Six Novel Species of Distoseptispora (Distoseptisporaceae, Distoseptisporales) and Helminthosporium (Massarinaceae, Pleosporales) Isolated from Terrestrial Habitats in Southern China
Abstract
1. Introduction
2. Materials and Methods
2.1. Sample Collection, Fungal Isolation and Morphological Observation
2.2. DNA Extraction, PCR Amplification and Sequencing
2.3. Phylogenetic Analyses
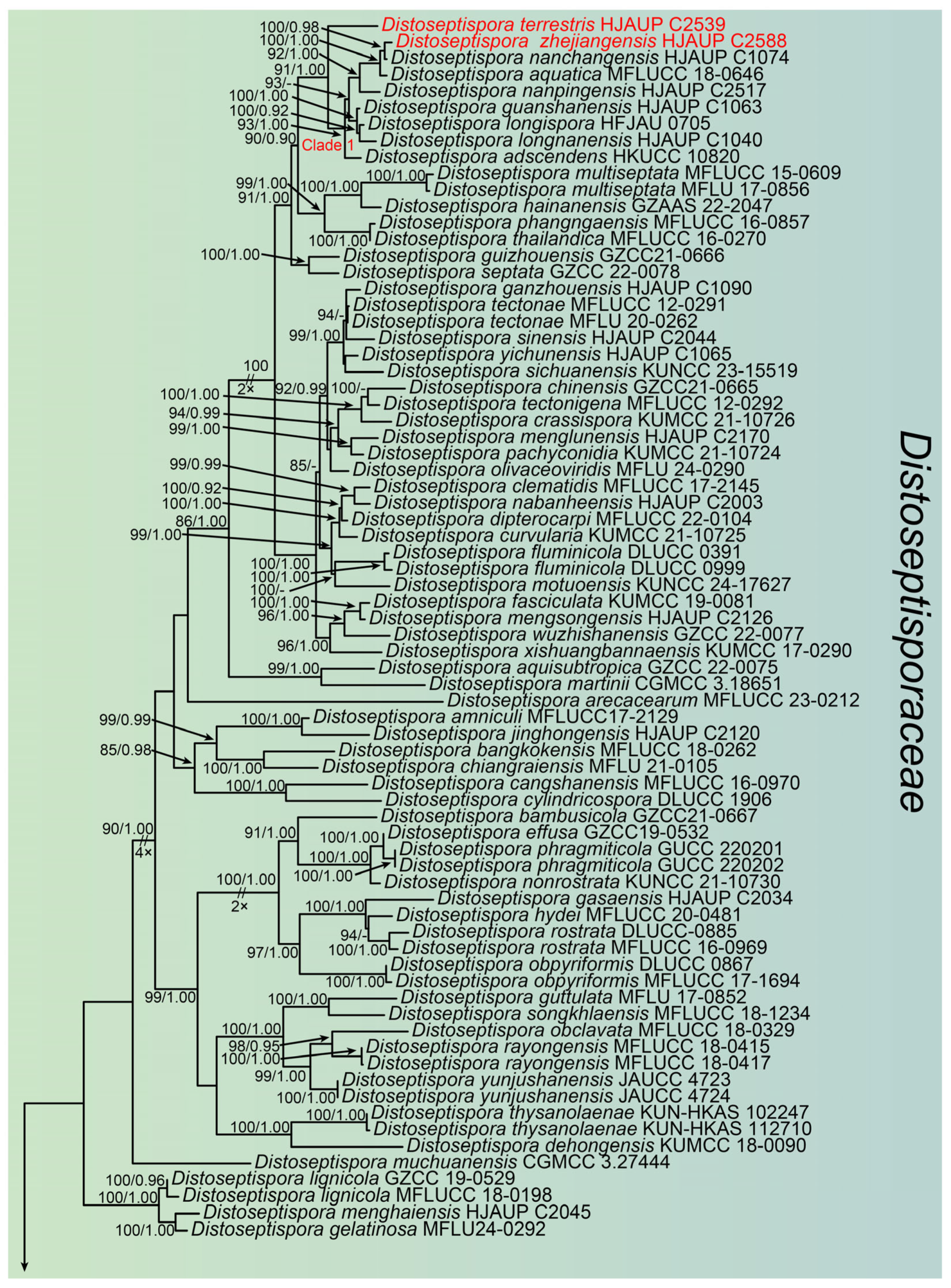
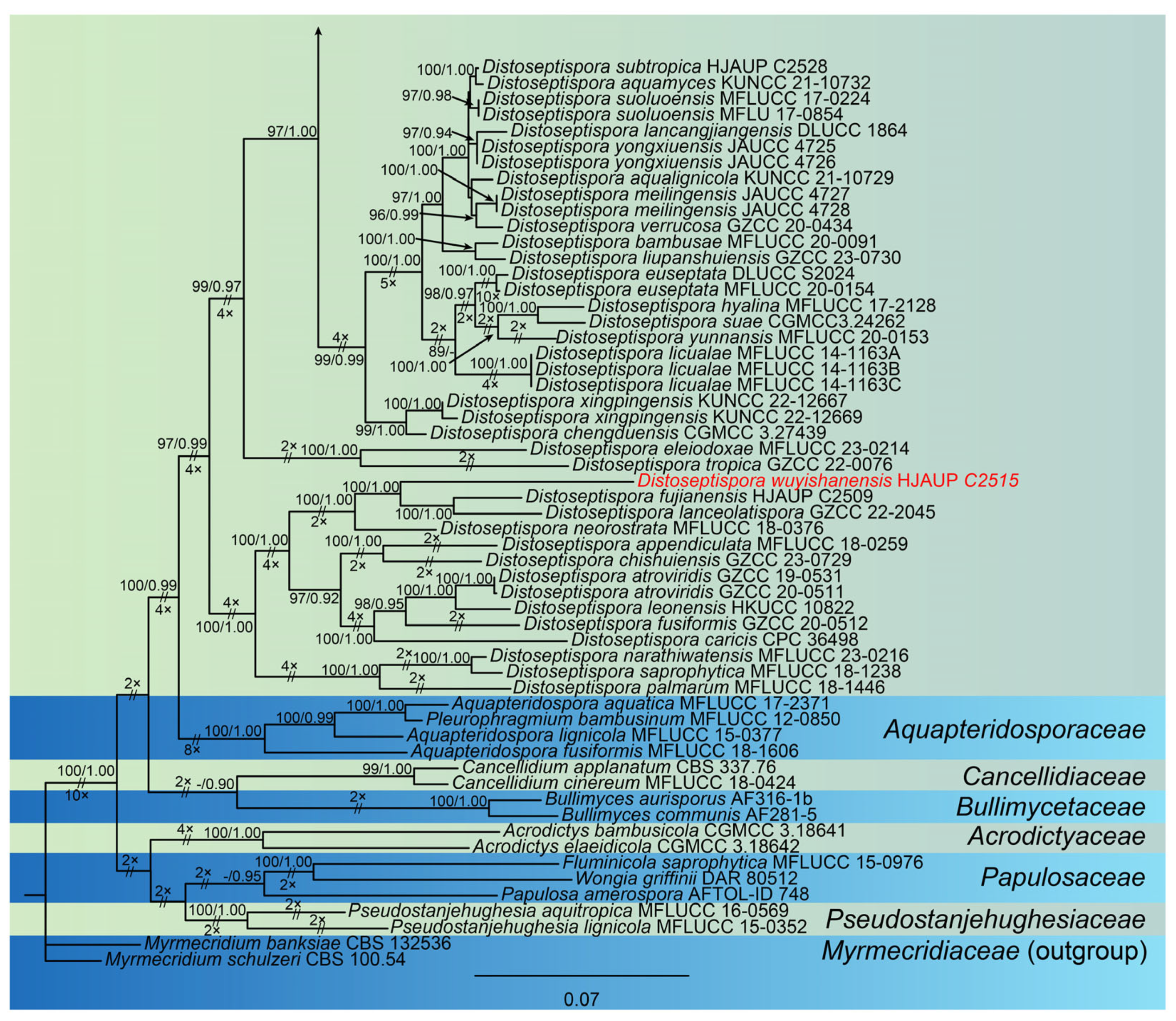
3. Results
3.1. Molecular Phylogeny
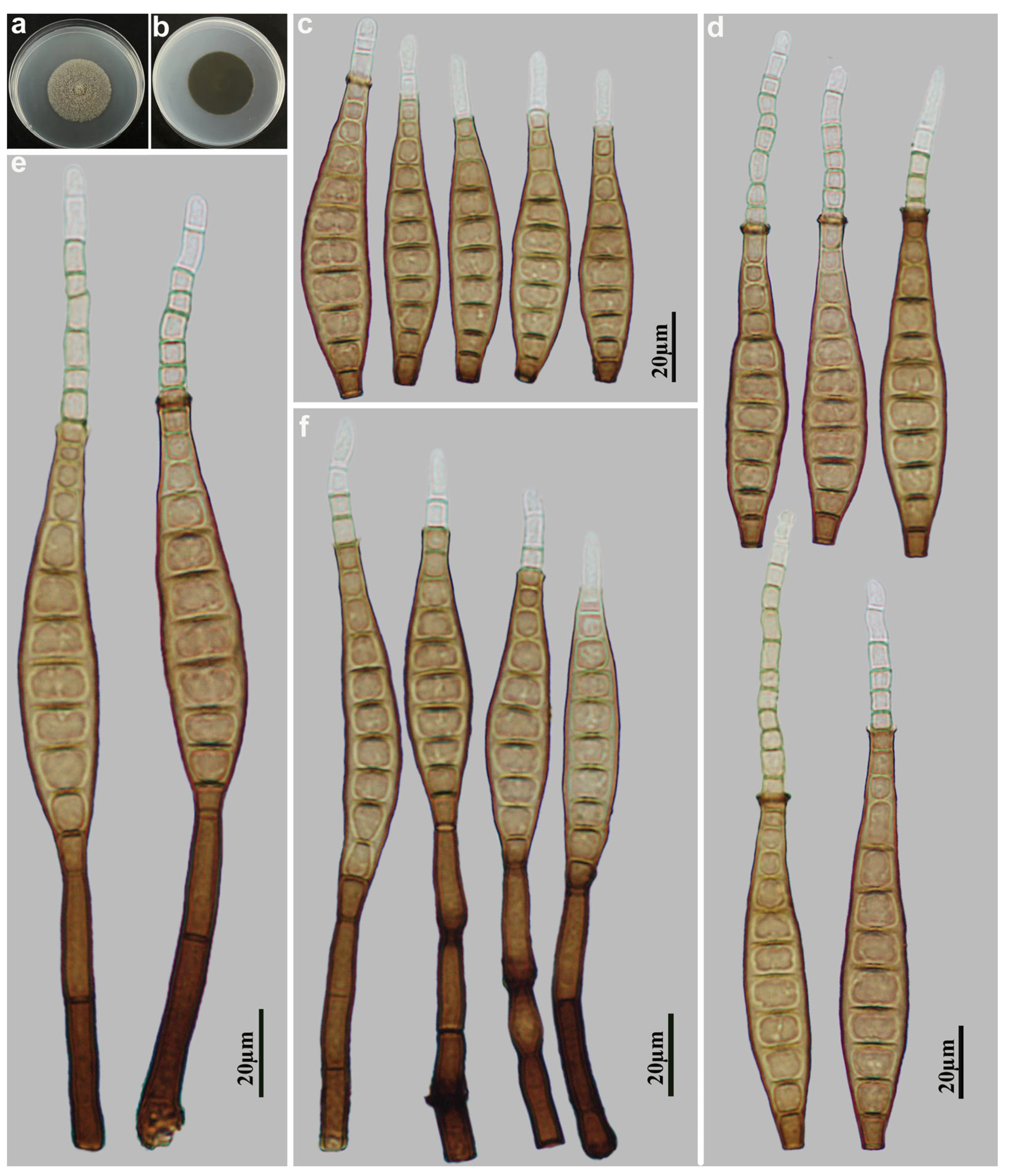
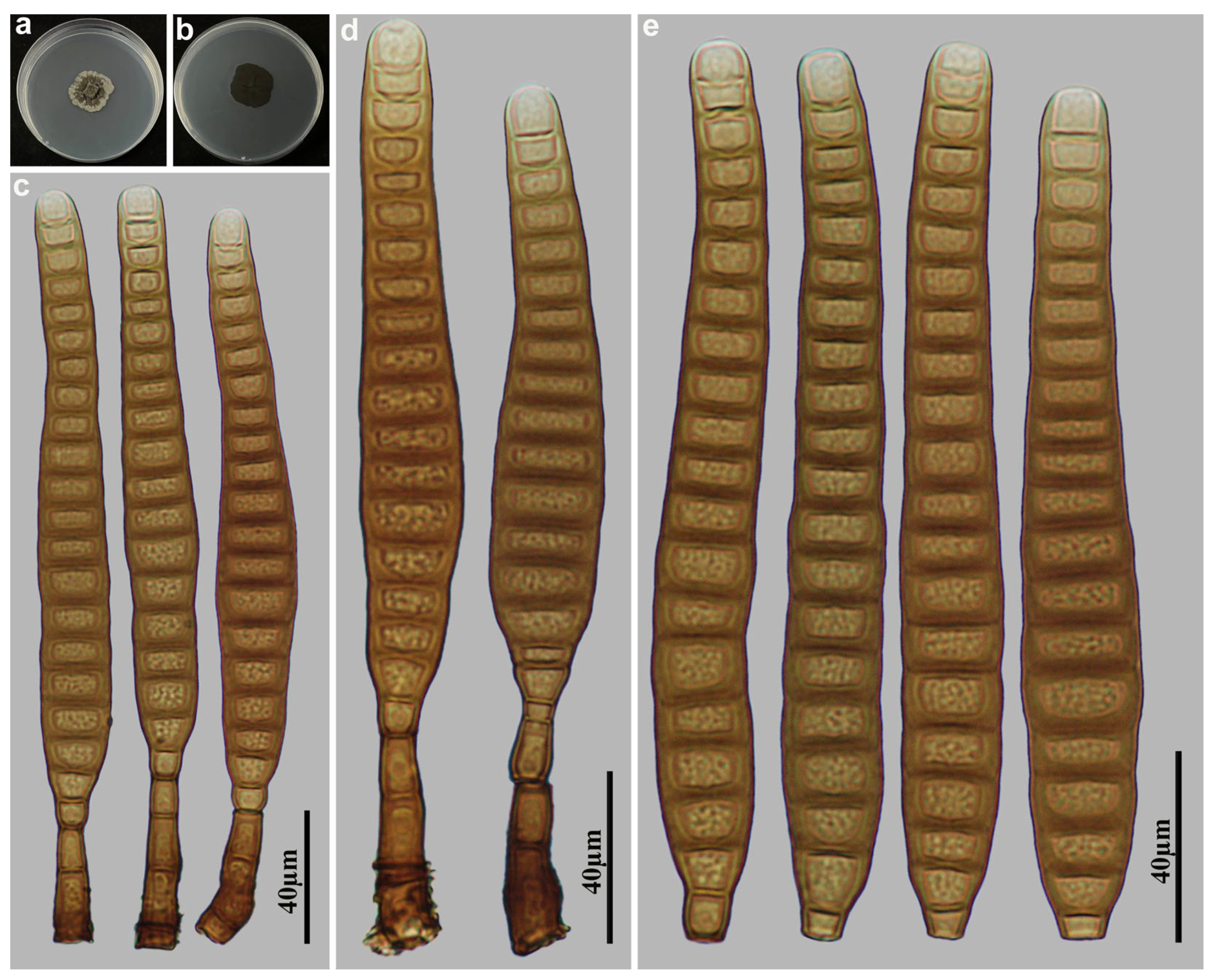
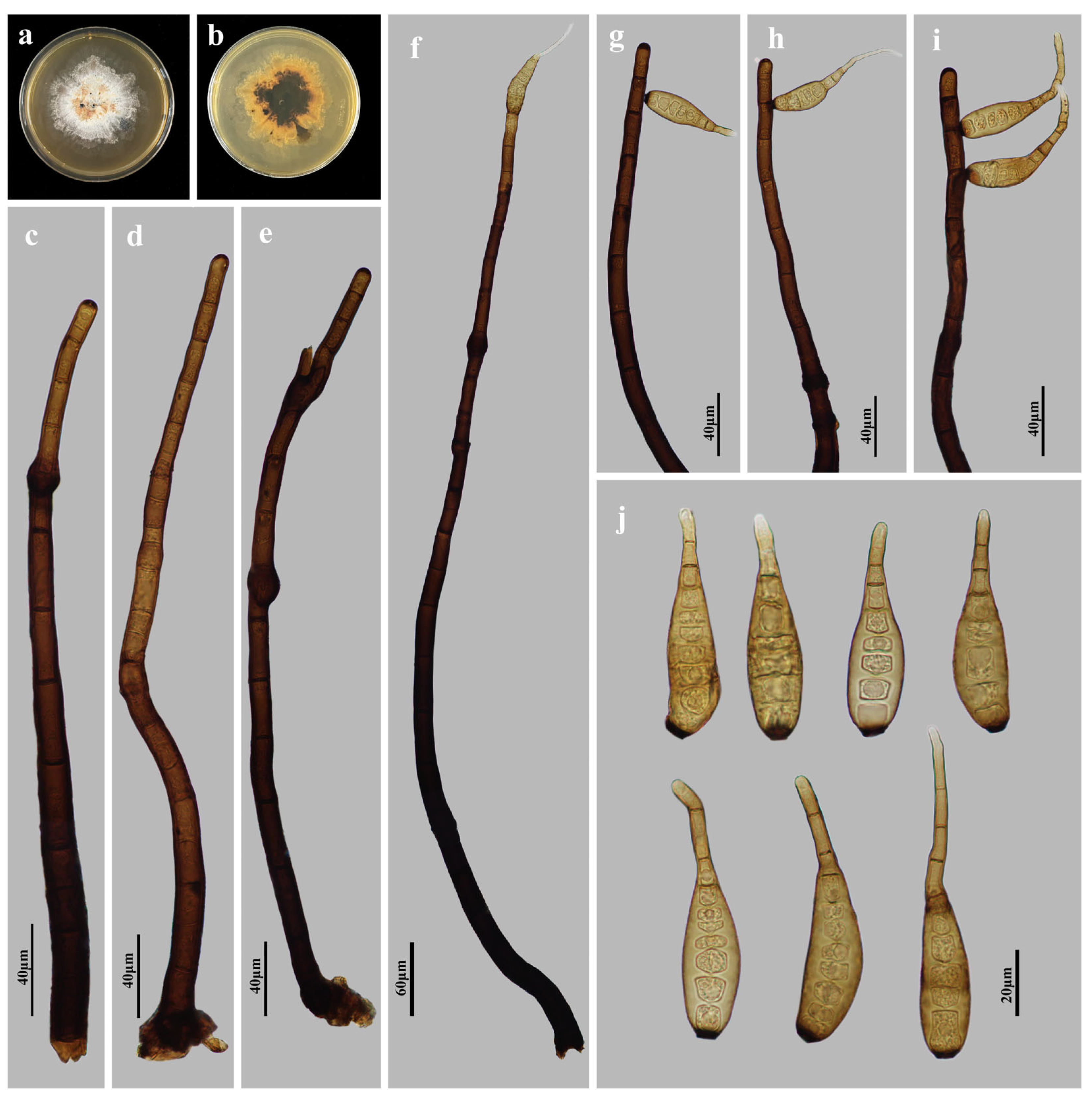
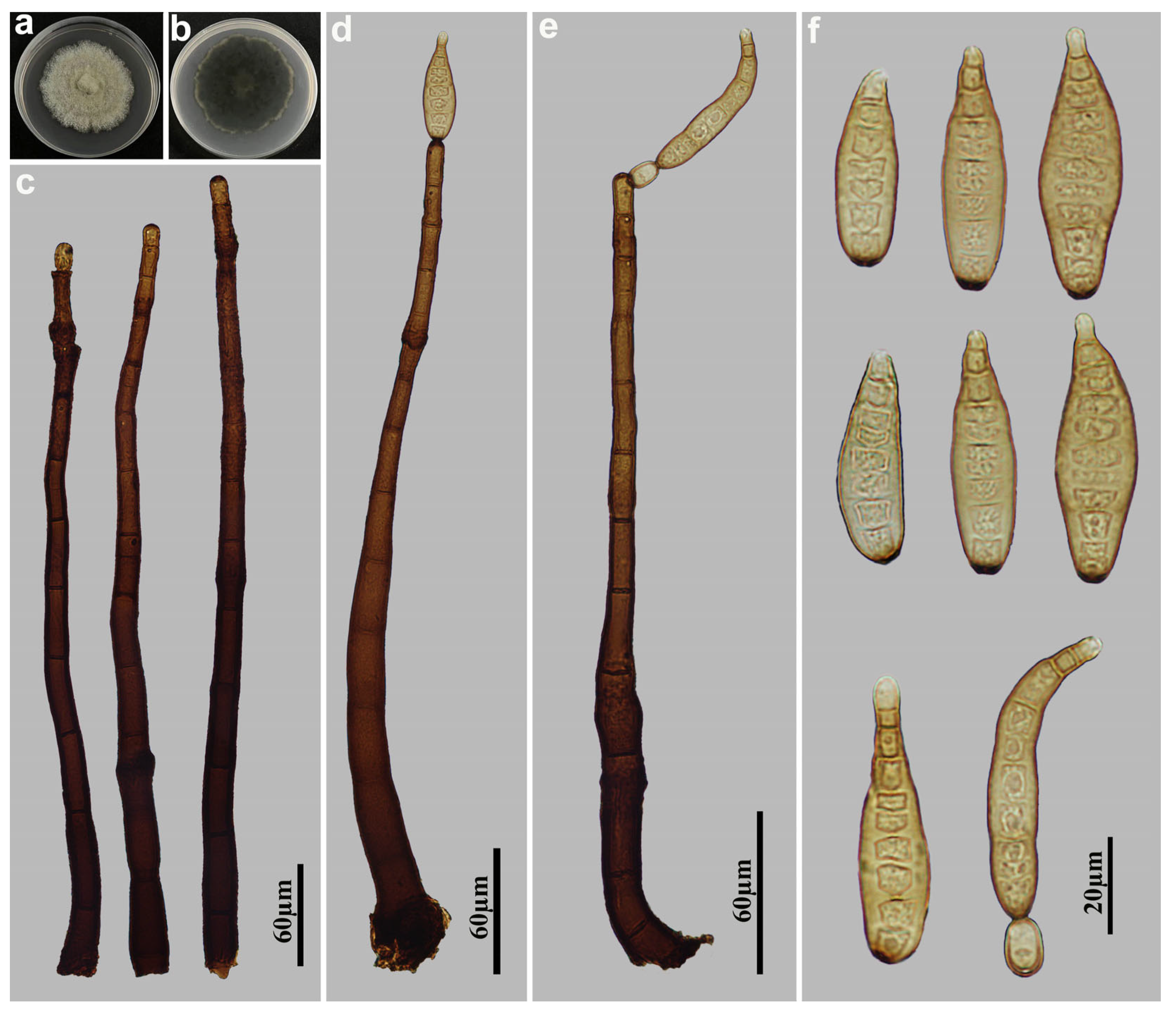
3.2. Taxonomy
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Schimann, H.; Bach, C.; Lengelle, J.; Louisanna, E.; Barantal, S.; Murat, C.; Buée, M. Diversity and structure of fungal communities in neotropical rainforest soils: The effect of host recurrence. Microb. Ecol. 2017, 73, 310–320. [Google Scholar] [CrossRef]
- Hawksworth, D.L. The fungal dimension of biodiversity: Magnitude, significance, and conservation. Mycol. Res. 1991, 95, 641–655. [Google Scholar] [CrossRef]
- Hawksworth, D.L.; Lücking, R. Fungal diversity revisited: 2.2 to 3.8 million species. Microbiol. Spectr. 2017, 5, 79–95. [Google Scholar] [CrossRef] [PubMed]
- Hyde, K.D. The numbers of fungi. Fungal Divers. 2022, 114, 1. [Google Scholar] [CrossRef]
