Morphological and Phylogenetic Characterization of Alternaria Section Undifilum Fungal Endophytes from Astragalus and Swainsona spp.
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Collections
2.2. Cultural and Morphological Characterization
2.3. DNA Extraction, PCR Amplification, and Sequencing
2.4. Phylogeny
3. Results
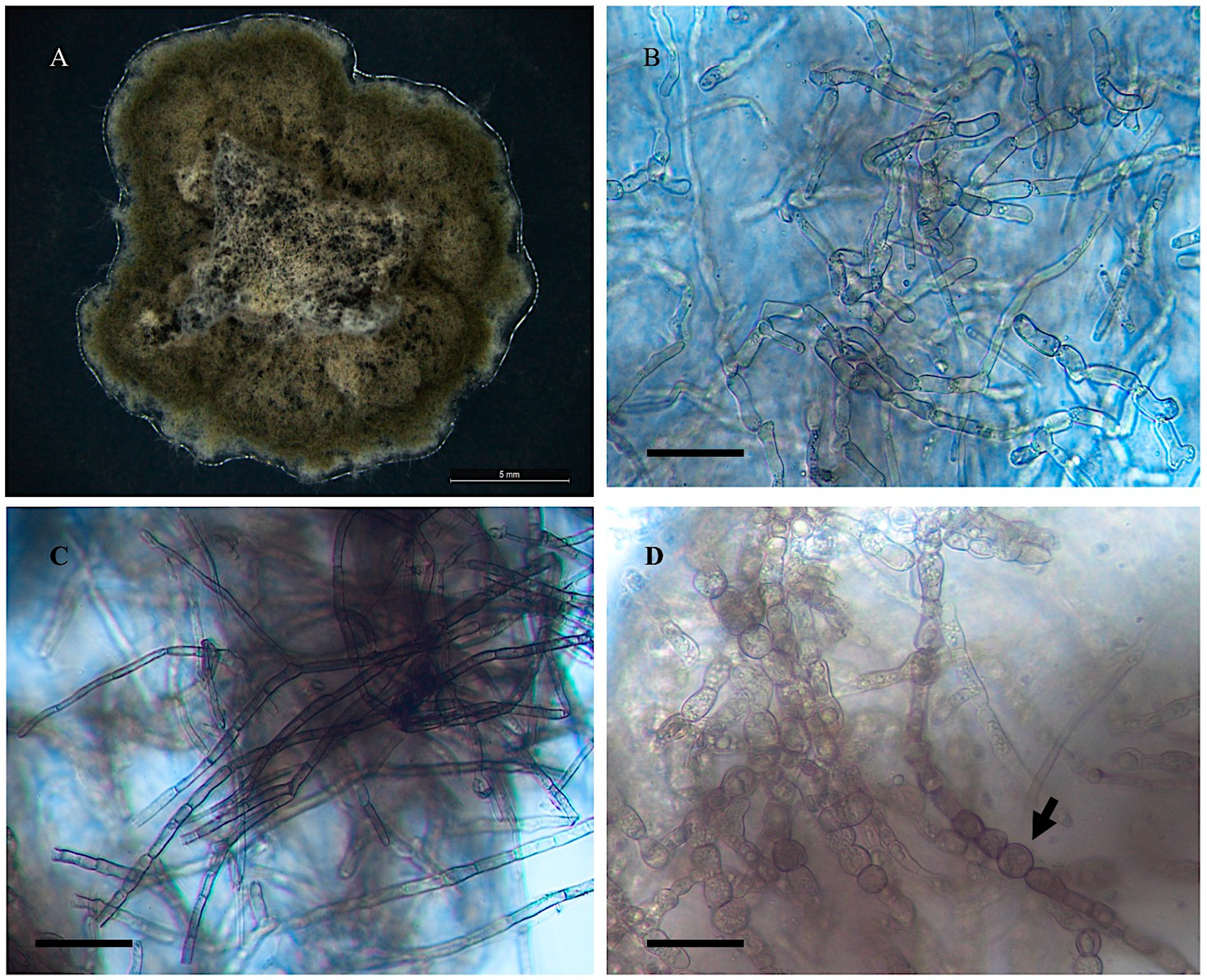
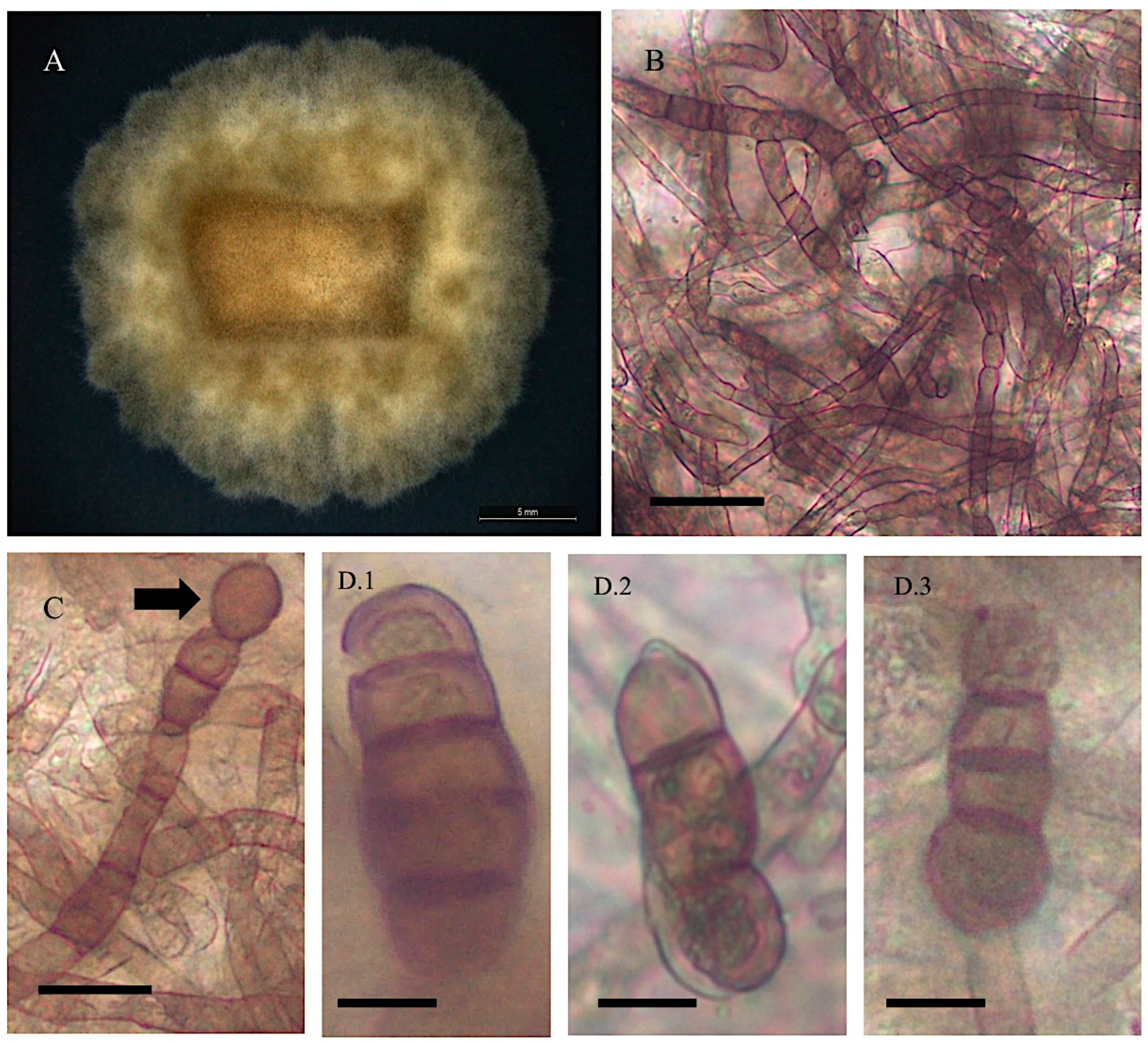
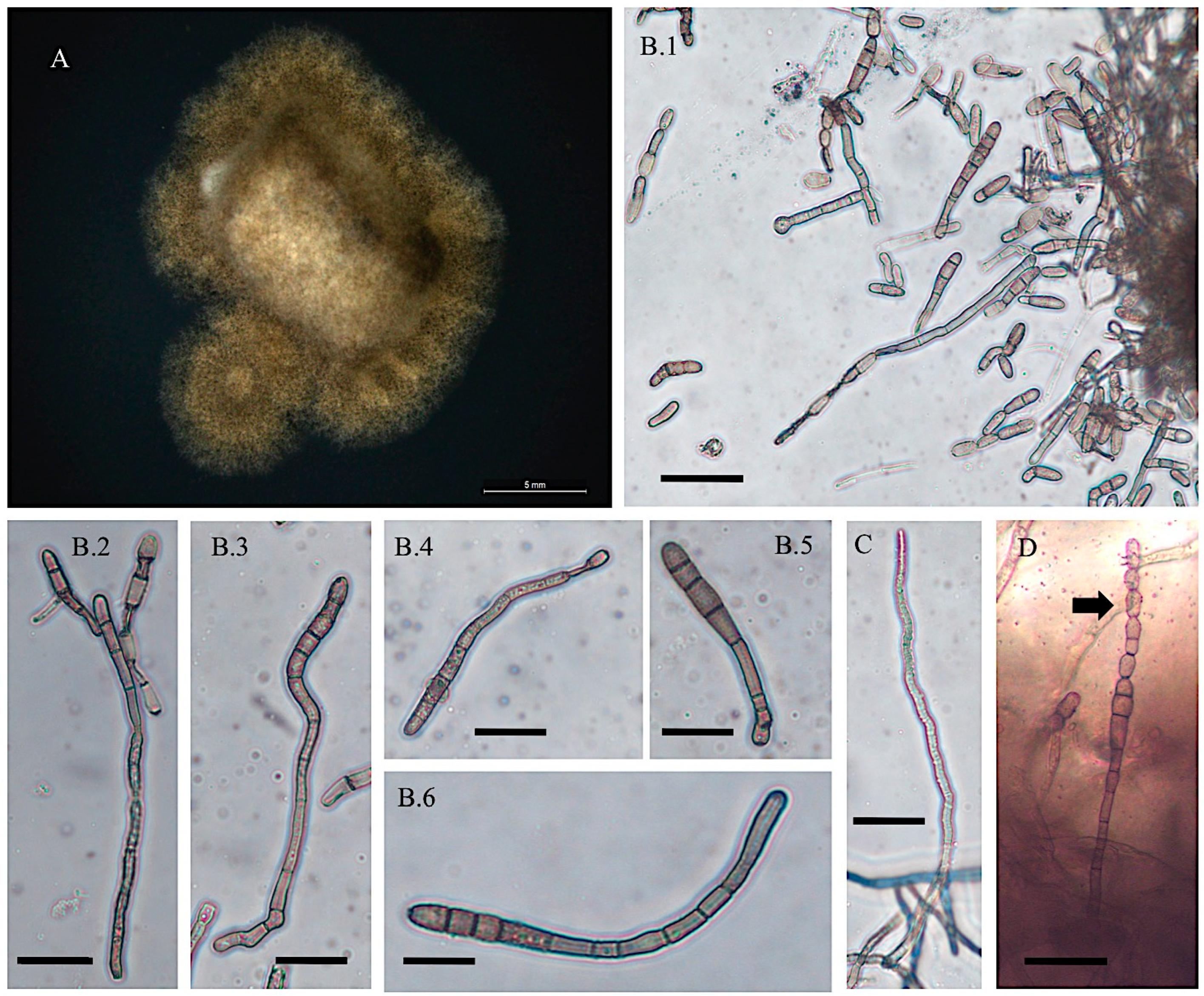
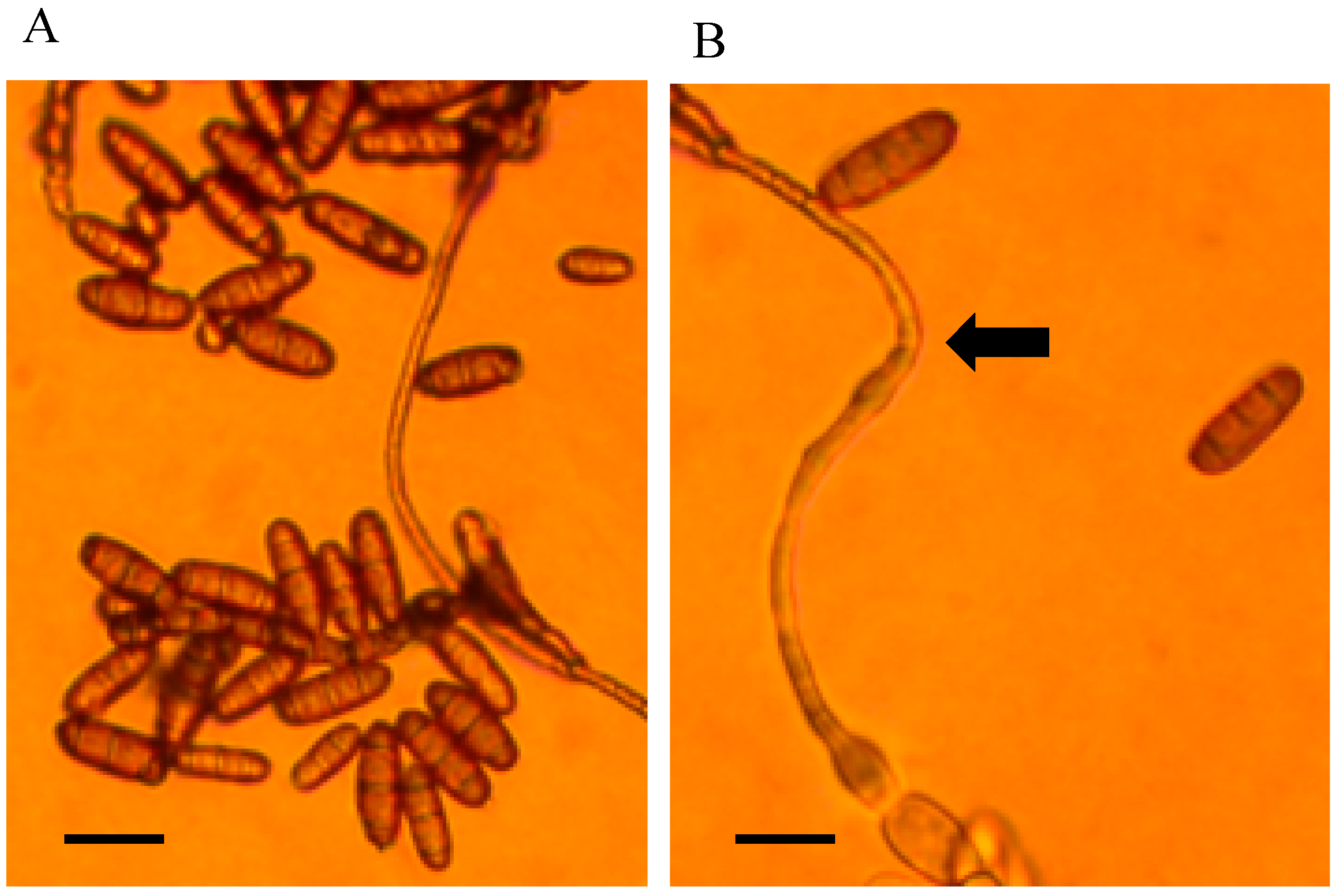
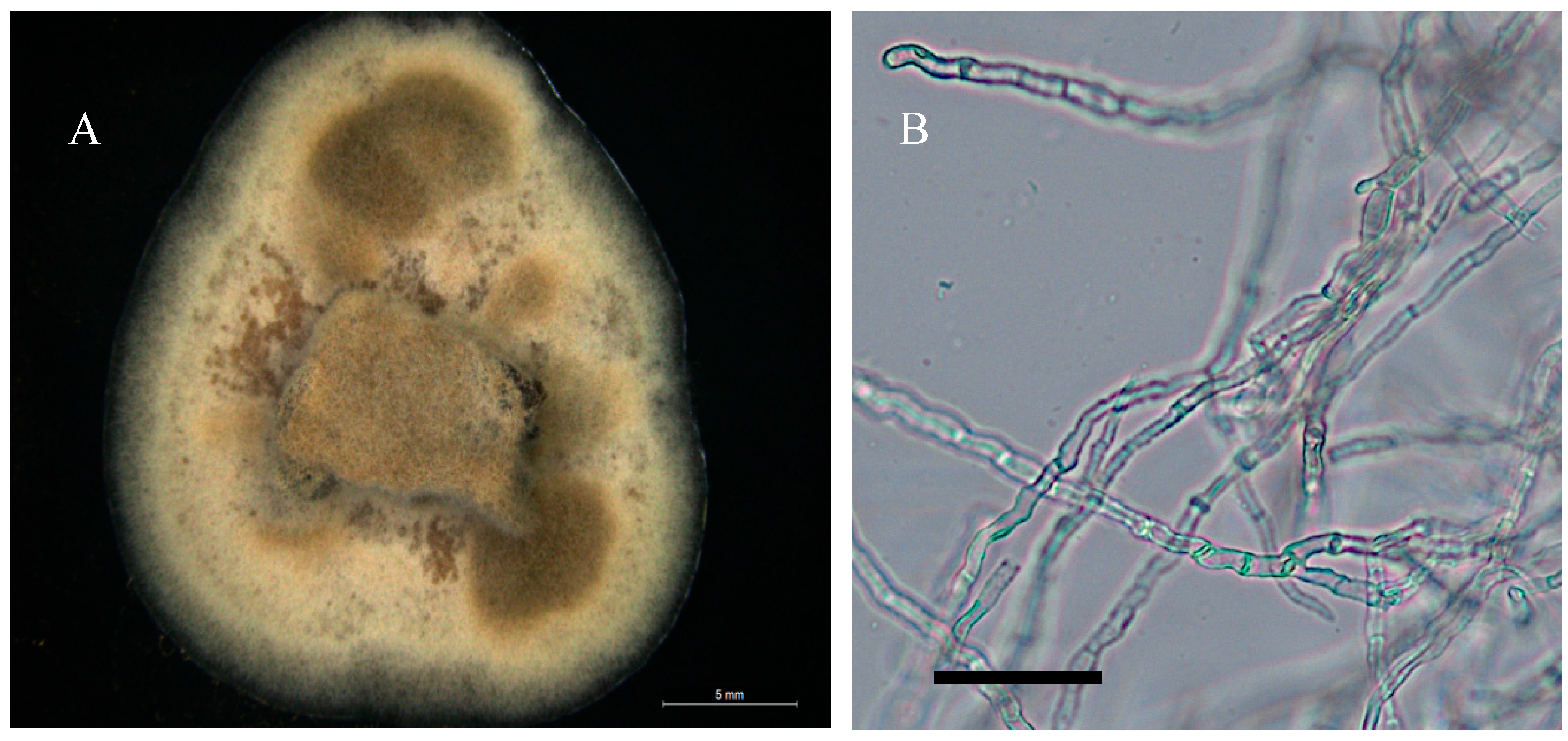
3.1. Cultural and Morphological Characteristics
- Alternaria wetherii
- Collection location
- Description
- 2.
- Alternaria pubentissima
- Collection location
- Description
- 3.
- Alternaria pubentissimoides
- Collection location
- Description
- 4.
- Alternaria swainsonii
- Collection location
- Description
- 5.
- Alternaria swainsonii
- Collection location
- Description
- 6.
- Alternaria swainsonii
- Collection location
- Description
- 7.
- Alternaria swainsonii
- Collection location
- Description
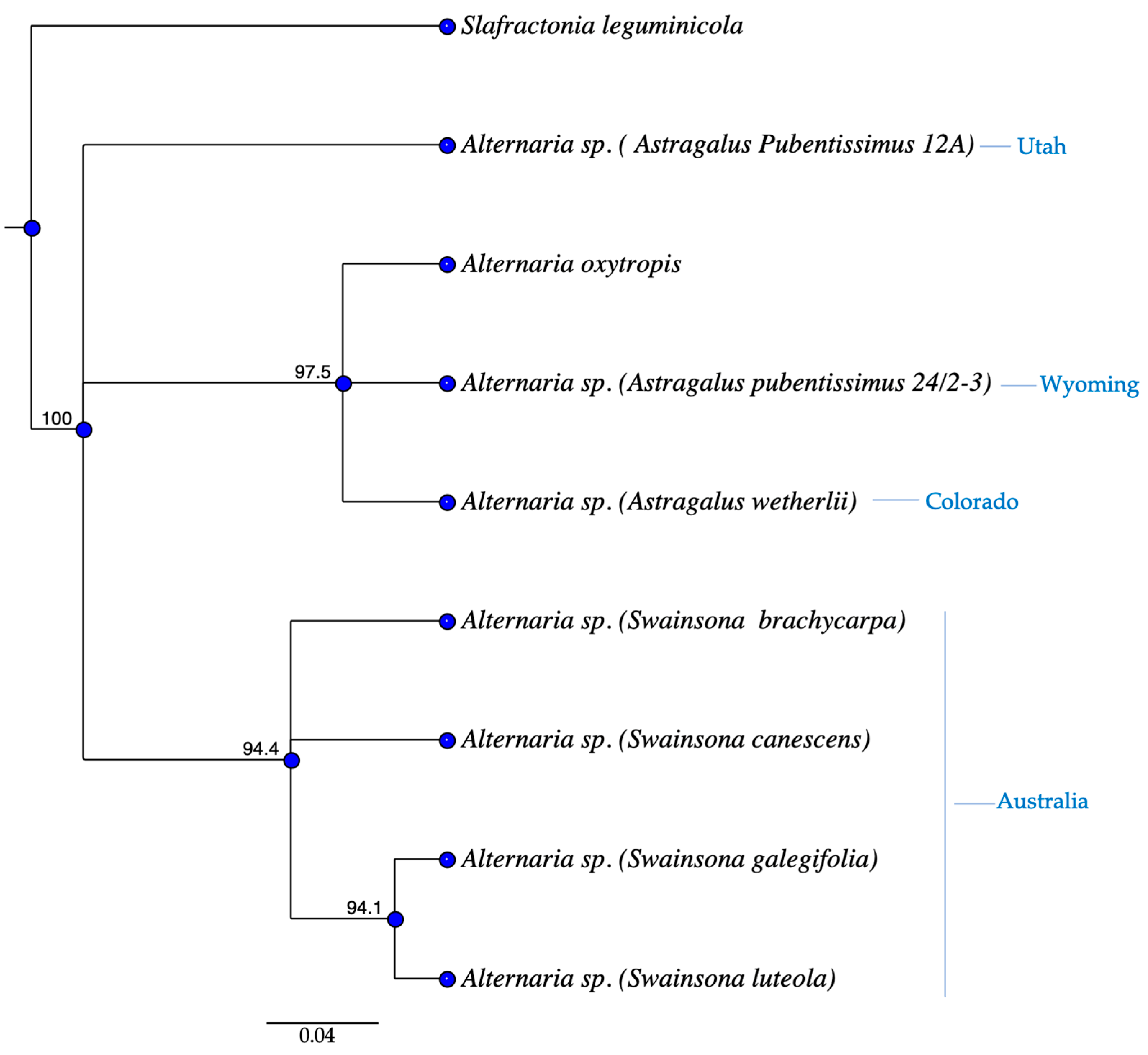
3.2. Phylogenetics
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Wojciechowski, M.F.; Lavin, M.; Sanderson, M.J. A phylogeny of legumes (Leguminosae) based on analysis of the plastid matK gene resolves many well-supported subclades within the family. Am. J. Bot. 2004, 91, 1846–1862. [Google Scholar] [CrossRef] [PubMed]
