The Transcription Factor SsSR Mediates Ergosterol Biosynthesis and Virulence in Sclerotinia sclerotiorum
Abstract
1. Introduction
2. Experimental Procedures
2.1. Strain Culture and Plant Culture Conditions
2.2. Gene Deletion and Genetic Complementation
2.3. Pathogenicity Assays
2.4. Analysis of Growth, Acid Production, and Mycelial Tip Morphology
2.5. Stress Response Assays
2.6. RNA Extraction and RT-qPCR
2.7. ChIP(Chromatin Immunoprecipitation)-qPCR
2.8. Yeast One-Hybrid (Y1H) Assays
2.9. Ergosterol Extraction and High-Performance Liquid Chromatography Analysis
2.10. Measurement of Membrane Permeability
2.11. Statistical Analysis
3. Results
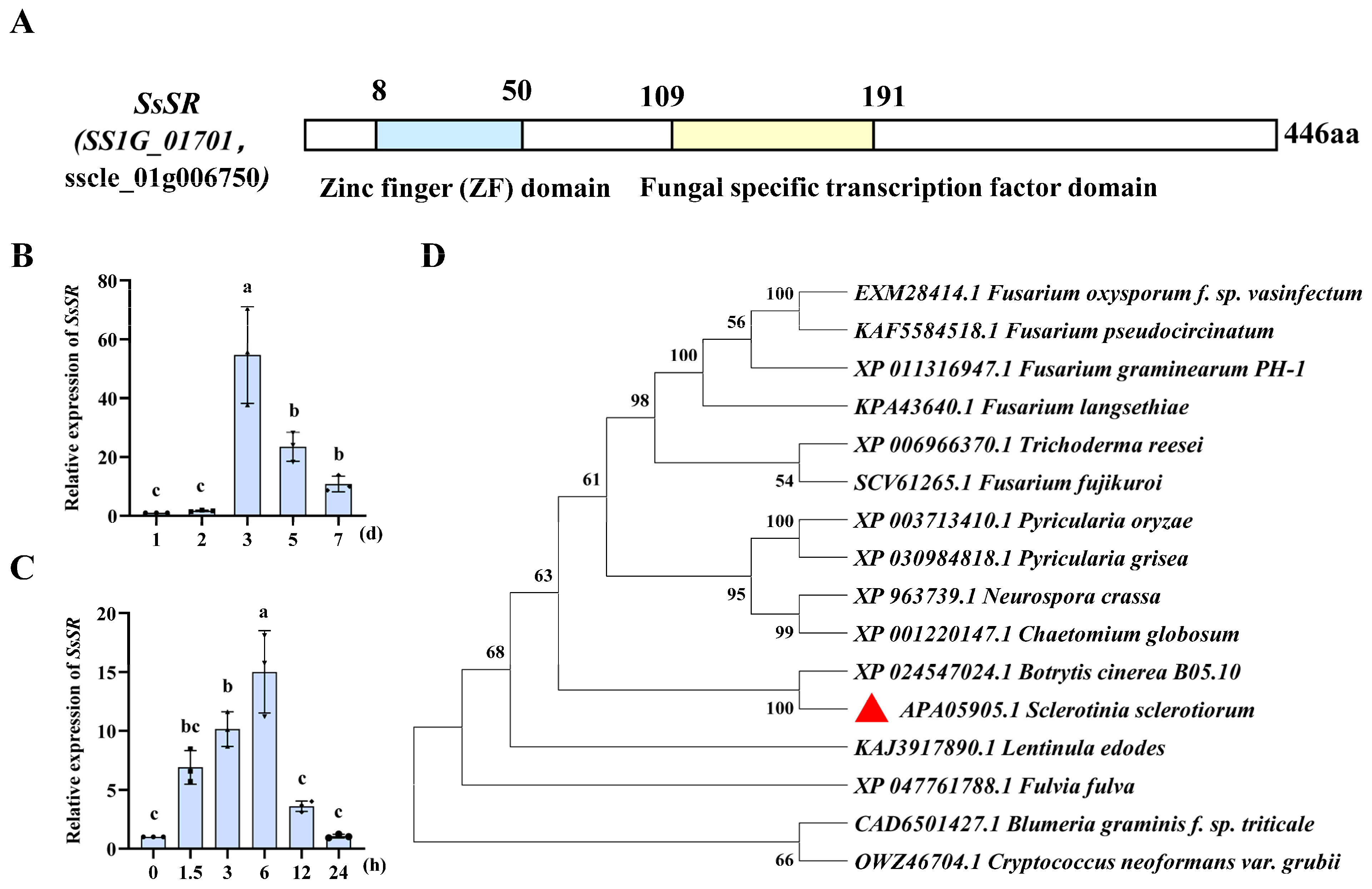
3.1. The Expression of the Transcription Factor SsSR Was Significantly Upregulated During Infection
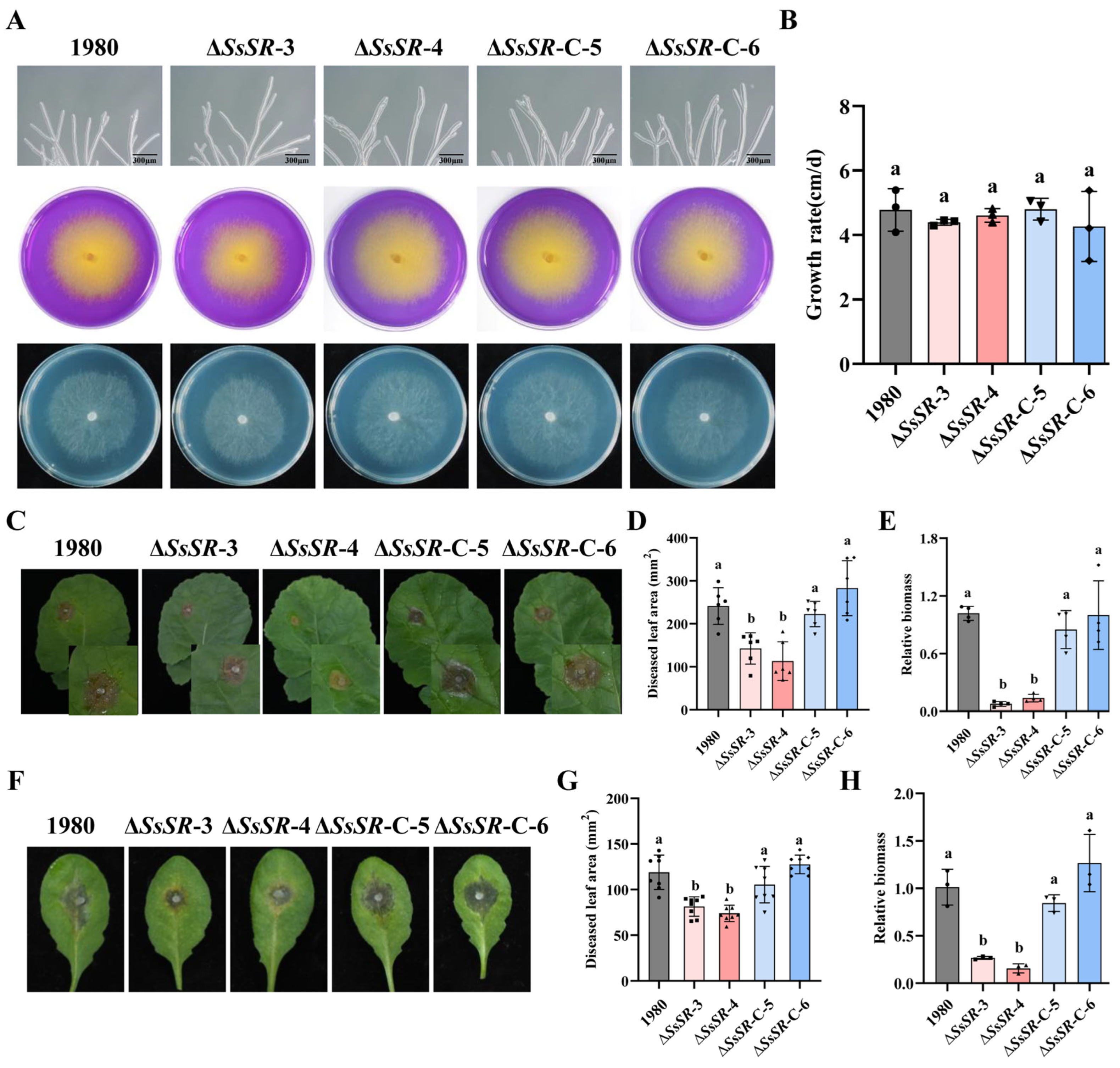
3.2. SsSR Is Essential for the Full Virulence of S. sclerotiorum
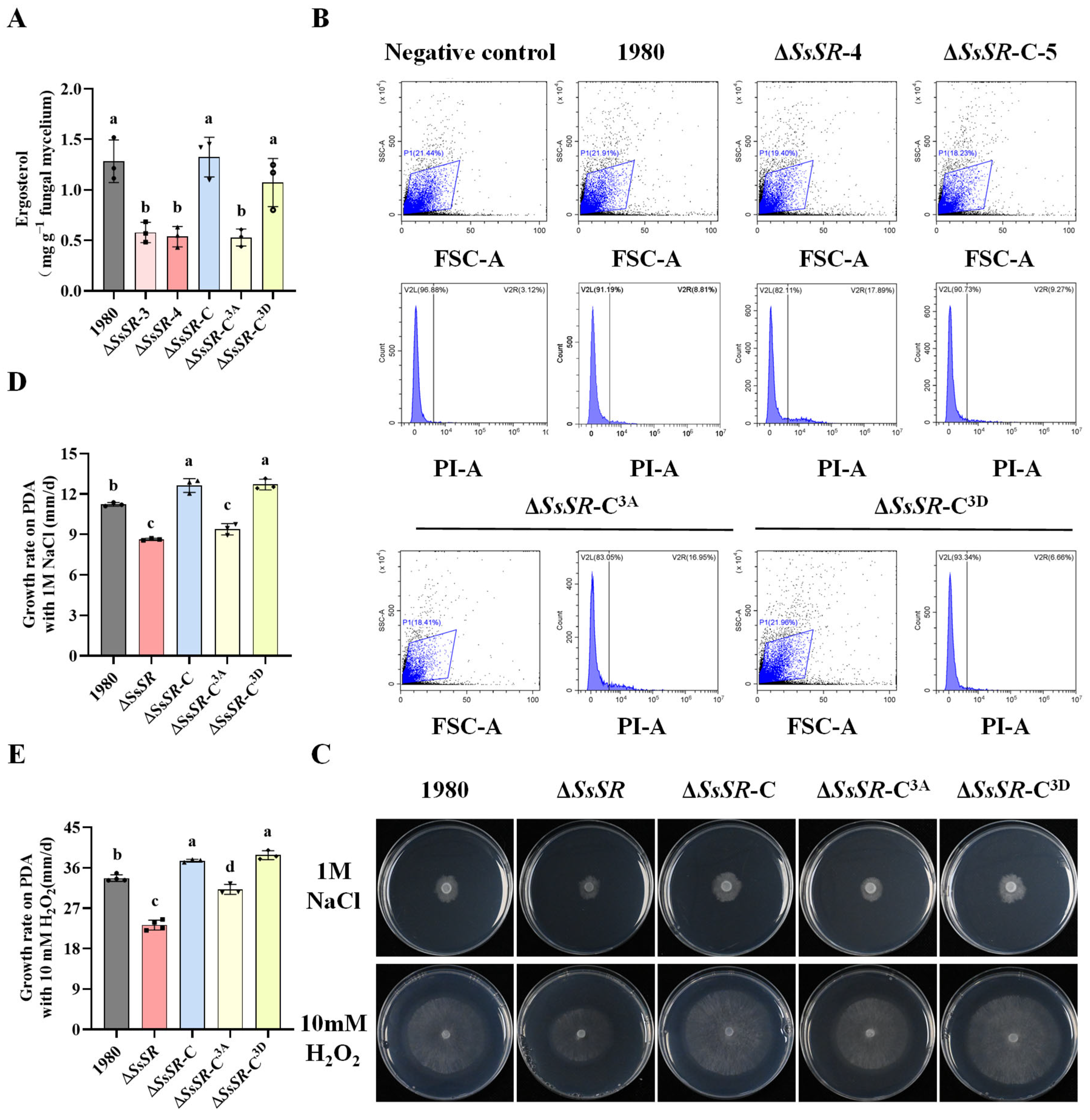
3.3. SsSR Promotes Infection by Regulating Ergosterol Synthesis
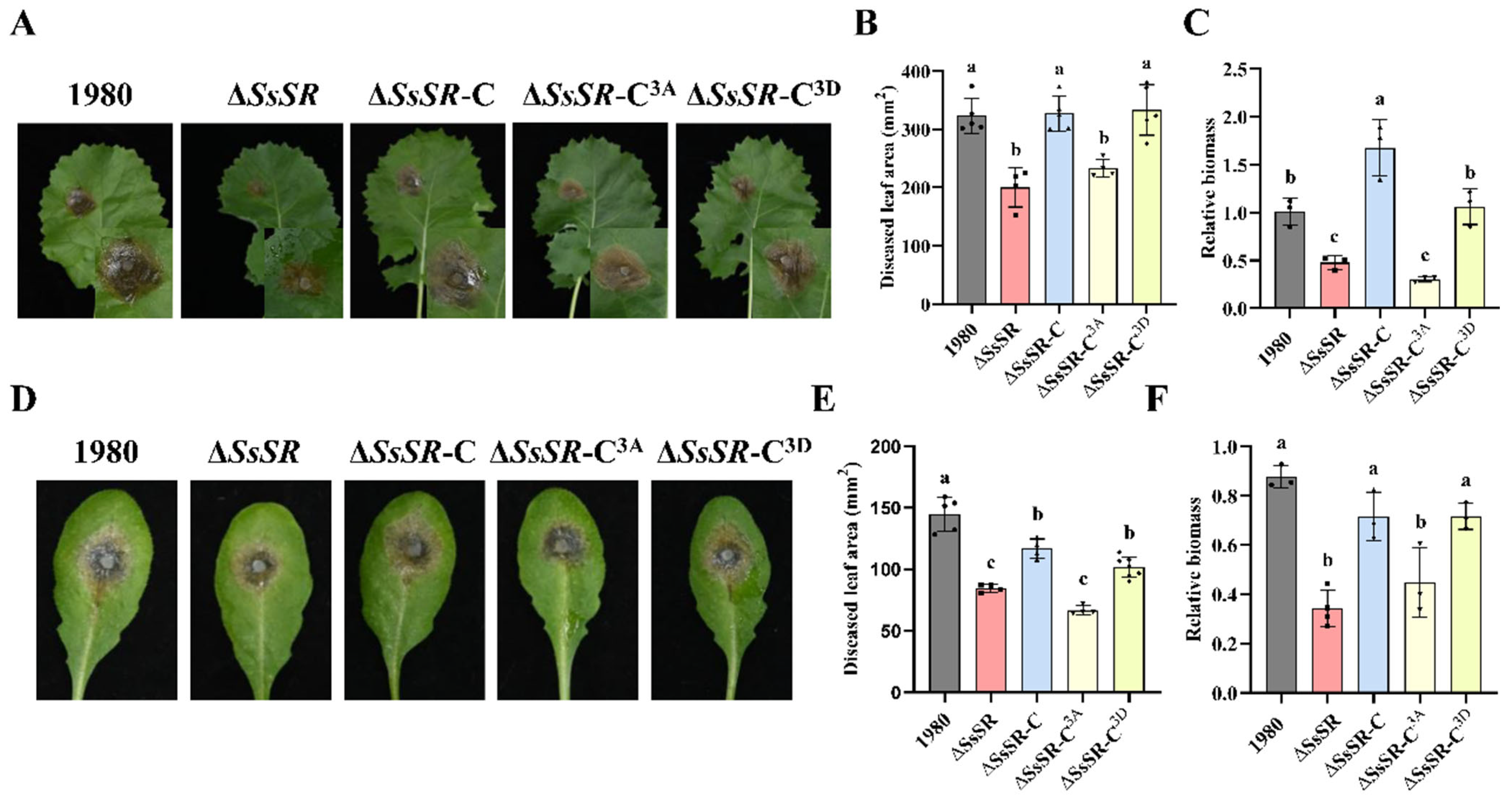
3.4. The Phosphorylation Sites of SsSR Were Indispensable for Virulence and Ergosterol Synthesis in Sclerotinia sclerotiorum
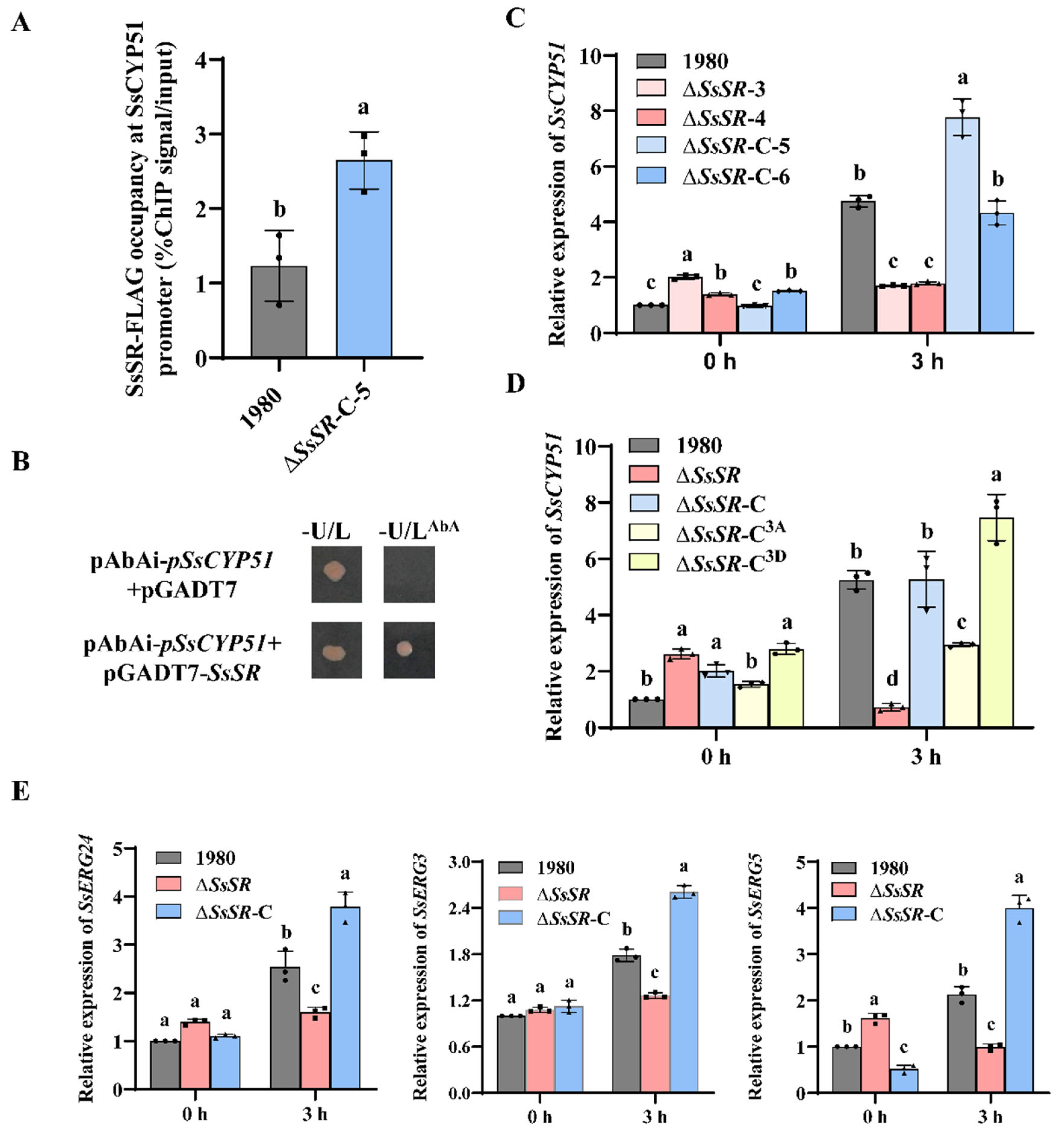
3.5. SsSR Regulates the Transcription of Genes Involved in Sterol Synthesis During Infection
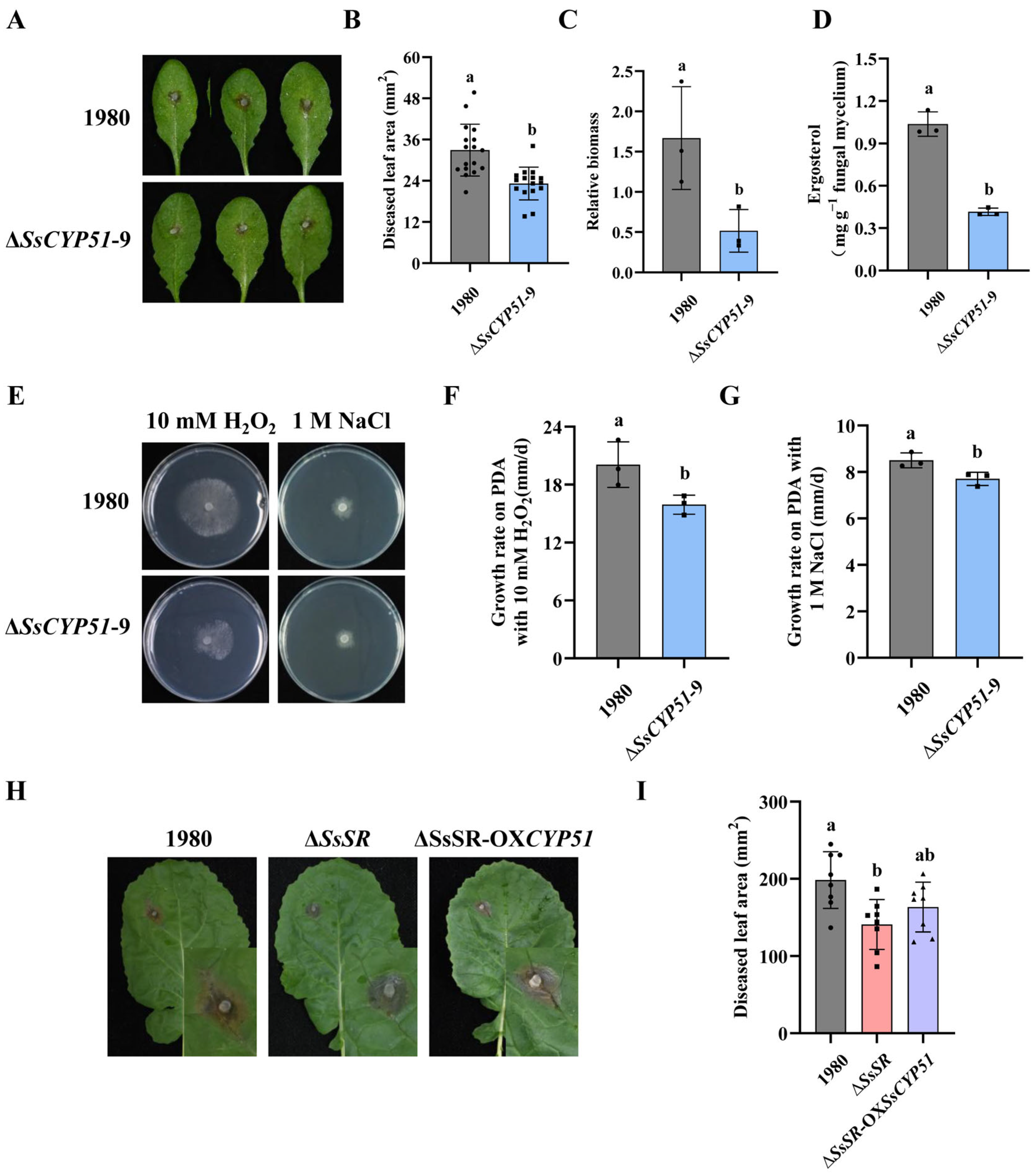
3.6. Regulation of SsCYP51 by SsSR Is Essential for Virulence and Ergosterol Biosynthesis in Sclerotinia sclerotiorum
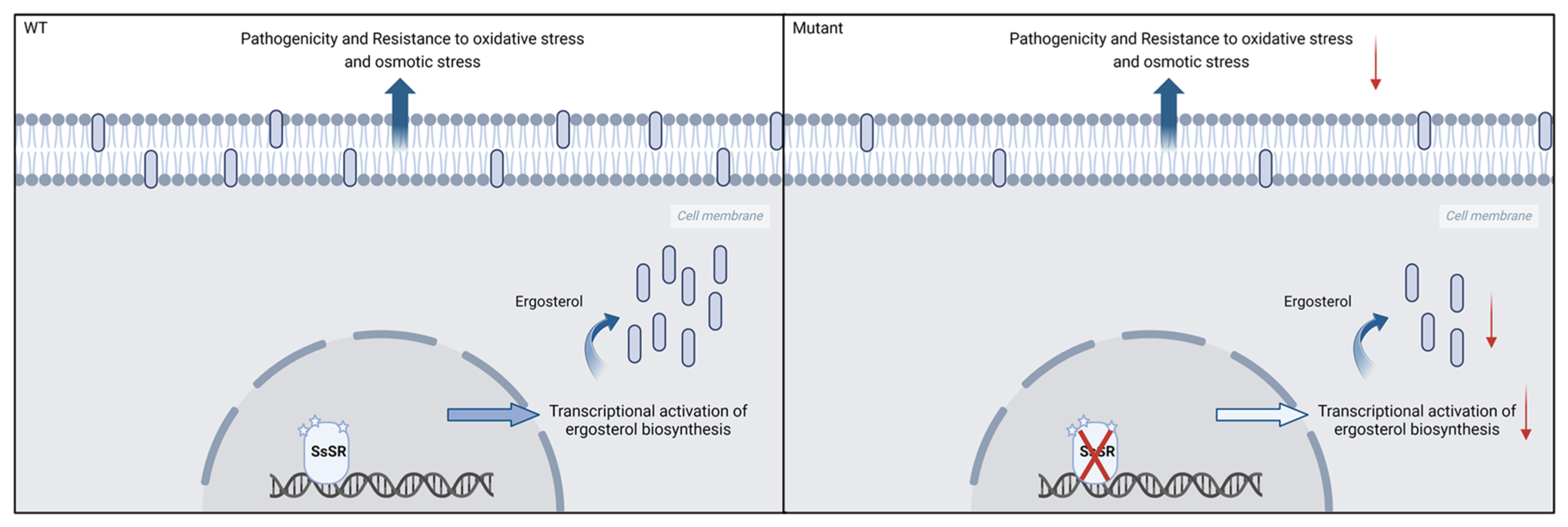
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Bolton, M.D.; Thomma, B.P.H.J.; Nelson, B.D. Sclerotinia sclerotiorum (Lib.) de Bary: Biology and molecular traits of a cosmopolitan pathogen. Mol. Plant Pathol. 2006, 7, 1–16. [Google Scholar] [CrossRef]
- Shang, Q.; Jiang, D.; Xie, J.; Cheng, J.; Xiao, X. The schizotrophic lifestyle of Sclerotinia sclerotiorum. Mol. Plant Pathol. 2024, 25, e13423. [Google Scholar] [CrossRef]