- Phukhamsakda, C.; Nilsson, R.H.; Bhunjun, C.S.; De Farias, A.R.G.; Sun, Y.-R.; Wijesinghe, S.N.; Raza, M.; Bao, D.-F.; Lu, L.; Tibpromma, S.; et al. The numbers of fungi: Contributions from traditional taxonomic studies and challenges of metabarcoding. Fungal Divers. 2022, 114, 327–386. [Google Scholar] [CrossRef]
- Bhunjun, C.S.; Niskanen, T.; Suwannarach, N.; Wannathes, N.; Chen, Y.-J.; McKenzie, E.H.C.; Maharachchikumbura, S.S.N.; Buyck, B.; Zhao, C.-L.; Fan, Y.-G.; et al. The numbers of fungi: Are the most speciose genera truly diverse? Fungal Divers. 2022, 114, 387–462. [Google Scholar] [CrossRef]
- Su, H.-Y.; Hyde, K.D.; Maharachchikumbura, S.S.N.; Ariyawansa, H.A.; Luo, Z.-L.; Promputtha, I.; Tian, Q.; Lin, C.-G.; Shang, Q.-J.; Zhao, Y.-C.; et al. The families Distoseptisporaceae fam. nov., Kirschsteiniotheliaceae, Sporormiaceae and Torulaceae, with new species from freshwater in Yunnan Province, China. Fungal Divers. 2016, 80, 375–409. [Google Scholar] [CrossRef]
- Xia, J.-W.; Ma, Y.-R.; Li, Z.; Zhang, X.-G. Acrodictys-like wood decay fungi from southern China, with two new families Acrodictyaceae and Junewangiaceae. Sci. Rep. 2017, 7, 7888. [Google Scholar] [CrossRef]
- Yang, J.; Maharachchikumbura, S.S.N.; Liu, J.-K.; Hyde, K.D.; Jones, E.B.G.; Al-Sadi, A.M.; Liu, Z.-Y. Pseudostanjehughesia aquitropica gen. et sp. nov. and Sporidesmium sensu lato species from freshwater habitats. Mycol. Prog. 2018, 17, 591–616. [Google Scholar] [CrossRef]
- Hyde, K.D.; Tennakoon, D.S.; Jeewon, R.; Bhat, D.J.; Maharachchikumbura, S.S.N.; Rossi, W.; Leonardi, M.; Lee, H.B.; Mun, H.Y.; Houbraken, J.; et al. Fungal diversity notes 1036–1150: Taxonomic and phylogenetic contributions on genera and species of fungal taxa. Fungal Divers. 2019, 96, 1–242. [Google Scholar] [CrossRef]