- Colegate, S.M.; Dorling, P.R.; Huxtable, C.R. A spectroscopic investigation of swainsonine: An α-mannosidase inhibitor isolated from Swainsona canescens. Aust. J. Chem. 1979, 32, 2257–2264. [Google Scholar] [CrossRef]
- Gardiner, M.R.; Linto, A.C.; Applin, T.E.H. Toxicity of Swainsona canescens for sheep in Western Australia. Aust. J. Agric. Res. 1969, 20, 87–97. [Google Scholar] [CrossRef]
- Marsh, C.D. The Loco-Weed Disease of the Plains; Bulletin 112; US Department of Agriculture, Bureau of Animal Industry: Washington, DC, USA, 1909. [Google Scholar]
- Kingsbury, J.M. Poisonous plants of the United States and Canada. Soil Sci. 1964, 98, 349. [Google Scholar] [CrossRef]
- Molyneux, R.J.; Gomez-Sosa, E. Presencia del alcaloide indolizidínico Swainsonine en Astragalus pehuenches (Leguminosae galegueae). Bol. Soc. Argent. Bot. 1991, 27, 59–64. [Google Scholar]
- Dorling, P.R.; Huxtable, C.R.; Colegate, S.M. Inhibition of lysosomal α-mannosidase by swainsonine, an indolizidine alkaloid isolated from Swainsona canescens. Biochem. J. 1980, 191, 649–651. [Google Scholar] [CrossRef] [PubMed]
- Tulsiani, D.R.; Harris, T.M.; Touster, O. Swainsonine inhibits the biosynthesis of complex glycoproteins by inhibition of Golgi mannosidase II. J. Biol. Chem. 1982, 257, 7936–7939. [Google Scholar] [CrossRef] [PubMed]