- Jørgensen, L.N.; Heick, T.M. Azole use in agriculture, horticulture, and wood preservation—Is it indispensable? Front. Cell. Infect. Microbiol. 2021, 11, 730297. [Google Scholar] [CrossRef]
- Riou, C.; Freyssinet, G.; Fevre, M. Production of Cell Wall-Degrading Enzymes by the Phytopathogenic Fungus Sclerotinia sclerotiorum. Appl. Environ. Microbiol. 1991, 57, 1478–1484. [Google Scholar] [CrossRef] [PubMed]
- Cessna, S.G.; Sears, V.E.; Dickman, M.B.; Low, P.S. Oxalic acid, a pathogenicity factor for Sclerotinia sclerotiorum, suppresses the oxidative burst of the host plant. Plant Cell 2000, 12, 2191–2200. [Google Scholar] [CrossRef] [PubMed]
- Xu, L.; Xiang, M.; White, D.; Chen, W. pH dependency of sclerotial development and pathogenicity revealed by using genetically defined oxalate-minus mutants of Sclerotinia sclerotiorum. Environ. Microbiol. 2015, 17, 2896–2909. [Google Scholar] [CrossRef] [PubMed]
- Kabbage, M.; Williams, B.; Dickman, M.B. Cell death control: The interplay of apoptosis and autophagy in the pathogenicity of Sclerotinia sclerotiorum. PLoS Pathog. 2013, 9, e1003287. [Google Scholar] [CrossRef]
- Liang, X.; Rollins, J.A. Mechanisms of broad host range necrotrophic pathogenesis in Sclerotinia sclerotiorum. Phytopathology 2018, 108, 1128–1140. [Google Scholar] [CrossRef]
- Williams, B.; Kabbage, M.; Kim, H.-J.; Britt, R.; Dickman, M.B. Tipping the balance: Sclerotinia sclerotiorum secreted oxalic acid suppresses host defenses by manipulating the host redox environment. PLoS Pathog. 2011, 7, e1002107. [Google Scholar] [CrossRef]
- Yang, G.; Tang, L.; Gong, Y.; Xie, J.; Fu, Y.; Jiang, D.; Li, G.; Collinge, D.B.; Chen, W.; Cheng, J. A cerato-platanin protein SsCP1 targets plant PR1 and contributes to virulence of Sclerotinia sclerotiorum. New Phytol. 2018, 217, 739–755. [Google Scholar] [CrossRef]
