The Study of Metschnikowia pulcherrima E1 in the Induction of Improved Gray Spot Disease Resistance in Loquat Fruit
Abstract
1. Introduction
2. Materials and Methods
2.1. Fruit
2.2. Fungal Pathogen
2.3. Antagonistic Yeast
2.4. Effect of Metschnikowia pulcherrima E1 on Control of Gray Spot of Loquat
2.5. RNA Extraction and Detection of Loquat Fruit Tissue Samples
2.5.1. Preparation of Loquat Fruit Tissue Samples
2.5.2. Extraction and Detection of RNA
2.6. Transcriptome Sequencing and Bioinformatics Analysis of Loquat Fruit
2.7. RT–qPCR Verification of DEGs
- (1)
- Ct values were normalized to the geometric mean of two reference genes (Actin and EF1α).
- (2)
- ΔΔCt values were derived by comparing treated samples (E1-inoculated) to controls (water-treated).
- (3)
- Fold changes were expressed as mean ± SD of three biological replicates (n = 3).
2.8. Statistical Analysis
3. Results
3.1. Isolation, Screening and Identification Results of the Pathogen and Antagonistic Yeast
3.2. Effect of M. pulcherrima E1 on Control of Gray Spot of Loquat
3.3. Sequencing Data and Quality Control
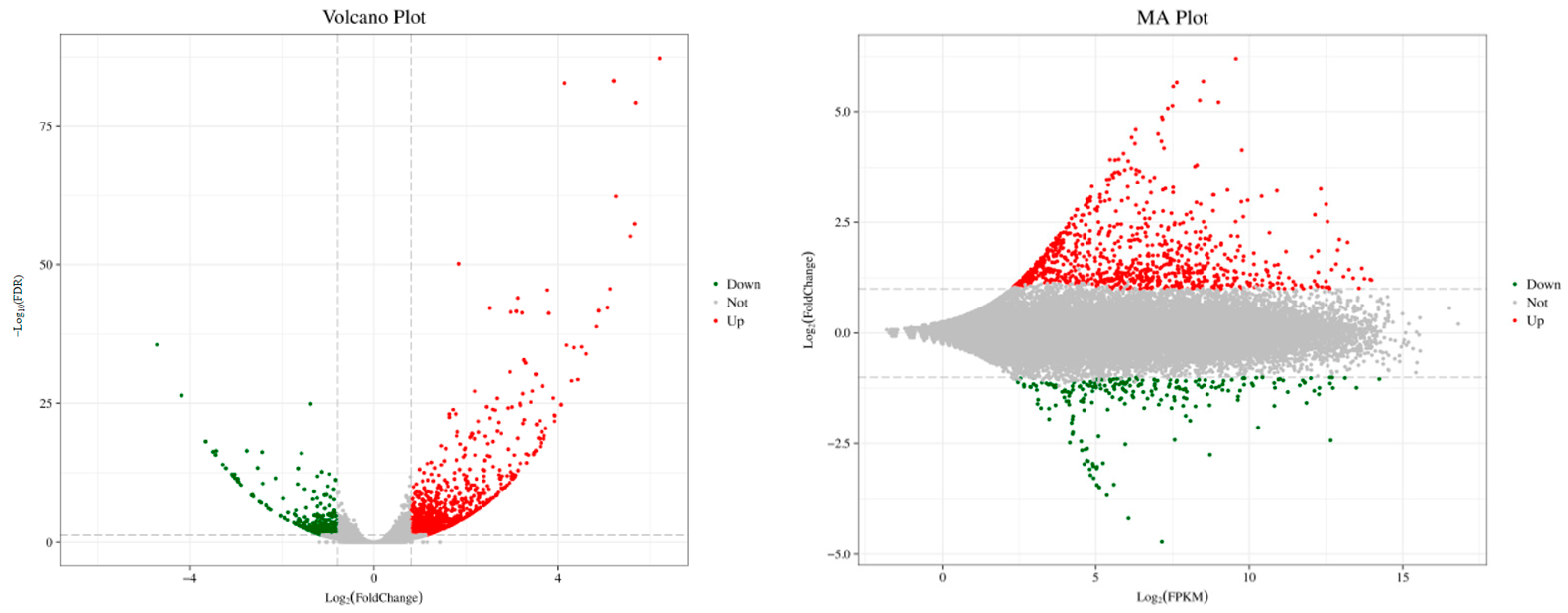
3.4. DEG Analysis
3.5. GO Enrichment Analysis of DEGs
3.6. KEGG Enrichment Analysis of DEGs
3.6.1. Effects on Plant–Pathogen Interaction Pathways
3.6.2. Effects on Secondary Metabolic Pathways
3.6.3. Effects on the Related Pathways of Plant Hormone Metabolism
3.6.4. Effects of Glutathione Metabolic Pathways
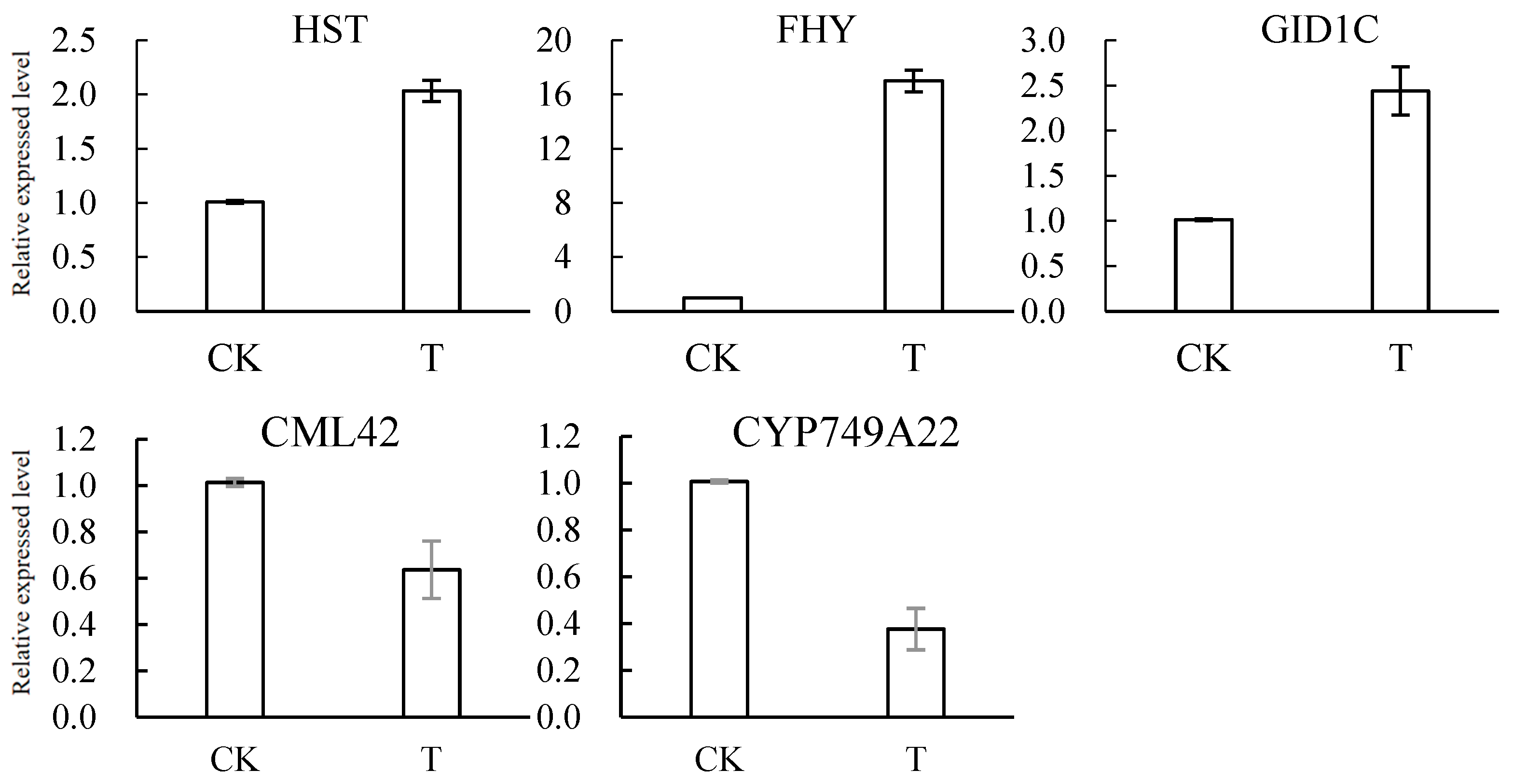
3.7. Results of RT–qPCR Verification of DEGs
4. Discussion
4.1. The Biocontrol Efficacy of Antagonistic Yeasts
4.2. KEGG Pathway Enrichment Analysis of DEGs
4.2.1. Analysis of Plant–Pathogen Interaction Pathways
4.2.2. Analysis of Secondary Metabolic Pathways
4.2.3. Analysis of the Related Pathways of Plant Hormone Metabolism
4.2.4. Analysis of Glutathione Metabolic Pathways
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Yan, F.; Zhang, D.; Wang, X.; Liu, C.; Zhang, F. Reduction of postharvest diseases of loquat fruit by serine protease and possible mechanisms involved. Sci. Hortic. 2022, 304, 111246. [Google Scholar] [CrossRef]
- Yan, F.; Zhang, D.; Ye, X.; Wu, Y.; Fang, T. Potential of Saturnispora diversa MA as a postharvest biocontrol agent against anthracnose in loquat fruit. Biol. Control 2022, 173, 105006. [Google Scholar] [CrossRef]
- Ye, W.Q.; Sun, Y.F.; Tang, Y.J.; Zhou, W.W. Biocontrol potential of a broad-spectrum antifungal strain Bacillus amyloliquefaciens B4 for postharvest loquat fruit storage. Postharvest Biol. Technol. 2021, 174, 111439. [Google Scholar] [CrossRef]