- Winchester, B.; al Daher, S.A.M.E.R.; Carpenter, N.C.; Cenci di Bello, I.; Choi, S.S.; Fairbanks, A.J.; Fleet, G.W.J. The structural basis of the inhibition of human α-mannosidases by azafuranose analogues of mannose. Biochem. J. 1993, 290, 743–749. [Google Scholar] [CrossRef] [PubMed]
- Molyneux, R.J.; James, L.F. Loco intoxication: Indolizidine alkaloids of spotted locoweed (Astragalus lentiginosus). Science 1982, 216, 190–191. [Google Scholar] [CrossRef] [PubMed]
- Molyneux, R.J.; James, L.F.; Panter, K.E.; Ralphs, M.H. Analysis and distribution of swainsonine and related polyhydroxyindolizidine alkaloids by thin layer chromatography. Phytochem. Anal. 1991, 2, 125–129. [Google Scholar] [CrossRef]
- James, L.F.; Panter, K.E. Locoweed poisoning in livestock. In Swainsonine and Related Glycosidase Inhibitors; James, L.F., Elbein, A.D., Molyneux, R.J., Warren, C.D., Eds.; Iowa State University Press: Ames, IA, USA, 1989; pp. 23–38. [Google Scholar]
- Braun, K.; Romero, J.; Liddell, C.; Creamer, R. Production of swainsonine by fungal endophytes of locoweed. Mycol. Res. 2003, 107, 980–988. [Google Scholar] [CrossRef] [PubMed]
- Cook, D.; Beaulieu, W.T.; Mott, I.W.; Riet-Correa, F.; Gardner, D.R.; Grum, D.; Pfister, J.A.; Clay, K.; Marcolongo-Pereira, C. Production of the alkaloid swainsonine by a fungal endosymbiont of the Ascomycete order Chaetothyriales in the host Ipomoea carnea. J. Agric. Food Chem. 2013, 61, 3797–3803. [Google Scholar] [CrossRef] [PubMed]