- Ma, M.; Tang, L.; Sun, R.; Lyu, X.; Xie, J.; Fu, Y.; Li, B.; Chen, T.; Lin, Y.; Yu, X.; et al. An effector SsCVNH promotes the virulence of Sclerotinia sclerotiorum through targeting class III peroxidase AtPRX71. Mol. Plant Pathol. 2024, 25, e13464. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Zhao, H.; Yuan, M.; Li, P.; Xie, J.; Fu, Y.; Li, B.; Yu, X.; Chen, T.; Lin, Y.; et al. An effector essential for virulence of necrotrophic fungi targets plant HIRs to inhibit host immunity. Nat. Commun. 2024, 15, 9391. [Google Scholar] [CrossRef] [PubMed]
- Wei, W.; Xu, L.; Peng, H.; Zhu, W.; Tanaka, K.; Cheng, J.; Sanguinet, K.A.; Vandemark, G.; Chen, W. A fungal extracellular effector inactivates plant polygalacturonase-inhibiting protein. Nat. Commun. 2022, 13, 2213. [Google Scholar] [CrossRef]
- John, E.; Singh, K.B.; Oliver, R.P.; Tan, K.-C. Transcription factor control of virulence in phytopathogenic fungi. Mol. Plant Pathol. 2021, 22, 858–881. [Google Scholar] [CrossRef] [PubMed]
- Gu, Q.; Wang, Y.; Zhao, X.; Yuan, B.; Zhang, M.; Tan, Z.; Zhang, X.; Chen, Y.; Wu, H.; Luo, Y.; et al. Inhibition of histone acetyltransferase GCN5 by a transcription factor FgPacC controls fungal adaption to host-derived iron stress. Nucleic Acids Res. 2022, 50, 6190–6210. [Google Scholar] [CrossRef]
- López-Berges, M.S.; Capilla, J.; Turrà, D.; Schafferer, L.; Matthijs, S.; Jöchl, C.; Cornelis, P.; Guarro, J.; Haas, H.; Di Pietro, A. HapX-Mediated Iron Homeostasis Is Essential for Rhizosphere Competence and Virulence of the Soilborne Pathogen Fusarium oxysporum. Plant Cell 2012, 24, 3805–3822. [Google Scholar] [CrossRef]
- Sun, K.; Li, Y.; Gai, Y.; Wang, J.; Jian, Y.; Liu, X.; Wu, L.; Shim, W.-B.; Lee, Y.-W.; Ma, Z.; et al. HapX-mediated H2B deub1 and SreA-mediated H2A.Z deposition coordinate in fungal iron resistance. Nucleic Acids Res. 2023, 51, 10238–10260. [Google Scholar] [CrossRef]