- Sun, X.W.; Xu, Y.; Chen, L.; Jin, X.M.; Ni, H. The salt-tolerant phenazine-1-carboxamide-producing bacterium Pseudomonas aeruginosa NF011 isolated from wheat rhizosphere soil in dry farmland with antagonism against Fusarium graminearum. Microbiol. Res. 2021, 245, 126673. [Google Scholar] [CrossRef]
- Ali, M.A.; Ren, H.; Ahmed, T.; Luo, J.; An, Q.; Qi, X.; Li, B. Antifungal effects of rhizospheric Bacillus species against bayberry twig blight pathogen Pestalotiopsis versicolor. Agronomy 2020, 10, 1811. [Google Scholar] [CrossRef]
- Saju, K.A.; Mech, S.; Deka, T.N.; Biswas, A.K. In vitro evaluation of biocontrol agents, botanicals and fungicides against Pestalotiopsis sp. infecting large cardamom (Amomum subulatum Roxb.). J. Spices Aromat. Crops 2012, 20, 89–92. [Google Scholar]
- Zhang, X.; Wu, F.; Gu, N.; Yan, X.; Wang, K.; Dhanasekaran, S.; Zhang, H.; Gu, X.; Zhao, L. Postharvest biological control of Rhizopus rot and the mechanisms involved in induced disease resistance of peaches by Pichia membranefaciens. Postharvest Biol. Technol. 2020, 163, 111146. [Google Scholar] [CrossRef]
- Zhang, X.; Yao, Y.; Dhanasekaran, S.; Li, J.; Ngea, G.L.N.; Gu, X.; Zhang, H. Controlling black spot of postharvest broccoli by Meyerozyma guilliermondii and its regulation on ROS metabolism of broccoli. Biol. Control 2022, 170, 104938. [Google Scholar] [CrossRef]
- Bonaterra, A.; Badosa, E.; Daranas, N.; Francés, J.; Roselló, G.; Montesinos, E. Bacteria as Biological Control Agents of Plant Diseases. Microorganisms 2022, 10, 1759. [Google Scholar] [CrossRef] [PubMed]
- Yang, H.H.; Wang, L.; Li, S.J.; Gao, X.B.; Wu, N.N.; Zhao, Y.F.; Sun, W.H. Control of postharvest grey spot rot of loquat fruit with Metschnikowia pulcherrima E1 and potential mechanisms of action. Biol. Control 2021, 152, 104406. [Google Scholar] [CrossRef]
- Zhao, L.; Shu, Y.; Xiao, J.; Lin, R.; Godana, E.A.; Zhang, X.; Zhang, H. Transcriptome analysis reveals mechanisms involved in the enhanced antagonistic efficacy of Sporidiobolus pararoseus Y16 treated by γ-aminobutyric acid. Biol. Control 2022, 176, 105089. [Google Scholar] [CrossRef]
- Di Francesco, A.; Martini, C.; Mari, M. Biological control of postharvest diseases by microbial antagonists: How many mechanisms of action? Eur. J. Plant Pathol. 2016, 145, 711–717. [Google Scholar] [CrossRef]
- Li, Q.; Zhang, H.; Yang, Q.; Lin, Z.; Cheng, Y.; Sun, Y. Progress in biocontrol yeast agents for preventing and treating diseases of fruits. Food Sci. 2018, 39, 291–296. [Google Scholar] [CrossRef]