- Noor, A.I.; Neyaz, M.; Cook, D.; Creamer, R. Molecular characterization of a fungal ketide synthase gene among swainsonine-producing Alternaria species in the USA. Curr. Microbiol. 2020, 77, 2554–2563. [Google Scholar] [CrossRef] [PubMed]
- Neyaz, M.; Gardner, D.R.; Creamer, R.; Cook, D. Localization of the swainsonine-producing Chaetothyriales symbiont in the seed and shoot apical meristem in its host Ipomoea carnea. Microorganisms 2022, 10, 545. [Google Scholar] [CrossRef] [PubMed]
- Grum, D.S.; Cook, D.; Baucom, D.; Mott, I.W.; Gardner, D.R.; Creamer, R.; Allen, J.G. Production of the alkaloid swainsonine by a fungal endophyte in the host Swainsona canescens. J. Nat. Prod. 2013, 76, 1984–1988. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Nagao, H.; Li, Y.; Wang, H.; Kakishima, M. Embellisia oxytropis, a new species isolated from Oxytropis kansuensis in China. Mycotaxon 2006, 95, 255–260. [Google Scholar]
- Pryor, B.M.; Creamer, R.; Shoemaker, R.A.; McLain-Romero, J.; Hambleton, S. Undifilum, a new genus for endophytic Embellisia oxytropis and parasitic Helminthosporium bornmuelleri on legumes. Botany 2009, 87, 178–194. [Google Scholar] [CrossRef]