- Wang, Z.; Ma, T.; Huang, Y.; Wang, J.; Chen, Y.; Kistler, H.C.; Ma, Z.; Yin, Y. A fungal ABC transporter FgAtm1 regulates iron homeostasis via the transcription factor cascade FgAreA-HapX. PLoS Pathog. 2019, 15, e1007791. [Google Scholar] [CrossRef]
- Xu, C.; Wang, J.; Zhang, Y.; Luo, Y.; Zhao, Y.; Chen, Y.; Ma, Z. The transcription factor FgStuA regulates virulence and mycotoxin biosynthesis via recruiting the SAGA complex in Fusarium graminearum. New Phytol. 2023, 240, 2455–2467. [Google Scholar] [CrossRef]
- Wang, Q.; Huang, Z.; Khan, I.A.; Li, Y.; Wang, J.; Wang, J.; Liu, X.-H.; Lin, F.; Lu, J. Key transcription factors required for outburst of rice blast disease in Magnaporthe oryzae. Phytopathol. Res. 2024, 6, 5. [Google Scholar] [CrossRef]
- Minh, D.N.; Tsukahara, Y.; Thach, D.A.; Ikeda, K.; Nakayashiki, H. MoSET1-dependent transcription factors regulate different stages of infection-related morphogenesis in Pyricularia oryzae. J. Gen. Plant Pathol. 2023, 89, 77–83. [Google Scholar] [CrossRef]
- Huang, P.; Wang, J.; Li, Y.; Wang, Q.; Huang, Z.; Qian, H.; Liu, X.-H.; Lin, F.-C.; Lu, J. Transcription factors Vrf1 and Hox7 coordinately regulate appressorium maturation in the rice blast fungus Magnaporthe oryzae. Microbiol. Res. 2022, 263, 127141. [Google Scholar] [CrossRef] [PubMed]
- Park, G.; Xue, C.; Zheng, L.; Lam, S.; Xu, J.-R. MST12 regulates infectious growth but not appressorium formation in the rice blast fungus Magnaporthe grisea. Mol. Plant-Microbe Interact. 2002, 15, 183–192. [Google Scholar] [CrossRef] [PubMed]
- Xiao, K.; Liu, L.; He, R.; Rollins, J.A.; Li, A.; Zhang, G.; He, X.; Wang, R.; Liu, J.; Zhang, X.; et al. The Snf5-Hsf1 transcription module synergistically regulates stress responses and pathogenicity by maintaining ROS homeostasis in Sclerotinia sclerotiorum. New Phytol. 2024, 241, 1794–1812. [Google Scholar] [CrossRef]