- Yang, J.; Liu, L.-L.; Jones, E.B.G.; Li, W.-L.; Hyde, K.D.; Liu, Z.-Y. Morphological variety in Distoseptispora and introduction of six novel species. J. Fungi 2021, 7, 945. [Google Scholar] [CrossRef] [PubMed]
- Konta, S.; Tibpromma, S.; Karunarathna, S.C.; Samarakoon, M.C.; Steven, L.S.; Mapook, A.; Boonmee, S.; Senwanna, C.; Balasuriya, A.; Eungwanichayapant, P.D.; et al. Morphology and multigene phylogeny reveal ten novel taxa in Ascomycota from terrestrial palm substrates (Arecaceae) in Thailand. Mycosphere 2023, 14, 107–152. [Google Scholar] [CrossRef]
- Manawasinghe, I.S.; Hyde, K.D.; Wanasinghe, D.N.; Karunarathna, S.C.; Maharachchikumbura, S.S.N.; Samarakoon, M.C.; Voglmayr, H.; Pang, K.-L.; Chiang, M.W.-L.; Jones, E.B.G.; et al. Fungal diversity notes 1818–1918: Taxonomic and phylogenetic contributions on genera and species of fungi. Fungal Divers. 2025, 130, 1–261. [Google Scholar] [CrossRef]
- Liu, J.-W.; Hu, Y.-F.; Luo, X.-X.; Xu, Z.-H.; Castañeda-Ruíz, R.F.; Xia, J.-W.; Zhang, X.-G.; Zhang, L.-H.; Cui, R.-Q.; Ma, J. Morphological and phylogenetic analyses reveal three new species of Distoseptispora (Distoseptisporaceae, Distoseptisporales) from Yunnan, China. J. Fungi 2023, 9, 470. [Google Scholar] [CrossRef] [PubMed]
- Index Fungorum. Available online: http://www.indexfungorum.org/Names/Names.asp (accessed on 10 March 2025).