- Woudenberg, J.H.C.; Groenewald, J.Z.; Binder, M.; Crous, P.W. Alternaria redefined. Stud. Mycol. 2013, 75, 171–212. [Google Scholar] [CrossRef] [PubMed]
- Schneider, M.; Ungemach, F.; Broquist, H.; Harris, T. (1S,2R,8R,8aR)-1,2,8-trihydroxyoctahydroindolizine (swainsonine), an α-mannosidase inhibitor from Rhizoctonia leguminicola. Tetrahedron 1983, 39, 29–32. [Google Scholar] [CrossRef]
- Alhawatema, M.S.; Sanogo, S.; Baucom, D.L.; Creamer, R. A search for the phylogenetic relationship of the ascomycete Rhizoctonia leguminicola using genetic analysis. Mycopathologia 2015, 179, 381–389. [Google Scholar] [CrossRef] [PubMed]
- Patrick, M.; Adlard, M.W.; Keshavarz, T. Production of an indolizidine alkaloid, swainsonine by the filamentous fungus, Metarhizium anisopliae. Biotechnol. Lett. 1993, 15, 997–1000. [Google Scholar] [CrossRef]
- Cook, D.; Donzelli, B.G.; Creamer, R.; Baucom, D.L.; Gardner, D.R.; Pan, J.; Moore, N.; Krasnoff, S.T.; Jaromczyk, J.W.; Schardl, C.L. Swainsonine biosynthesis genes in diverse symbiotic and pathogenic fungi. G3 Genes Genomes Genet. 2017, 7, 1791–1797. [Google Scholar] [CrossRef] [PubMed]
- Baucom, D.L.; Romero, M.; Belfon, R.; Creamer, R. Two new species of Undifilum, fungal endophytes of Astragalus (locoweeds) in the United States. Botany 2012, 90, 866–875. [Google Scholar] [CrossRef] [PubMed]