- Zhang, X.; Zhou, Y.; Li, J.; Gu, X.; Zhao, L.; Li, B.; Zhang, H. Pichia caribbica improves disease resistance of cherry tomatoes by regulating ROS metabolism. Biol. Control 2022, 169, 104870. [Google Scholar] [CrossRef]
- Costa, V.; Angelini, C.; De Feis, I.; Ciccodicola, A. Uncovering the Complexity of Transcriptomes with RNA-Seq. J. Biomed. Biotechnol. 2010, 1, 853916. [Google Scholar] [CrossRef]
- Strauss, T.; van Poecke, R.M.P.; Strauss, A.; Romer, P.; Minsavage, G.V.; Singh, S.; Wolf, C.; Strauss, A.; Kim, S.; Lee, H.A.; et al. RNA-seq pinpoints a Xanthomonas TAL-effector activated resistance gene in a large-crop genome. Proc. Natl. Acad. Sci. USA 2012, 109, 19480–19485. [Google Scholar] [CrossRef] [PubMed]
- Qi, Y.W.; Lei, Q.; Zhang, Y.J.; Liu, X.R.; Zhou, B.; Liu, C.H.; Ren, X.L. Comparative transcripome data for commercial maturity and physiological maturity of ‘Royal Gala’ apple fruit under room temperature storage condition. Sci. Hortic. 2017, 225, 386–393. [Google Scholar] [CrossRef]
- Kurtzman, C.; Fell, J.W. The Yeasts—A Taxonomic Study; Elsevier Science: Amsterdam, The Netherlands, 1998. [Google Scholar]
- Hsu, S.C.; Tsen, H.Y. PCR primers designed from malic acid dehydrogenase gene and their use for detection of Escherichia coli in water and milk samples. Int. J. Food Microbiol. 2001, 64, 1–11. [Google Scholar] [CrossRef]
- Redshaw, N.; Wilkes, T.; Whale, A.; Cowen, S.; Huggett, J.; Foy, C.A. A comparison of miRNA isolation and RT-qPCR technologies and their effects on quantification accuracy and repeatability. Biotechniques 2013, 54, 155–164. [Google Scholar] [CrossRef]
- Puyo, M.; Simonin, S.; Bach, B.; Klein, G.; Alexandre, H.; Tourdot-Maréchal, R. Bio-protection in oenology by Metschnikowia pulcherrima: From field results to scientific inquiry. Front. Microbiol. 2023, 14, 1252973. [Google Scholar] [CrossRef]
- Zhou, Y.; Zhao, L.; Chen, Y.; Dhanasekaran, S.; Chen, X.; Zhang, X.; Zhang, H. Study on the control effect and physiological mechanism of Wickerhamomyces anomalus on primary postharvest diseases of peach fruit. Int. J. Food Microbiol. 2024, 413, 110575. [Google Scholar] [CrossRef]
- Zhou, Y.; Zhao, L.; Chen, Y.; Dhanasekaran, S.; Chen, X.; Zhang, X.; Zhang, H. Characterization of a Bacillus velezensis strain as a potential biocontrol agent against soft rot of eggplant fruits. Int. J. Food Microbiol. 2024, 410, 110480. [Google Scholar] [CrossRef]
- Wang, W.; Feng, B.; Zhou, J.-M.; Tang, D. Plant immune signaling: Advancing on two frontiers. J. Integr. Plant Biol. 2020, 62, 2–24. [Google Scholar] [CrossRef] [PubMed]