- Qu, X.; Yu, B.; Liu, J.; Zhang, X.; Li, G.; Zhang, D.; Li, L.; Wang, X.; Wang, L.; Chen, J.; et al. MADS-Box transcription factor SsMADS is involved in regulating growth and virulence in Sclerotinia sclerotiorum. Int. J. Mol. Sci. 2014, 15, 8049–8062. [Google Scholar] [CrossRef]
- Liu, L.; Wang, Q.; Sun, Y.; Zhang, Y.; Zhang, X.; Liu, J.; Yu, G.; Pan, H. Sssfh1, a gene encoding a putative component of the RSC chromatin remodeling Complex, is involved in hyphal growth, reactive oxygen species accumulation, and pathogenicity in Sclerotinia sclerotiorum. Front. Microbiol. 2018, 9, 1828. [Google Scholar] [CrossRef]
- Cong, J.; Xiao, K.; Jiao, W.; Zhang, C.; Zhang, X.; Liu, J.; Zhang, Y.; Pan, H. The coupling between cell wall integrity mediated by MAPK kinases and SsFkh1 is involved in sclerotia formation and pathogenicity of Sclerotinia sclerotiorum. Front. Microbiol. 2022, 13, 816091. [Google Scholar] [CrossRef]
- Catlett, N.L.; Lee, B.-N.; Yoder, O.C.; Turgeon, B.G. Split-marker recombination for efficient targeted deletion of fungal genes. Fungal Genet. Rep. 2003, 50, 9–11. [Google Scholar] [CrossRef]
- Rollins, J.A. The Sclerotinia sclerotiorum pac1 gene is required for sclerotial development and virulence. Mol. Plant Microbe Interact. 2003, 16, 785–795. [Google Scholar] [CrossRef]
- Gutierrez, L.; Mauriat, M.; Guénin, S.; Pelloux, J.; Lefebvre, J.-F.; Louvet, R.; Rusterucci, C.; Moritz, T.; Guerineau, F.; Bellini, C.; et al. The lack of a systematic validation of reference genes: A serious pitfall undervalued in reverse transcription-polymerase chain reaction (RT-PCR) analysis in plants. Plant Biotechnol. J. 2008, 6, 609–618. [Google Scholar] [CrossRef]
- Schmittgen, T.D.; Zakrajsek, B.A.; Mills, A.G.; Gorn, V.; Singer, M.J.; Reed, M.W. Quantitative reverse transcription-polymerase chain reaction to study mRNA decay: Comparison of endpoint and real-time methods. Anal. Biochem. 2000, 285, 194–204. [Google Scholar] [CrossRef]