- Ralphs, M.H.; Creamer, R.; Baucom, D.; Gardner, D.R.; Welsh, S.L.; Graham, J.D.; Hart, C.; Cook, D.; Stegelmeier, B.L. Relationship between the endophyte Embellisia spp., and the toxic alkaloid swainsonine in major locoweed species (Astragalus and Oxytropis). J. Chem. Ecol. 2008, 34, 32–38. [Google Scholar] [CrossRef] [PubMed]
- Oldrup, E.; McLain-Romero, J.; Padilla, A.; Moya, A.; Gardner, D.; Creamer, R. Localization of endophytic Undifilum fungi in locoweed seed and influence of environmental parameters on a locoweed in vitro culture system. Botany 2010, 88, 512–521. [Google Scholar] [CrossRef]
- Cook, D.; Grum, D.S.; Gardner, D.R.; Welch, K.D.; Pfister, J.A. Influence of endophyte genotype on swainsonine concentrations in Oxytropis sericea. Toxicon 2013, 61, 105–111. [Google Scholar] [CrossRef] [PubMed]
- Lawrence, D.P.; Gannibal, P.B.; Peever, T.L.; Pryor, B.M. The sections of Alternaria: Formalizing species-group concepts. Mycologia 2013, 105, 530–546. [Google Scholar] [CrossRef] [PubMed]
- Creamer, R.; Hille, D.B.; Neyaz, M.; Nusayr, T.; Schardl, C.L.; Cook, D. Genetic relationships in the toxin-producing fungal endophyte, Alternaria oxytropis using polyketide synthase and non-ribosomal peptide synthase genes. J. Fungi 2021, 7, 538. [Google Scholar] [CrossRef] [PubMed]