- Ma, Z.; Bykova, N.V.; Igamberdiev, A.U. Cell signaling mechanisms and metabolic regulation of germination and dormancy in barley seeds. Crop J. 2017, 5, 459–477. [Google Scholar] [CrossRef]
- Dietz, K.-J.; Mittler, R.; Noctor, G. Recent Progress in Understanding the Role of Reactive Oxygen Species in Plant Cell Signaling. Plant Physiol. 2016, 171, 1535–1539. [Google Scholar] [CrossRef] [PubMed]
- Miller, G.; Shulaev, V.; Mittler, R. Reactive oxygen signaling and abiotic stress. Physiol. Plant. 2008, 133, 481–489. [Google Scholar] [CrossRef]
- Feng, J.; Chen, L.; Zuo, J. Protein S-Nitrosylation in plants: Current progresses and challenges. J. Integr. Plant Biol. 2019, 61, 1206–1223. [Google Scholar] [CrossRef]
- Noman, A.; Aqeel, M.; Qari, S.H.; Al Surhanee, A.A.; Yasin, G.; Alamri, S.; Hashem, M.; Al-Saadi, A.M. Plant hypersensitive response vs. pathogen ingression: Death of few gives life to others. Microb Pathog 2020, 145, 104224. [Google Scholar] [CrossRef]
- Suzuki, N.; Katano, K. Coordination Between ROS Regulatory Systems and Other Pathways Under Heat Stress and Pathogen Attack. Front. Plant Sci. 2018, 9, 490. [Google Scholar] [CrossRef]
- Lucas, W.J.; Groover, A.; Lichtenberger, R.; Furuta, K.; Yadav, S.-R.; Helariutta, Y.; He, X.-Q.; Fukuda, H.; Kang, J.; Brady, S.M.; et al. The Plant Vascular System: Evolution, Development and Functions. J. Integr. Plant Biol. 2013, 55, 294–388. [Google Scholar] [CrossRef]
- Zhang, Q.L.; Li, Y.; Zhang, Y.; Wu, C.B.; Wang, S.N.; Hao, L.; Wang, S.Y.; Li, T.Z. Md-miR156ab and Md-miR395 Target WRKY Transcription Factors to Influence Apple Resistance to Leaf Spot Disease. Front. Plant Sci. 2017, 8, 526. [Google Scholar] [CrossRef] [PubMed]
- Ballester, A.R.; Marcet-Houben, M.; Levin, E.; Sela, N.; Selma-Lazaro, C.; Carmona, L.; Wisniewski, M.; Droby, S.; Gonzalez-Candelas, L.; Gabaldon, T. Genome, Transcriptome, and Functional Analyses of Penicillium expansum Provide New Insights into Secondary Metabolism and Pathogenicity. Mol. Plant Microbe Interact. 2015, 28, 232–248. [Google Scholar] [CrossRef]
- Zhao, Q.; Dixon, R.A. Transcriptional networks for lignin biosynthesis: More complex than we thought? Trends Plant Sci. 2011, 16, 227–233. [Google Scholar] [CrossRef] [PubMed]
- Pacifici, E.; Polverari, L.; Sabatini, S. Plant hormone cross-talk: The pivot of root growth. J. Exp. Bot. 2015, 66, 1113–1121. [Google Scholar] [CrossRef]