- Dong, W.; Hyde, K.D.; Jeewon, R.; Doilom, M.; Yu, X.-D.; Wang, G.-N.; Liu, N.-G.; Hu, D.-M.; Nalumpang, S.; Zhang, H. Towards a natural classification of annulatascaceae-like taxa II: Introducing five new genera and eighteen new species from freshwater. Mycosphere 2021, 12, 1–88. [Google Scholar] [CrossRef]
- Link, H.F. Observationes in Ordines plantarum naturales. Dissertatio prima complectens Anandrarum ordines Epiphytas, Mucedines, Gastromycos et Fungos. Ges. Natur. 1809, 3, 3–42. [Google Scholar]
- Voglmayr, H.; Jaklitsch, W.M. Corynespora, Exosporium and Helminthosporium revisited—New species and generic reclassification. Stud. Mycol. 2017, 87, 43–76. [Google Scholar] [CrossRef]
- Ellis, M.B. Dematiaceous hyphomycetes. III. Mycol. Pap. 1961, 82, 55. [Google Scholar]
- Luttrell, E.S. Systematics of Helminthosporium and related genera. Mycologia 1964, 56, 119–132. [Google Scholar] [CrossRef]
- Tanaka, K.; Hirayama, K.; Yonezawa, H.; Sato, G.; Toriyabe, A.; Kudo, H.; Hashimoto, A.; Matsumura, M.; Harada, Y.; Kurihara, Y.; et al. Revision of the Massarineae (Pleosporales, Dothideomycetes). Stud. Mycol. 2015, 82, 75–136. [Google Scholar] [CrossRef]
- Siboe, G.M.; Kirk, P.M.; Cannon, P.F. New dematiaceous hyphomycetes from Kenyan rare plants. Mycotaxon 1999, 73, 283–302. [Google Scholar]
- Hu, Y.-F.; Liu, J.-W.; Xu, Z.-H.; Castañeda-Ruíz, R.F.; Zhang, K.; Ma, J. Morphology and multigene phylogeny revealed three new species of Helminthosporium (Massarinaceae, Pleosporales) from China. J. Fungi 2023, 9, 280. [Google Scholar] [CrossRef]
- Hyde, K.D.; Norphanphoun, C.; Ma, J.; Yang, H.D.; Zhang, J.-Y.; Du, T.-Y.; Gao, Y.; De Farias, A.R.G.; He, S.-C.; He, Y.-K.; et al. Mycosphere notes 387–412–novel species of fungal taxa from around the world. Mycosphere 2023, 14, 663–744. [Google Scholar] [CrossRef]
- Hyde, K.D.; Wijesinghe, S.N.; Afshari, N.; Aumentado, H.D.; Bhunjun, C.S.; Boonmee, S.; Camporesi, E.; Chethana, K.W.T.; Doilom, M.; Dong, W.; et al. Mycosphere Notes 469–520. Mycosphere 2024, 15, 1294–1454. [Google Scholar] [CrossRef]
- Ilyukhin, E.; Markovskaja, S. Helminthosporium paraoligosporum sp. nov., a new ascomycetous fungus from Ontario, Canada. Phytotaxa 2023, 607, 151–159. [Google Scholar] [CrossRef]
- Tian, W.-H.; Jin, Y.; Liao, Y.-C.; Faraj, T.K.; Guo, X.-Y.; Maharachchikumbura, S.S.N. Phylogenetic insights reveal new taxa in Thyridariaceae and Massarinaceae. J. Fungi 2024, 10, 542. [Google Scholar] [CrossRef] [PubMed]
- Zou, M.; Al-Otibi, F.; Hyde, K.D.; Wang, Y.; Pan, X.-J. New Helminthosporium (Massarinaceae, Dothideomycetes) and Nigrospora (Incertae sedis, Sordariomycetes) species associated with walnut (Juglans regia L.) in China. MycoKeys 2024, 109, 265–284. [Google Scholar] [CrossRef]
- Lu, L.; Karunarathna, S.C.; Rajeshkumar, K.C.; Elgorban, A.M.; Jayawardena, R.S.; Hongsanan, S.; Suwannarach, N.; Kumla, J.; Xiong, Y.-R.; Hyde, K.D.; et al. Unveiling fungal diversity associated with coffee trees in China using a polyphasic approach and a global review of coffee saprobic fungi. IMA Fungus 2025, 16, e144874. [Google Scholar] [CrossRef]
- Sun, Y.-R.; Hyde, K.D.; Liu, N.-G.; Jayawardena, R.S.; Wijayawardene, N.N.; Ma, J.; Zhang, Q.; Al-Otibi, F.; Wang, Y. Micro-fungi in southern China and northern Thailand: Emphasis on medicinal plants. Fungal Divers. 2025, 131, 99–299. [Google Scholar] [CrossRef]
- Errampalli, D.; Saunders, J.M.; Holley, J.D. Emergence of silver scurf (Helminthosporium solani) as an economically important disease of potato. Plant Pathol. 2001, 50, 141–153. [Google Scholar] [CrossRef]
- Ma, J.; Wang, Y.; Ma, L.-G.; Zhang, Y.-D.; Castañeda-Ruíz, R.F.; Zhang, X.G. Three new species of Neosporidesmium from Hainan, China. Mycol. Prog. 2011, 10, 157–162. [Google Scholar] [CrossRef]
- White, T.J.; Bruns, T.D.; Lee, S.B.; Taylor, J.W. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In PCR Protocols: A Guide to Methods and Applications; Innis, M.A., Gelfand, D.H., Sninsky, J.J., White, T.J., Eds.; Academic Press: New York, NY, USA, 1990; pp. 315–322. [Google Scholar] [CrossRef]
- Sung, G.H.; Sung, J.M.; Hywel-Jones, N.L.; Spatafora, J.W. A multi-gene phylogeny of Clavicipitaceae (Ascomycota, Fungi): Identification of localized incongruence using a combinational bootstrap approach. Mol. Phylogenet. Evol. 2007, 44, 1204–1223. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.-J.; Whelen, S.; Hall, B.D. Phylogenetic relationships among ascomycetes: Evidence from an RNA polymerse II subunit. Mol. Biol. Evol. 1999, 16, 1799–1808. [Google Scholar] [CrossRef]
- Rehner, S.A.; Buckley, E. A Beauveria phylogeny inferred from nuclear ITS and EF1-alpha sequences: Evidence for cryptic diversification and links to Cordyceps teleomorphs. Mycologia 2005, 97, 84–98. [Google Scholar] [CrossRef]
- Zhao, N.; Luo, Z.-L.; Hyde, K.D.; Su, H.-Y.; Bhat, D.J.; Liu, J.-K.; Bao, D.-F.; Hao, Y.-E. Helminthosporium submersum sp. nov. (Massarinaceae) from submerged wood in north-western Yunnan Province. China. Phytotaxa 2018, 348, 269–278. [Google Scholar] [CrossRef]
- Katoh, K.; Standley, D.M. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef] [PubMed]
- Zhang, K.; Zhang, H.; Li, D.-W.; Castañeda-Ruíz, R.F. Mirohelminthosporium gen. nov. for an atypical Helminthosporium species and H. matsushimae nom. nov. Mycotaxon 2020, 135, 777–783. [Google Scholar] [CrossRef]
- Kalyaanamoorthy, S.; Minh, B.Q.; Wong, T.K.F.; von Haeseler, A.; Jermiin, L.S. ModelFinder: Fast model selection for accurate phylogenetic estimates. Nat. Methods 2017, 14, 587–589. [Google Scholar] [CrossRef]
- Nguyen, L.T.; Schmidt, H.A.; von Haeseler, A.; Minh, B.Q. IQ-TREE: A fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 2015, 32, 268–274. [Google Scholar] [CrossRef]
- Minh, B.Q.; Nguyen, M.A.; Von Haeseler, A. Ultrafast approximation for phylogenetic bootstrap. Mol. Biol. Evol. 2013, 30, 1188–1195. [Google Scholar] [CrossRef]