- Cook, D.; Gardner, D.R.; Lee, S.T.; Pfister, J.A.; Stonecipher, C.A.; Welsh, S.L. A swainsonine survey of North American Astragalus and Oxytropis taxa implicated as locoweeds. Toxicon 2016, 118, 104–111. [Google Scholar] [CrossRef] [PubMed]
- Cook, D.; Gardner, D.R.; Pfister, J.A.; Lee, S.T.; Welch, K.D.; Welsh, S.L. A screen for swainsonine in select North American Astragalus species. Chem. Biodivers. 2017, 14, e1600364. [Google Scholar] [CrossRef] [PubMed]
- Cook, D.; Gardner, D.R.; Welch, K.D.; Allen, J.G. A survey of swainsonine content in Swainsona species. Rangel. J. 2017, 39, 213–218. [Google Scholar] [CrossRef]
- Cook, D.; Gardner, D.R.; Pfister, J.A. Swainsonine-containing plants and their relationship to endophytic fungi. J. Agric. Food Chem. 2014, 62, 7326–7334. [Google Scholar] [CrossRef] [PubMed]
- Ray, J.D.; Burgess, T.; Malajczuk, N.; Hardy, G.E.; St, J. First report of Alternaria blight of Paulownia spp. Aust. Plant Pathol. 2005, 34, 107–109. [Google Scholar] [CrossRef]
- White, T.J.; Bruns, T.; Lee, S.J.W.T.; Taylor, J. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In PCR Protocols: A Guide to Methods and Applications; Academic Press: New York, NY, USA, 1990; Volume 18, pp. 315–322. [Google Scholar]