- Gong, Z.; Xiong, L.; Shi, H.; Yang, S.; Herrera-Estrella, L.R.; Xu, G.; Chao, D.-Y.; Li, J.; Wang, P.-Y.; Qin, F.; et al. Plant abiotic stress response and nutrient use efficiency. Sci. China Life Sci. 2020, 63, 635–674. [Google Scholar] [CrossRef]
- Pieterse, C.M.J.; Van der Does, D.; Zamioudis, C.; Leon-Reyes, A.; Van Wees, S.C.M. Hormonal Modulation of Plant Immunity. In Annual Review of Cell and Developmental Biology; Schekman, R., Ed.; Annual Reviews: San Mateo, CA, USA, 2012; Volume 28, pp. 489–521. [Google Scholar]
- Robert-Seilaniantz, A.; Grant, M.; Jones, J.D.G. Hormone Crosstalk in Plant Disease and Defense: More Than Just JASMONATE-SALICYLATE Antagonism. In Annual Review of Phytopathology; VanAlfen, N.K., Bruening, G., Leach, J.E., Eds.; Annual Reviews: Palo Alto, CA, USA, 2011; Volume 49, pp. 317–343. [Google Scholar]
- Thakur, M.; Sohal, B.S. Role of Elicitors in Inducing Resistance in Plants against Pathogen Infection: A Review. ISRN Biochem. 2013, 2013, 762412. [Google Scholar] [CrossRef] [PubMed]
- Shigenaga, A.M.; Argueso, C.T. No hormone to rule them all: Interactions of plant hormones during the responses of plants to pathogens. Semin. Cell Dev. Biol. 2016, 56, 174–189. [Google Scholar] [CrossRef]
- Yu, X.Y.; Hu, K.X.; Geng, X.Y.; Cao, L.F.; Zhou, T.T.; Lin, X.X.; Liu, H.C.; Chen, J.R.; Luo, C.G.; Qu, S.C. The Mh-miR393a-TIR1 module regulates Alternaria alternata resistance of Malus hupehensis mainly by modulating the auxin signaling. Plant Sci. 2024, 341, 112008. [Google Scholar] [CrossRef]
- Peng, H.; Neff, M.M. CIRCADIAN CLOCK ASSOCIATED 1 and ATAF2 differentially suppress cytochrome P450-mediated brassinosteroid inactivation. J. Exp. Bot. 2020, 71, 970–985. [Google Scholar] [CrossRef]
- Venturas, M.D.; Sperry, J.S.; Hacke, U.G. Plant xylem hydraulics: What we understand, current research, and future challenges. J. Integr. Plant Biol. 2017, 59, 356–389. [Google Scholar] [CrossRef] [PubMed]
- Lecourieux, D.; Kappel, C.; Claverol, S.; Pieri, P.; Feil, R.; Lunn, J.E.; Bonneu, M.; Wang, L.; Gomes, E.; Delrot, S.; et al. Proteomic and metabolomic profiling underlines the stage- and time-dependent effects of high temperature on grape berry metabolism. J. Integr. Plant Biol. 2020, 62, 1132–1158. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J.; Dixon, R.A. The ‘ins’ and ‘outs’ of flavonoid transport. Trends Plant Sci. 2010, 15, 72–80. [Google Scholar] [CrossRef] [PubMed]
- Kong, F.; Deng, Y.; Wang, G.; Wang, J.; Liang, X.; Meng, Q. LeCDJ1, a chloroplast DnaJ protein, facilitates heat tolerance in transgenic tomatoes. J. Integr. Plant Biol. 2014, 56, 63–74. [Google Scholar] [CrossRef] [PubMed]
