | |||||||||
| 3 | - | PSZ | 1 | 2 | - | - | - | - | ||||||||||
| Aspergillus unguis (1) | 1 | - | VRZ | 1 | - | - | - | - | ||||||||||
| 1 | - | ITZ | 1 | - | - | - | - | |||||||||||
| 1 | - | PSZ | 1 | - | - | - | - | |||||||||||
| Yeast Species | MIC (µg/mL) | |||
|---|---|---|---|---|
| *FLZ | **ITZ | £VRZ | ¥PSZ | |
| C. parapsilosis | 0.5 | 0.25 | 0.063 | 0.032 |
| 1 | 0.25 | 0.063 | 0.032 | |
| 0.25 | 0.032 | 0.125 | 0.032 | |
| 0.5 | 0.125 | 0.125 | 0.063 | |
| 1 | 0.125 | 0.25 | 0.032 | |
| 0.5 | 0.125 | 0.25 | 0.063 | |
| N. glabratus | 8 | 0.25 | 0.25 | 0.063 |
| 16 | 1 | 0.25 | 0.063 | |
| 32 | 1 | 0.25 | 0.032 | |
| Rhodotorula sp. | 64 | 2 | 1 | 2 |
| 16 | 2 | 1 | 1 | |
| ≥64 | 4 | 1 | 1 | |
| 64 | 4 | 2 | 1 | |
| 64 | 4 | 1 | 1 | |
| ≥64 | 4 | 2 | 2 | |
| ≥64 | 8 | 2 | 2 | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Saravani, A.; Brandão, J.; Ahmadi, B.; Rezaei-Matehkolaei, A.; Hedayati, M.T.; Abastabar, M.; Zarrinfar, H.; Nabili, M.; Faeli, L.; Javidnia, J.; et al. Insights into Persian Gulf Beach Sand Mycobiomes: Promises and Challenges in Fungal Diversity. J. Fungi 2025, 11, 554. https://doi.org/10.3390/jof11080554
Saravani A, Brandão J, Ahmadi B, Rezaei-Matehkolaei A, Hedayati MT, Abastabar M, Zarrinfar H, Nabili M, Faeli L, Javidnia J, et al. Insights into Persian Gulf Beach Sand Mycobiomes: Promises and Challenges in Fungal Diversity. Journal of Fungi. 2025; 11(8):554. https://doi.org/10.3390/jof11080554
Chicago/Turabian StyleSaravani, Abolfazl, João Brandão, Bahram Ahmadi, Ali Rezaei-Matehkolaei, Mohammad Taghi Hedayati, Mahdi Abastabar, Hossein Zarrinfar, Mojtaba Nabili, Leila Faeli, Javad Javidnia, and et al. 2025. "Insights into Persian Gulf Beach Sand Mycobiomes: Promises and Challenges in Fungal Diversity" Journal of Fungi 11, no. 8: 554. https://doi.org/10.3390/jof11080554
APA StyleSaravani, A., Brandão, J., Ahmadi, B., Rezaei-Matehkolaei, A., Hedayati, M. T., Abastabar, M., Zarrinfar, H., Nabili, M., Faeli, L., Javidnia, J., Parsay, S., Abtahian, Z., Moazeni, M., & Badali, H. (2025). Insights into Persian Gulf Beach Sand Mycobiomes: Promises and Challenges in Fungal Diversity. Journal of Fungi, 11(8), 554. https://doi.org/10.3390/jof11080554