- Woudenberg, J.H.C.; Seidl, M.F.; Groenewald, J.Z.; deVries, M.; Stielow, J.B.; Thomma, B.P.H.J.; Crous, P.W. Alternaria section Alternaria: Species, formae speciales, or pathotypes? Stud. Mycol. 2015, 82, 1–21. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J.; Shan, T.; Mou, Y.; Zhou, L. Plant-derived bioactive compounds produced by endophytic fungi. Mini-Rev. Med. Chem. 2011, 11, 159–168. [Google Scholar] [CrossRef] [PubMed]




Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Neyaz, M.; Adebisi, O.; Cook, D.; Creamer, R. Morphological and Phylogenetic Characterization of Alternaria Section Undifilum Fungal Endophytes from Astragalus and Swainsona spp. J. Fungi 2025, 11, 541. https://doi.org/10.3390/jof11070541
Neyaz M, Adebisi O, Cook D, Creamer R. Morphological and Phylogenetic Characterization of Alternaria Section Undifilum Fungal Endophytes from Astragalus and Swainsona spp. Journal of Fungi. 2025; 11(7):541. https://doi.org/10.3390/jof11070541
Chicago/Turabian StyleNeyaz, Marwa, Olabisi Adebisi, Daniel Cook, and Rebecca Creamer. 2025. "Morphological and Phylogenetic Characterization of Alternaria Section Undifilum Fungal Endophytes from Astragalus and Swainsona spp." Journal of Fungi 11, no. 7: 541. https://doi.org/10.3390/jof11070541
APA StyleNeyaz, M., Adebisi, O., Cook, D., & Creamer, R. (2025). Morphological and Phylogenetic Characterization of Alternaria Section Undifilum Fungal Endophytes from Astragalus and Swainsona spp. Journal of Fungi, 11(7), 541. https://doi.org/10.3390/jof11070541