- Jiang, J.J.; Ma, S.H.; Ye, N.H.; Jiang, M.; Cao, J.S.; Zhang, J.H. WRKY transcription factors in plant responses to stresses. J. Integr. Plant Biol. 2017, 59, 86–101. [Google Scholar] [CrossRef]
- Guo, B.; Chen, Z.-Y.; Lee, R.D.; Scully, B.T. Drought stress and preharvest aflatoxin contamination in agricultural commodity: Genetics, genomics and proteomics. J. Integr. Plant Biol. 2008, 50, 1281–1291. [Google Scholar] [CrossRef]
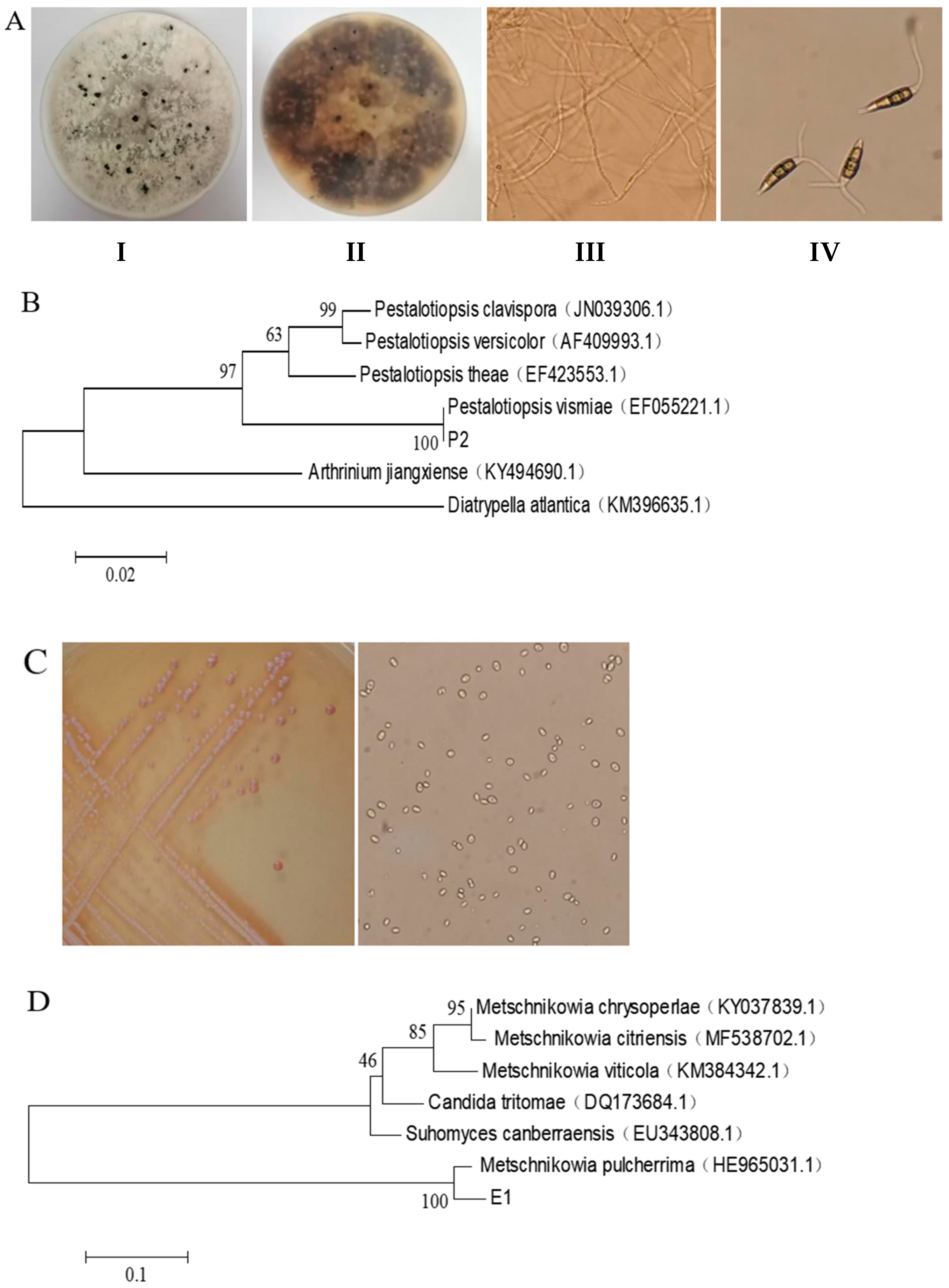




| Experiment | Incident Rate (%) | Spot Diameter (cm) |
| treatment group | 22.73 | 0.95 ± 0.06 b |
| control group | 100 | 1.59 ± 0.16 a |
| Samples | JSHY-ID | ReadSum | BaseSum | GC (%) | Q30 (%) |
|---|---|---|---|---|---|
| CK1 | T01 | 24562436 | 7368730800 | 46.56 | 92.22 |
| CK2 | T02 | 25701745 | 7710523500 | 46.03 | 92.84 |
| CK3 | T03 | 23718990 | 7115697000 | 46.5 | 92.95 |
| T1 | T04 | 28165336 | 8449600800 | 46.44 | 94.37 |
| T2 | T05 | 24068412 | 7220523600 | 46.48 | 94.24 |
| T3 | T06 | 23280409 | 6984122700 | 46.26 | 92.62 |
| Gene ID | Gene Name | log2FC | Definition |
|---|---|---|---|
| TRINITY_DN14842_c0_g1 | RBOHC | 0.96 | respiratory burst oxidase |
| TRINITY_DN12258_c0_g1 | CML5 | 1.40 | calcium-binding protein |
| TRINITY_DN14083_c0_g1 | HSP82 | −1.16 | molecular chaperone HtpG |
| TRINITY_DN15797_c0_g1 | CML23 | 1.56 | calcium-binding protein |
| TRINITY_DN15820_c0_g1 | CML27 | 1.57 | calcium-binding protein |
| TRINITY_DN15820_c0_g3 | CML27 | 1.17 | calcium-binding protein |
| TRINITY_DN16219_c0_g1 | CML19 | 2.00 | calcium-binding protein |
| TRINITY_DN16219_c0_g2 | CML19 | 0.97 | calcium-binding protein |
| TRINITY_DN17371_c0_g3 | EDS1 | 1.53 | enhanced disease susceptibility 1 protein |
| TRINITY_DN17724_c5_g2 | CP1 | 1.20 | Calcium-binding protein |
| TRINITY_DN17724_c5_g8 | CP1 | 1.74 | Calcium-binding protein |
| TRINITY_DN18123_c0_g1 | CML42 | −1.64 | calcium-binding protein |
| TRINITY_DN18265_c0_g4 | CML36 | 1.04 | calcium-binding protein |
| TRINITY_DN24035_c0_g1 | RPS2 | 1.06 | disease resistance protein |
| TRINITY_DN20679_c0_g1 | WRKY29 | 0.86 | WRKY transcription factor 29 |
| TRINITY_DN23615_c1_g2 | PTI5 | 0.93 | pathogenesis-related genes transcriptional activator |
| Gene ID | Gene Name | log2FC | Definition |
|---|---|---|---|
| Phenylpropanoid biosynthesis | |||
| TRINITY_DN15146_c0_g1 | PER52 | 1.82 | peroxidase |
| TRINITY_DN15166_c0_g1 | - | 0.97 | caffeoyl-CoA O-methyltransferase |
| TRINITY_DN15653_c0_g1 | PER52 | 1.23 | peroxidase |