- Ronquist, F.; Teslenko, M.; van der Mark, P.; Ayres, D.L.; Darling, A.; Höhna, S.; Larget, B.; Liu, L.; Suchard, M.A.; Huelsenbeck, J.P. MrBayes 3.2: Efficient bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 2012, 61, 539–542. [Google Scholar] [CrossRef] [PubMed]
- Hu, Y.-F.; Liu, J.-W.; Luo, X.-X.; Xu, Z.-H.; Xia, J.-W.; Zhang, X.-G.; Castañeda-Ruíz, R.F.; Ma, J. Multi-locus phylogenetic analyses reveal eight novel species of Distoseptispora from southern China. Microbiol. Spectr. 2023, 11, e0246823. [Google Scholar] [CrossRef]
- Liao, M.-G.; Luo, X.-X.; Hu, Y.-F.; Castañeda-Ruíz, R.F.; Xu, Z.-H.; Ma, J. Morphological and phylogenetic analyses reveal four novel species of Distoseptispora (Distoseptisporaceae, Distoseptisporales) from southern China. MycoKeys 2025, 113, 31–55. [Google Scholar] [CrossRef]
- Chen, X.-M.; Tang, X.; Ma, J.; Liu, N.-G.; Tibpromma, S.; Karunarathna, S.C.; Xiao, Y.-P.; Lu, Y.-Z. Identification of two new species and a new host record of Distoseptispora (Distoseptisporaceae, Distoseptisporales, Sordariomycetes) from terrestrial and freshwater habitats in Southern China. MycoKeys 2024, 102, 83–105. [Google Scholar] [CrossRef]
- De Bory, S.V.; De Durieu, M. Exploration Scientifique de l’Algérie Pendant les Années 1840, 1841, 1842 Publiée par Ordre du Gouvernement et avec le Concours d’une Commission Académique; Sciences physiques; Botanique par MM Bory de St-.Vincent et Durieu de Maisonneuve membres de la Commission scientifique d’Algérie; Imprimerie Nationale: Paris, France, 1846–1869. [Google Scholar]
- Zhang, Z.-X.; Shang, Y.-X.; Liu, Q.-Y.; Li, D.-H.; Yin, X.-Y.; Liu, X.-Y.; Tao, M.-F.; Jiang, Y.; Wang, Y.-X.; Zhang, M.-Y.; et al. Deciphering the evolutionary and taxonomic complexity of Diaporthales (Sordariomycetes, Ascomycota) through integrated phylogenomic and divergence time estimation. Fungal Divers. 2025, 132, 1–125. [Google Scholar] [CrossRef]
- Liu, N.-G.; Hyde, K.D.; Sun, Y.-R.; Bhat, D.J.; Jones, E.B.G.; Jumpathong, J.J.; Lin, C.-G.; Lu, Y.-Z.; Yang, J.; Liu, J.-J.; et al. Notes, outline, taxonomy and phylogeny of brown-spored hyphomycetes. Fungal Divers. 2024, 129, 1–281. [Google Scholar] [CrossRef]
- Liu, J.-W.; Hu, Y.-F.; Luo, X.-X.; Castañeda-Ruíz, R.F.; Xia, J.-W.; Xu, Z.-H.; Cui, R.-Q.; Shi, X.-G.; Zhang, L.-H.; Ma, J. Molecular Phylogeny and Morphology Reveal Four Novel Species of Corynespora and Kirschsteiniothelia (Dothideomycetes, Ascomycota) from China: A Checklist for Corynespora Reported Worldwide. J. Fungi 2023, 9, 107. [Google Scholar] [CrossRef] [PubMed]
- Tian, W.-H.; Jin, Y.; Liao, Y.-C.; Faraj, T.K.; Guo, X.-Y.; Maharachchikumbura, S.S.N. New and Interesting Pine-Associated Hyphomycetes from China. J. Fungi 2024, 10, 546. [Google Scholar] [CrossRef]
- Luo, X.-X.; Liao, M.-G.; Hu, Y.-F.; Zhang, X.-G.; Xu, Z.-H.; Ma, J. Identification of three novel species and one new record of Kirschsteiniothelia (Kirschsteiniotheliaceae, Kirschsteiniotheliales) from Jiangxi, China. MycoKeys 2025, 112, 277–306. [Google Scholar] [CrossRef]
- Luo, Z.-L.; Hyde, K.D.; Liu, J.-K.; Maharachchikumbura, S.S.N.; Jeewon, R.; Bao, D.-F.; Bhat, D.J.; Lin, C.-G.; Li, W.-L.; Yang, J.; et al. Freshwater Sordariomycetes. Fungal Divers. 2019, 99, 451–660. [Google Scholar] [CrossRef]
- Sun, Y.; Goonasekara, I.D.; Thambugala, K.M.; Jayawardena, R.S.; Wang, Y.; Hyde, K.D. Distoseptispora bambusae sp. nov. (Distoseptisporaceae) on bamboo from China and Thailand. Biodivers. Data J. 2020, 8, e53678. [Google Scholar] [CrossRef] [PubMed]
- Shen, H.-W.; Bao, D.-F.; Boonmee, S.; Lu, Y.-Z.; Su, X.-J.; Li, Y.-X.; Luo, Z.-L. Diversity of Distoseptispora (Distoseptisporaceae) taxa on submerged decaying wood from the Red River in Yunnan, China. MycoKeys 2024, 102, 1–28. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Tian, W.; Guo, Y.; Madrid, H.; Maharachchikumbura, S.S.N. Synhelminthosporium gen. et sp. nov. and Two New Species of Helminthosporium (Massarinaceae, Pleosporales) from Sichuan Province, China. J. Fungi 2022, 8, 712. [Google Scholar] [CrossRef]
- Hassani, M.A.; Durán, P.; Hacquard, S. Microbial interactions within the plant holobiont. Microbiome 2018, 6, 58. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Yan, C.-L.; Tang, Y.-Q.; Ma, X.; Chen, Y.-H.; Chen, S.-B.; Lin, M.; Liu, Z. Endophytic bacterial and fungal microbiota in different cultivars of cassava (Manihot esculenta Crantz). J. Microbiol. 2020, 58, 614–623. [Google Scholar] [CrossRef]
- Ivanyuk, V.; Zezyulina, G. Helminthosporium solani causing silver scab is reported from Belarus. Zashch. Rastenii 1991, 3, 45. [Google Scholar]
- El Immane-Collet, R.; Elakel, M.; Jouan, B. Comparative study of the agronomical incidence of the silver scurf disease of potato in Morocco and in France. Al Awamia 1995, 91, 1–8. [Google Scholar]




| Locus | Initial Denaturation | Denaturation | Annealing | Elongation | Final Extension |
|---|---|---|---|---|---|
| ITS | 94 °C/3 min | 94 °C/15 s | 54 °C/15 s | 72 °C/30 s | 72 °C/10 min |
| LSU | 94 °C/3 min | 94 °C/15 s | 54 °C/15 s | 72 °C/30 s | 72 °C/10 min |
| RPB2 | 94 °C/3 min | 94 °C/15 s | 59 °C/50 s | 72 °C/30 s | 72 °C/10 min |
| SSU | 94 °C/3 min | 94 °C/15 s | 54 °C/15 s | 72 °C/30 s | 72 °C/10 min |
| TEF1 | 94 °C/3 min | 94 °C/15 s | 54 °C/15 s | 72 °C/30 s | 72 °C/10 min |
| Taxon | Strain Number | GenBank Accession Numbers | |||
|---|---|---|---|---|---|
| ITS | LSU | RPB2 | TEF1 | ||
| Acrodictys bambusicola | CGMCC 3.18641 T | KU999973 | KX033564 | — | — |
| A. elaeidicola | CGMCC 3.18642 | KU999978 | KX033569 | — | — |
| Aquapteridospora aquatica | MFLUCC 17–2371 T | MW286493 | MW287767 | — | — |
| A. fusiformis | MFLUCC 18–1606 T | MK828652 | MK849798 | — | MN194056 |
| A. lignicola | MFLUCC 15–0377 T | MZ868774 | KU221018 | MZ892986 | MZ892980 |
| Bullimyces aurisporus | AF316–1b T | — | JF775590 | — | — |
| B. communis | AF281–5 | — | JF775587 | — | — |
| Cancellidium applanatum | CBS 337.76 T | MH860985 | MH872755 | — | — |