- Kim, T.H.; Dekker, J. ChIP. In Cold Spring Harbor Protocols; CSH Press: Long Island, NY, USA, 2018. [Google Scholar] [CrossRef]
- Liu, Z.; Jian, Y.; Chen, Y.; Corby-Kistler, H.; He, P.; Ma, Z.; Yin, Y. A phosphorylated transcription factor regulates sterol biosynthesis in Fusarium graminearum. Nat. Commun. 2019, 10, 1228. [Google Scholar] [CrossRef] [PubMed]
- Kim, T.H.; Dekker, J. ChIP–Quantitative Polymerase Chain Reaction (ChIP-qPCR). In Cold Spring Harbor Protocols; CSH Press: Long Island, NY, USA, 2018. [Google Scholar] [CrossRef]
- Crowley, L.C.; Scott, A.P.; Marfell, B.J.; Boughaba, J.A.; Chojnowski, G.; Waterhouse, N.J. Measuring Cell Death by Propidium Iodide Uptake and Flow Cytometry. In Cold Spring Harbor Protocols; CSH Press: Long Island, NY, USA, 2016. [Google Scholar] [CrossRef]
- Lyu, X.; Shen, C.; Fu, Y.; Xie, J.; Jiang, D.; Li, G.; Cheng, J. Comparative genomic and transcriptional analyses of the carbohydrate-active enzymes and secretomes of phytopathogenic fungi reveal their significant roles during infection and development. Sci. Rep. 2015, 5, 15565. [Google Scholar] [CrossRef]
- Sinkala, M.; Nkhoma, P.; Mulder, N.; Martin, D.P. Integrated molecular characterisation of the MAPK pathways in human cancers reveals pharmacologically vulnerable mutations and gene dependencies. Commun. Biol. 2021, 4, 9. [Google Scholar] [CrossRef]
- Zhang, R.; Wang, Y.; Wu, A.; Wang, J.; Zhang, J. Strategies of targeting CYP51 for IFIs therapy: Emerging prospects, opportunities and challenges. Eur. J. Med. Chem. 2023, 259, 115658. [Google Scholar] [CrossRef] [PubMed]
- Zuriegat, Q.; Zheng, Y.; Liu, H.; Wang, Z.; Yun, Y. Current progress on pathogenicity-related transcription factors in Fusarium oxysporum. Mol. Plant Pathol. 2021, 22, 882–895. [Google Scholar] [CrossRef] [PubMed]