| TRINITY_DN16201_c0_g1 | MEE23 | −1.06 | cinnamyl-alcohol dehydrogenase |
| TRINITY_DN17214_c1_g1 | ACT | 1.55 | shikimate O-hydroxycinnamoyltransferase |
| TRINITY_DN18626_c3_g2 | PER63 | 0.92 | peroxidase |
| TRINITY_DN22855_c1_g4 | HST | 1.06 | shikimate O-hydroxycinnamoyltransferase |
| TRINITY_DN23721_c0_g2 | F6' H1 | −1.52 | feruloyl-CoA 6-hydroxylase |
| Flavonoid biosynthesis | |||
| TRINITY_DN15166_c0_g1 | - | 0.97 | caffeoyl-CoA O-methyltransferase |
| TRINITY_DN17214_c1_g1 | ACT | 1.55 | shikimate O-hydroxycinnamoyltransferase |
| TRINITY_DN21813_c0_g1 | PKS5 | 0.88 | chalcone synthase |
| TRINITY_DN21935_c3_g2 | SALAT | −0.88 | shikimate O-hydroxycinnamoyltransferase |
| TRINITY_DN22199_c0_g1 | UGT88A1 | 1.10 | phlorizin synthase |
| TRINITY_DN22855_c1_g4 | HST | 1.06 | shikimate O-hydroxycinnamoyltransferase |
| TRINITY_DN9789_c0_g1 | FLS | 1.26 | flavonol synthase |
| Diterpenoid biosynthesis | |||
| TRINITY_DN15372_c0_g1 | GA2OX8 | 1.36 | gibberellin 2beta-dioxygenase |
| TRINITY_DN15922_c0_g2 | GA2OX1 | 1.80 | gibberellin 2beta-dioxygenase |
| TRINITY_DN20240_c0_g1 | GA2OX2 | 0.97 | gibberellin 2beta-dioxygenase |
| TRINITY_DN6982_c0_g1 | GES | 1.05 | geranyllinalool synthase |
| Sesquiterpenoid and triterpenoid biosynthesis | |||
| TRINITY_DN12696_c0_g1 | AFS1 | 2.38 | alpha-farnesene synthase |
| TRINITY_DN22548_c2_g1 | - | −1.01 | squalene monooxygenase |
| Riboflavin metabolism | |||
| TRINITY_DN10940_c0_g1 | PAP3 | 2.91 | tartrate-resistant acid phosphatase type 5 |
| TRINITY_DN12491_c0_g1 | FHY | 2.59 | riboflavin kinase/FMN hydrolase |
| TRINITY_DN14884_c0_g1 | PAP17 | 3.41 | tartrate-resistant acid phosphatase type 5 |
| Stilbenoid, diarylheptanoid and gingerol biosynthesis | |||
| TRINITY_DN15166_c0_g1 | - | 0.97 | caffeoyl-CoA O-methyltransferase |
| TRINITY_DN17214_c1_g1 | ACT | 1.55 | shikimate O-hydroxycinnamoyltransferase |
| TRINITY_DN21935_c3_g2 | SALAT | −0.88 | shikimate O-hydroxycinnamoyltransferase |
| TRINITY_DN22855_c1_g4 | HST | 1.06 | shikimate O-hydroxycinnamoyltransferase |
| Gene ID | Gene Name | log2FC | Definition |
|---|---|---|---|
| Auxin metabolism | |||
| TRINITY_DN26772_c0_g1 | IAA17 | 1.14 | auxin-responsive protein IAA |
| TRINITY_DN14799_c0_g1 | SAUR32 | −0.84 | SAUR family protein |
| Cytokinine metabolism | |||
| TRINITY_DN21313_c0_g1 | AHP1 | 0.81 | histidine-containing phosphotransfer protein |
| TRINITY_DN17952_c2_g1 | ARR3 | 0.94 | two-component response regulator ARR-A |
| Gibberellin metabolism | |||