| Cancellidium cinereum | MFLUCC 18–0424 T | MT370353 | MT370363 | MT370486 | MT370488 |
| Distoseptispora adscendens | HKUCC 10820 | — | DQ408561 | DQ435092 | — |
| D. amniculi | MFLUCC 17–2129 T | MZ868770 | MZ868761 | MZ892982 | — |
| D. appendiculata | MFLUCC 18–0259 T | MN163009 | MN163023 | — | MN174866 |
| D. aqualignicola | KUNCC 21–10729 T | OK341186 | ON400845 | OP413474 | OP413480 |
| D. aquamyces | KUNCC 21–10732 T | OK341187 | OK341199 | OP413476 | OP413482 |
| D. aquatica | MFLUCC 18–0646 | MK828648 | MK849793 | — | MN194052 |
| D. aquisubtropica | GZCC 22–0075 T | ON527933 | ON527941 | ON533685 | ON533677 |
| D. arecacearum | MFLUCC 23–0212 | OR354399 | OR510860 | OR481048 | OR481045 |
| D. atroviridis | GZCC 20–0511 T | MZ868772 | MZ868763 | MZ892984 | MZ892978 |
| D. atroviridis | GZCC 19–0531 | MW133915 | MZ227223 | — | MZ206155 |
| D. bambusae | MFLUCC 20–0091 T | MT232713 | MT232718 | MT232881 | MT232880 |
| D. bambusicola | GZCC 21–0667 T | MZ474873 | MZ474872 | — | OM272845 |
| D. bangkokensis | MFLUCC 18–0262 T | MZ518205 | MZ518206 | — | OK067246 |
| D. cangshanensis | MFLUCC 16–0970 T | MG979754 | MG979761 | — | MG988419 |
| D. caricis | CPC 36498 T | MN562124 | MN567632 | MN556805 | — |
| D. chinensis | GZCC 21–0665 T | MZ474871 | MZ474867 | — | MZ501609 |
| D. clematidis | MFLUCC 17–2145 T | MT310661 | MT214617 | MT394721 | — |
| D. chengduensis | CGMCC 3.27439 T | PQ067828 | PQ067744 | — | PQ278565 |
| D. chiangraiensis | MFLU 21-0105 T | MZ890145 | MZ890139 | — | MZ892970 |
| D. chishuiensis | GZCC 23-0729 T | PP663310 | PP584670 | — | PP584767 |
| D. crassispora | KUMCC 21–10726 T | OK310698 | OK341196 | OP413473 | OP413479 |
| D. curvularia | KUMCC 21–10725 T | OK310697 | OK341195 | OP413472 | OP413478 |
| D. cylindricospora | DLUCC 1906 T | OK491122 | OK513523 | — | OK524220 |
| D. dehongensis | KUMCC 18–0090 T | MK085061 | MK079662 | — | MK087659 |
| D. dipterocarpi | MFLUCC 22–0104 T | OP600053 | OP600052 | OP595140 | — |
| D. effusa | GZCC 19–0532 T | MW133916 | MZ227224 | — | MZ206156 |
| D. eleiodoxae | MFLUCC 23–0214 | OR354398 | OR510859 | OR481047 | OR481044 |
| D. euseptata | MFLUCC 20–0154 T | MW081539 | MW081544 | MW151860 | — |
| D. euseptata | DLUCC S2024 | MW081540 | MW081545 | MW084996 | MW084994 |
| D. fasciculata | KUMCC 19–0081 T | MW286501 | MW287775 | — | MW396656 |
| D. fluminicola | DLUCC 0391 | MG979755 | MG979762 | — | MG988420 |
| D. fluminicola | DLUCC 0999 | MG979756 | MG979763 | — | MG988421 |
| D. fujianensis | HJAUP C2509 T | PQ211095 | PQ211103 | PQ303679 | PQ303682 |
| D. fusiformis | GZCC 20–0512 T | MZ868773 | MZ868764 | MZ892985 | MZ892979 |
| D. ganzhouensis | HJAUP C1090 T | PQ211100 | PQ211108 | — | PQ303687 |
| D. gasaensis | HJAUP C2034 T | OQ942896 | OQ942891 | — | OQ944455 |
| D. guanshanensis | HJAUP C1063 T | OQ942894 | OQ942898 | OQ944458 | OQ944452 |
| D. guizhouensis | GZCC 21–0666 T | MZ474868 | MZ474869 | MZ501611 | MZ501610 |
| D. guttulata | MFLU 17–0852 T | MF077543 | MF077554 | — | MF135651 |
| D. hainanensis | GZCC 22-2047 T | OR427328 | OR438894 | OR449119 | OR449122 |
| D. hyalina | MFLUCC 17–2128 T | MZ868769 | MZ868760 | MZ892981 | MZ892976 |
| D. hydei | MFLUCC 20–0481 T | MT734661 | MT742830 | MT767128 | — |
| D. jinghongensis | HJAUP C2120 T | OQ942897 | OQ942893 | — | OQ944456 |
| D. lancangjiangensis | DLUCC 1864 T | MW723055 | MW879522 | — | — |
| D. lanceolatispora | GZCC 22-2045 T | OR427329 | OR43BB95 | OR449120 | OR449123 |
| D. leonensis | HKUCC 10822 | — | DQ408566 | DQ435089 | — |
| D. licualae | MFLUCC 14–1163A T | ON650686 | ON650675 | — | ON734007 |
| D. licualae | MFLUCC 14–1163B T | ON650687 | ON650676 | — | ON734008 |
| D. licualae | MFLUCC 14–1163C T | ON650688 | ON650677 | — | — |
| D. lignicola | MFLUCC 18–0198 T | MK828651 | MK849797 | — | — |
| D. lignicola | GZCC 19–0529 | MW133911 | MZ227219 | — | MZ206152 |
| D. liupanshuiensis | GZCC 23-0730 T | PP663309 | PP584669 | — | PP584766 |
| D. longispora | HFJAU 0705 T | MH555359 | MH555357 | — | — |
| D. longnanensis | HJAUP C1040 T | OQ942887 | OQ942886 | — | OQ944451 |
| D. martinii | CGMCC 3.18651 T | KU999975 | KX033566 | — | — |
| D. meilingensis | JAUCC 4727 T | OK562390 | OK562396 | — | OK562408 |
| D. meilingensis | JAUCC 4728 T | OK562391 | OK562397 | — | OK562409 |
| D. menghaiensis | HJAUP C2045 T | OQ942890 | OQ942900 | — | — |
| D. menglunensis | HJAUP C2170 T | OQ942899 | OQ942888 | OQ944461 | OQ944457 |
| D. mengsongensis | HJAUP C2126 T | OP787876 | OP787874 | — | OP961937 |
| D. motuoensis | KUNCC24-17628 T | PP600327 | PP621731 | — | PP639546 |
| D. muchuanensis | CGMCC 3.27444 T | PQ067834 | PQ067750 | — | PQ278571 |
| D. multiseptata | MFLUCC 15–0609 T | KX710145 | KX710140 | — | MF135659 |
| D. multiseptata | MFLU 17–0856 | MF077544 | MF077555 | MF135644 | MF135652 |
| D. nabanheensis | HJAUP C2003 T | OP787873 | OP787877 | — | OP961935 |
| D. nanchangensis | HJAUP C1074 T | OQ942889 | OQ942895 | OQ944460 | OQ944454 |
| D. nanpingensis | HJAUP C2517 T | PQ211096 | PQ211104 | PQ303678 | — |
| D. narathiwatensis | MFLUCC 23–0216 | OR354400 | OR510861 | OR481049 | OR481046 |
| D. neorostrata | MFLUCC 18–0376 T | MN163008 | MN163017 | — | — |
| D. nonrostrata | KUNCC 21–10730 T | OK310699 | OK341198 | OP413475 | OP413481 |
| D. obclavata | MFLUCC 18–0329 T | MN163012 | MN163010 | — | — |
| D. obpyriformis | MFLUCC 17–1694 T | — | MG979764 | MG988415 | MG988422 |
| D. obpyriformis | DLUCC 0867 | MG979757 | MG979765 | MG988416 | MG988423 |
| D. pachyconidia | KUMCC 21–10724 T | OK310696 | OK341194 | OP413471 | OP413477 |
| D. palmarum | MFLUCC 18–1446 T | MK085062 | MK079663 | MK087670 | MK087660 |
| D. phangngaensis | MFLUCC 16–0857 T | MF077545 | MF077556 | — | MF135653 |
| D. phragmiticola | GUCC 220201 T | OP749887 | OP749880 | OP752699 | OP749891 |
| D. phragmiticola | GUCC 220202 T | OP749888 | OP749881 | OP752700 | OP749892 |