- Xie, J.; Rybak, J.M.; Martin-Vicente, A.; Guruceaga, X.; Thorn, H.I.; Nywening, A.V.; Ge, W.; Parker, J.E.; Kelly, S.L.; Rogers, P.D.; et al. The sterol C-24 methyltransferase encoding gene, erg6, is essential for viability of Aspergillus species. Nat. Commun. 2024, 15, 4261. [Google Scholar] [CrossRef]
- Yan, X.; Ma, W.-B.; Li, Y.; Wang, H.; Que, Y.-W.; Ma, Z.-H.; Talbot, N.J.; Wang, Z.-Y. A sterol 14α-demethylase is required for conidiation, virulence and for mediating sensitivity to sterol demethylation inhibitors by the rice blast fungus Magnaporthe oryzae. Fungal Genet. Biol. 2011, 48, 144–153. [Google Scholar] [CrossRef]
- Liu, X.; Jiang, J.; Yin, Y.; Ma, Z. Involvement of FgERG4 in ergosterol biosynthesis, vegetative differentiation and virulence in Fusarium graminearum. Mol. Plant Pathol. 2012, 14, 71–83. [Google Scholar] [CrossRef]
- Yang, S.; Yan, D.; Li, M.; Li, D.; Zhang, S.; Fan, G.; Peng, L.; Pan, S. Ergosterol depletion under bifonazole treatment induces cell membrane damage and triggers a ROS-mediated mitochondrial apoptosis in Penicillium expansum. Fungal Biol. 2022, 126, 1–10. [Google Scholar] [CrossRef]
- Schuster, M.; Kilaru, S.; Steinberg, G. Azoles activate type I and type II programmed cell death pathways in crop pathogenic fungi. Nat. Commun. 2024, 15, 4357. [Google Scholar] [CrossRef] [PubMed]
- Waszczak, C.; Carmody, M.; Kangasjarvi, J. Reactive oxygen species in plant signaling. Annu. Rev. Plant Biol. 2018, 69, 209–236. [Google Scholar] [CrossRef]
- Elias, D.; Tóth Hervay, N.; Bujdos, M.; Gbelska, Y. Essential role of CgErg6p in maintaining oxidative stress tolerance and iron homeostasis in Candida glabrata. J. Fungi 2023, 9, 579. [Google Scholar] [CrossRef] [PubMed]