| TRINITY_DN13496_c0_g1 | GID1C | 1.00 | gibberellin receptor GID1 |
| Brassinosteroid biosynthesis and metabolism | |||
| TRINITY_DN18076_c3_g4 | CYP749A22 | −1.00 | PHYB activation tagged suppressor 1 |
| TRINITY_DN20436_c1_g2 | CYP749A22 | −1.11 | PHYB activation tagged suppressor 1 |
| TRINITY_DN17619_c0_g1 | XTH23 | 2.95 | xyloglucan:xyloglucosyl transferase TCH4 |
| TRINITY_DN19437_c1_g1 | XTH22 | 1.51 | xyloglucan:xyloglucosyl transferase TCH4 |
| TRINITY_DN21110_c1_g4 | XTH23 | 3.79 | xyloglucan:xyloglucosyl transferase TCH4 |
| TRINITY_DN21863_c1_g1 | XTH22 | 2.63 | xyloglucan:xyloglucosyl transferase TCH4 |
| Jasmonic acid | |||
| TRINITY_DN16088_c0_g2 | TIFY10A | 1.86 | jasmonate ZIM domain-containing protein |
| TRINITY_DN17853_c2_g4 | TIFY9 | 1.58 | jasmonate ZIM domain-containing protein |
| Salicylic acid | |||
| TRINITY_DN14251_c0_g1 | NPR1 | −1.29 | regulatory protein NPR1 |
| Gene ID | Gene Name | log2FC | Definition |
|---|---|---|---|
| TRINITY_DN17157_c0_g3 | - | 0.94 | glutathione S-transferase |
| TRINITY_DN17817_c0_g1 | - | 0.82 | glutathione S-transferase |
| TRINITY_DN14083_c0_g1 | - | 2.18 | glutathione S-transferase |
| TRINITY_DN21103_c2_g4 | - | 0.97 | glutathione S-transferase |
| TRINITY_DN21103_c2_g5 | - | 3.23 | glutathione S-transferase |
| TRINITY_DN21174_c0_g1 | - | 0.82 | glutathione S-transferase |
| TRINITY_DN22846_c1_g1 | CDKD-1 | 0.94 | glutathione S-transferase |
| TRINITY_DN24039_c0_g1 | GSTU10 | 3.60 | glutathione S-transferase |
| TRINITY_DN11198_c0_g1 | APX3 | 1.58 | L-ascorbate peroxidase |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, X.; Wu, K.; Li, X.; Zhao, Y.; Sun, W. The Study of Metschnikowia pulcherrima E1 in the Induction of Improved Gray Spot Disease Resistance in Loquat Fruit. J. Fungi 2025, 11, 497. https://doi.org/10.3390/jof11070497
Li X, Wu K, Li X, Zhao Y, Sun W. The Study of Metschnikowia pulcherrima E1 in the Induction of Improved Gray Spot Disease Resistance in Loquat Fruit. Journal of Fungi. 2025; 11(7):497. https://doi.org/10.3390/jof11070497
Chicago/Turabian StyleLi, Xiaoya, Kunkun Wu, Xin Li, Yuhao Zhao, and Weihong Sun. 2025. "The Study of Metschnikowia pulcherrima E1 in the Induction of Improved Gray Spot Disease Resistance in Loquat Fruit" Journal of Fungi 11, no. 7: 497. https://doi.org/10.3390/jof11070497
APA StyleLi, X., Wu, K., Li, X., Zhao, Y., & Sun, W. (2025). The Study of Metschnikowia pulcherrima E1 in the Induction of Improved Gray Spot Disease Resistance in Loquat Fruit. Journal of Fungi, 11(7), 497. https://doi.org/10.3390/jof11070497