| D. rayongensis | MFLUCC 18–0415 T | MH457172 | MH457137 | MH463255 | MH463253 |
| D. rayongensis | MFLUCC 18–0417 | MH457173 | MH457138 | MH463256 | MH463254 |
| D. rostrata | MFLUCC 16–0969 T | MG979758 | MG979766 | MG988417 | MG988424 |
| D. rostrata | DLUCC 0885 | MG979759 | MG979767 | — | MG988425 |
| D. saprophytica | MFLUCC 18–1238 T | MW286506 | MW287780 | MW504069 | MW396651 |
| D. septata | GZCC 22–0078 T | ON527939 | ON527947 | ON533690 | ON533683 |
| D. sinensis | HJAUP C2044 T | OP787878 | OP787875 | — | OP961936 |
| D. sichuanensis | KUNCC 23-15519 T | PP584672 | PP584769 | — | PP663312 |
| D. songkhlaensis | MFLUCC 18–1234 T | MW286482 | MW287755 | — | MW396642 |
| D. suae | CGMCC 3.24262 T | OQ874968 | OQ732679 | OQ870341 | OR367670 |
| D. subtropica | HJAUP C2528 T | PQ211099 | PQ211107 | PQ303677 | PQ303684 |
| D. suoluoensis | MFLUCC 17–0224 T | MF077546 | MF077557 | — | MF135654 |
| D. suoluoensis | MFLUCC 17–0854 | MF077547 | MF077558 | MZ945510 | — |
| D. tectonae | MFLUCC 12–0291 T | KX751711 | KX751713 | KX751708 | KX751710 |
| D. tectonae | MFLU 20–0262 | MT232714 | MT232719 | — | — |
| D. tectonigena | MFLUCC 12–0292 T | KX751712 | KX751714 | KX751709 | — |
| D. terrestris | HJAUP C2539 T | PV448667 | PV450538 | — | PV469764 |
| D. thailandica | MFLUCC 16–0270 T | MH275060 | MH260292 | — | MH412767 |
| D. thysanolaenae | KUN–HKAS 112710 | MW723057 | MW879524 | — | MW729783 |
| D. thysanolaenae | KUN–HKAS 102247 T | MK045851 | MK064091 | — | MK086031 |
| D. tropica | GZCC 22–0076 T | ON527935 | ON527943 | ON533687 | ON533679 |
| D. verrucosa | GZCC 20–0434 T | MZ868771 | MZ868762 | MZ892983 | MZ892977 |
| D. wuyishanensis | HJAUP C2515 T | PV448666 | PV450537 | PV469759 | PV469763 |
| D. wuzhishanensis | GZCC 22–0077 T | ON527938 | ON527946 | — | ON533682 |
| D. xingpingensis | KUNCC 22–12669 | OQ874969 | OQ732680 | — | — |
| D. xingpingensis | KUNCC 22–12667 T | OQ874970 | OQ732681 | OQ870340 | OR367671 |
| D. xishuangbannaensis | KUMCC 17–0290 T | MH275061 | MH260293 | MH412754 | MH412768 |
| D. yichunensis | HJAUP C1065 T | OQ942885 | OQ942892 | OQ944459 | OQ944453 |
| D. yongxiuensis | JAUCC 4725 T | OK562388 | OK562394 | — | OK562406 |
| D. yongxiuensis | JAUCC 4726 T | OK562389 | OK562395 | — | OK562407 |
| D. yunjushanensis | JAUCC 4723 T | OK562392 | OK562398 | — | OK562410 |
| D. yunjushanensis | JAUCC 4724 T | OK562393 | OK562399 | — | OK562411 |
| D. yunnansis | MFLUCC 20–0153 T | MW081541 | MW081546 | MW151861 | MW084995 |
| D. zhejiangensis | HJAUP C2588 T | PV448668 | PV450539 | — | PV469765 |
| Fluminicola saprophytica | MFLUCC 15–0976 T | MF374358 | MF374367 | MF370954 | MF370956 |
| Myrmecridium banksiae | CBS 132536 T | JX069871 | JX069855 | — | — |
| M. schulzeri | CBS 100.54 | EU041769 | EU041826 | — | — |
| Papulosa amerospora | AFTOL–ID 748 | — | DQ470950 | DQ470901 | DQ471069 |
| Pleurophragmium bambusinum | MFLUCC 12–0850 | KU940161 | KU863149 | — | KU940213 |
| Pseudostanjehughesia aquitropica | MFLUCC 16–0569 T | MF077548 | MF077559 | — | MF135655 |
| P. lignicol | MFLUCC 15–0352 T | MK828643 | MK849787 | MN124534 | MN194047 |
| Wongia griffinii | DAR 80512 T | KU850473 | KU850471 | — | — |
| Taxon | Strain Number | GenBank Accession Numbers | ||||
|---|---|---|---|---|---|---|
| ITS | LSU | RPB2 | SSU | TEF1 | ||
| Byssothecium circinans | CBS 675.92 | OM337536 | GU205217 | DQ767646 | GU205235 | GU349061 |
| Corynespora cassiicola | CBS 100822 | — | GU301808 | GU371742 | GU296144 | GU349052 |
| C. smithii | L120 | KY984297 | KY984297 | KY984361 | — | KY984435 |
| C. smithii | L130 | KY984298 | KY984298 | KY984362 | KY984419 | KY984436 |
| Cyclothyriella rubronotata | CBS 121892 | KX650541 | KX650541 | KX650571 | — | KX650516 |
| C. rubronotata | CBS 141486 T | KX650544 | KX650544 | KX650574 | KX650507 | KX650519 |
| Helminthosporium aquaticum | MFLUCC 15–0357 T | KU697302 | KU697306 | — | KU697310 | — |
| H. austriacum | CBS 139924 T | KY984301 | KY984301 | KY984365 | KY984420 | KY984437 |
| H. austriacum | CBS 142388 | KY984303 | KY984303 | KY984367 | — | KY984439 |
| H. caespitosum | CBS 484.77 T | JQ044429 | JQ044448 | KY984370 | KY984421 | KY984440 |
| H. caespitosum | L141 | KY984305 | KY984305 | KY984368 | — | — |
| H. chengduense | UESTC 22.0024 T | ON557751 | ON557745 | ON563073 | ON557757 | ON600598 |
| H. chengduense | UESTC 22.0025 | ON557750 | ON557744 | ON563072 | ON557756 | ON600597 |
| H. chiangraiense | MFLUCC 21–0087 T | MZ538504 | MZ538538 | — | — | — |
| H. chinense | CGMCC 3.23570 T | ON557754 | ON557748 | — | ON557760 | ON600601 |
| H. chlorophorae | BRIP 14521 | AF120259 | — | — | — | — |
| H. dalbergiae | MAFF 243853 | LC014555 | AB807521 | — | AB797231 | AB808497 |
| H. endiandrae | CBS 138902 T | KP004450 | KP004478 | — | — | — |
| H. erythrinicola | CBS 145569 T | NR_165563 | MK876432 | MK876486 | — | — |
| H. filamentosum | UESTCC 24.0132 T | PP835322 | PP835316 | PP844886 | PP835319 | PP844888 |
| H. ganzhouense | HJAUP C1086 T | PV448665 | PV450532 | PP501323 | PV450540 | PP501325 |
| H. genistae | CBS 139922 | KY984309 | KY984309 | KY984373 | KY984423 | — |
| H. genistae | CBS 142597 T | KY984310 | KY984310 | KY984374 | — | — |
| H. genistae | CBS 139927 | KY984311 | KY984311 | KY984375 | — | — |
| H. genistae | CBS 139928 | KY984312 | KY984312 | KY984376 | — | — |
| H. genistae | CBS 139929 | KY984315 | KY984315 | KY984379 | — | — |
| H. genistae | CBS 139930 | KY984316 | KY984316 | KY984380 | — | — |
| H. guanshanense | HJAUP C1022 T | OQ172249 | OQ172239 | OQ234978 | OQ172247 | OQ256247 |
| H. guizhouense | GUCC24–0011 T | PP915799 | PP949847 | PP947940 | PP949912 | — |
| H. guizhouense | HKAS 130313 | PP981907 | PP981904 | — | PP977159 | PQ041228 |
| H. hispanicum | CBS 136917 T | KY984318 | KY984318 | KY984381 | KY984424 | KY984441 |
| H. italicum | MFLUCC 17–0241 T | KY797638 | KY815015 | — | — | KY815021 |
| H. jiangxiense | HJAUP C1325 T | PV448662 | PV450534 | – | PV450542 | PV469760 |