- Xiao, Y.; Li, S.; Niu, S.; Ma, L.; Zhang, G.; Zhang, H.; Zhang, G.; Ju, J.; Zhang, C. Characterization of Tiacumicin B Biosynthetic Gene Cluster Affording Diversified Tiacumicin analogues and Revealing a Tailoring dihalogenase. J. Am. Chem. Soc. 2010, 133, 1092–1105. [Google Scholar] [CrossRef] [PubMed]
- Lyu, H.-N.; Liu, H.-W.; Keller, N.P.; Yin, W.-B. Harnessing diverse transcriptional regulators for natural product discovery in fungi. Nat. Prod. Rep. 2020, 37, 6–16. [Google Scholar] [CrossRef]
- Zeng, J.; Cao, Y.; Guo, Y.; Xiang, D.; Wang, J.; Xu, Q.; Lang, X.; Xu, H.; Cao, Y. Regulation of phenotype and secondary metabolic silencing gene clusters in Aspergillus sydowii by velvet transcription factors. Fungal Biol. 2025, 129, 101605. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhao, H.; Liu, X.; Jiang, J.; Xie, J.; Fu, Y.; Lin, Y.; Chen, T.; Li, B.; Yu, X.; Xiao, X.; et al. The Transcription Factor SsSR Mediates Ergosterol Biosynthesis and Virulence in Sclerotinia sclerotiorum. J. Fungi 2025, 11, 509. https://doi.org/10.3390/jof11070509
Zhao H, Liu X, Jiang J, Xie J, Fu Y, Lin Y, Chen T, Li B, Yu X, Xiao X, et al. The Transcription Factor SsSR Mediates Ergosterol Biosynthesis and Virulence in Sclerotinia sclerotiorum. Journal of Fungi. 2025; 11(7):509. https://doi.org/10.3390/jof11070509
Chicago/Turabian StyleZhao, Huihui, Xiaofan Liu, Jintao Jiang, Jiatao Xie, Yanping Fu, Yang Lin, Tao Chen, Bo Li, Xiao Yu, Xueqiong Xiao, and et al. 2025. "The Transcription Factor SsSR Mediates Ergosterol Biosynthesis and Virulence in Sclerotinia sclerotiorum" Journal of Fungi 11, no. 7: 509. https://doi.org/10.3390/jof11070509
APA StyleZhao, H., Liu, X., Jiang, J., Xie, J., Fu, Y., Lin, Y., Chen, T., Li, B., Yu, X., Xiao, X., Lyu, X., Chen, W., Jiang, D., & Cheng, J. (2025). The Transcription Factor SsSR Mediates Ergosterol Biosynthesis and Virulence in Sclerotinia sclerotiorum. Journal of Fungi, 11(7), 509. https://doi.org/10.3390/jof11070509