| H. jiulianshanense | HJAUP C1057 T | OQ172245 | OQ172253 | OQ234979 | — | — |
| H. juglandinum | CBS 136912 | KY984319 | KY984319 | KY984382 | — | KY984442 |
| H. juglandinum | CBS 136913 | KY984320 | KY984320 | — | — | — |
| H. juglandinum | CBS 136922 T | KY984321 | KY984321 | KY984383 | — | KY984443 |
| H. juglandinum | CBS 136911 | KY984322 | KY984322 | — | KY984425 | — |
| H. leucadendri | CBS 135133 T | KF251150 | KF251654 | KF252159 | — | KF253110 |
| H. lignicola | MFLUCC 22–0118 T | — | OP740252 | OP757656 | OP740253 | OP757657 |
| H. livistonae | CBS144413 T | NR_160348 | NG_064539 | — | — | — |
| H. magnisporum | MAFF 239278 T | AB811452 | AB807522 | — | AB797232 | AB808498 |
| H. massarinum | JCM 13094 | AB809628 | AB807523 | — | AB797233 | AB808499 |
| H. massarinum | CBS 139690 T | AB809629 | AB807524 | — | AB797234 | AB808500 |
| H. meilingense | HJAUP C1076 T | OQ172244 | OQ172238 | OQ234980 | OQ172246 | OQ234981 |
| H. microsorum | L94 | KY984327 | KY984327 | KY984388 | KY984426 | KY984446 |
| H. microsorum | L95 | KY984328 | KY984328 | KY984389 | — | KY984447 |
| H. microsorum | CBS 136910 T | KY984329 | KY984329 | KY984390 | KY984427 | KY984448 |
| H. nabanhense | HJAUP C2054 T | OP555394 | OP555398 | — | OP555400 | OP961931 |
| H. nanjingensis | HHAUF020380 | KF192322 | — | — | — | — |
| H. oligosporum | CBS 136908 | KY984332 | KY984332 | KY984393 | KY984428 | KY984450 |
| H. oligosporum | CBS 136909 T | KY984333 | KY984333 | KY984394 | — | KY984451 |
| H. paraoligosporum | EI–418 | ON206009 | OP888100 | — | — | — |
| H. puerensis | KUNCC 24–18347 T | OR428391 | OR428413 | OR515515 | OR428371 | OR509745 |
| H. pini | HKAS 135177 T | PP835323 | PP835317 | PP844887 | PP835320 | PP844889 |
| H. quercinum | CBS 112393 | KY984334 | KY984334 | KY984395 | — | KY984452 |
| H. quercinum | CBS 136915 | KY984336 | KY984336 | KY984397 | — | — |
| H. quercinum | CBS 136921 T | KY984339 | KY984339 | KY984400 | KY984429 | KY984453 |
| H. saprophyticum | HJAUP C2572 T | PV448663 | PV450536 | PV469758 | PV450544 | PV469761 |
| H. saprophyticum | HJAUP C2573 | PV448664 | PV450535 | – | PV450543 | PV469762 |
| H. shangrilaense | KUNCC 22–12540 T | OP767128 | OP767126 | — | OP767127 | — |
| H. sinense | HJAUP C2121 T | OP555393 | OP555397 | — | OP555399 | OP961932 |
| H. solani | CBS 365.75 | KY984341 | KY984341 | KY984402 | KY984430 | KY984455 |
| H. solani | CBS 640.85 | KY984342 | KY984342 | KY984403 | — | — |
| H. submersum | MFLUCC 16–1360 T | — | MG098787 | — | MG098796 | MG098586 |
| H. submersum | MFLUCC 16–1290 | MG098780 | MG098788 | MG098592 | MG098797 | MG098587 |
| H. syzygii | CBS 145570 T | NR_165564 | MK876433 | MK876487 | — | — |
| H. thailandicum | MFLU 24-0275 T | PQ325264 | PQ578298 | — | PQ651973 | — |
| H. tiliae | CBS 136906 | KY984344 | KY984344 | KY984405 | — | — |
| H. tiliae | CBS 136907 T | KY984345 | KY984345 | KY984406 | KY984431 | KY984457 |
| H. velutinum | H 4626 | LC014556 | AB807530 | — | AB797240 | AB808505 |
| H. velutinum | CBS 136924 | KY984347 | KY984347 | KY984408 | — | KY984458 |
| H. velutinum | CBS 139923 T | KY984352 | KY984352 | KY984413 | KY984432 | KY984463 |
| H. velutinum | L98 | KY984359 | KY984359 | KY984417 | KY984433 | KY984466 |
| H. velutinum | HJAUP C1289 | PV448661 | PV450533 | PP501324 | PV450541 | PP501326 |
| H. yunnanense | HJAUP C2071 T | OP555395 | OP555396 | OP961934 | OP555392 | OP961933 |
| Massarina cisti | CBS 266.62 T | LC014568 | AB807539 | — | AB797249 | AB808514 |
| M. eburnea | CBS 473.64 | AF383959 | GU301840 | GU371732 | AF164367 | GU349040 |
| M. eburnea | CBS 139697 | LC014569 | AB521735 | — | AB521718 | AB808517 |
| Periconia byssoides | MAFF243872 | LC014581 | AB807570 | — | AB797280 | AB808546 |
| P. digitata | CBS 510.77 | LC014584 | AB807561 | — | AB797271 | AB808537 |
| P. macrospinosa | CBS 135663 | KP183999 | KP184038 | — | KP184080 | — |
| P. pseudodigitata | CBS139699 | NR_153490 | NG_059396 | — | NG_064850 | AB808540 |
| Pseudosplanchnonema phorcioides | CBS 122935 | KY984360 | KY984360 | KY984418 | KY984434 | KY984467 |
| Stagonospora paludosa | CBS 135088 | KF251257 | KF251760 | KF252262 | — | KF253207 |
| S. perfecta | MAFF 239609 | AB809642 | AB807579 | — | AB797289 | AB808555 |
| S. pseudoperfecta | CBS 120236 T | AB809641 | AB807577 | — | AB797287 | AB808553 |
| S. tainanensis | MAFF 243860 | AB809643 | AB807580 | — | AB797290 | AB808556 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liao, M.-G.; Luo, X.-X.; Xia, J.-W.; Hu, Y.-F.; Zhang, X.-G.; Zhang, L.-H.; Zhang, X.-P.; Xu, Z.-H.; Ma, J. Six Novel Species of Distoseptispora (Distoseptisporaceae, Distoseptisporales) and Helminthosporium (Massarinaceae, Pleosporales) Isolated from Terrestrial Habitats in Southern China. J. Fungi 2025, 11, 494. https://doi.org/10.3390/jof11070494
Liao M-G, Luo X-X, Xia J-W, Hu Y-F, Zhang X-G, Zhang L-H, Zhang X-P, Xu Z-H, Ma J. Six Novel Species of Distoseptispora (Distoseptisporaceae, Distoseptisporales) and Helminthosporium (Massarinaceae, Pleosporales) Isolated from Terrestrial Habitats in Southern China. Journal of Fungi. 2025; 11(7):494. https://doi.org/10.3390/jof11070494
Chicago/Turabian StyleLiao, Ming-Gen, Xing-Xing Luo, Ji-Wen Xia, Ya-Fen Hu, Xiu-Guo Zhang, Lian-Hu Zhang, Xian-Peng Zhang, Zhao-Huan Xu, and Jian Ma. 2025. "Six Novel Species of Distoseptispora (Distoseptisporaceae, Distoseptisporales) and Helminthosporium (Massarinaceae, Pleosporales) Isolated from Terrestrial Habitats in Southern China" Journal of Fungi 11, no. 7: 494. https://doi.org/10.3390/jof11070494
APA StyleLiao, M.-G., Luo, X.-X., Xia, J.-W., Hu, Y.-F., Zhang, X.-G., Zhang, L.-H., Zhang, X.-P., Xu, Z.-H., & Ma, J. (2025). Six Novel Species of Distoseptispora (Distoseptisporaceae, Distoseptisporales) and Helminthosporium (Massarinaceae, Pleosporales) Isolated from Terrestrial Habitats in Southern China. Journal of Fungi, 11(7), 494. https://doi.org/10.3390/jof11070494








