Usefulness of Serum as a Non-Invasive Sample for the Detection of Histoplasma capsulatum Infections: Retrospective Comparative Analysis of Different Diagnostic Techniques and Quantification of Host Biomarkers
Abstract
1. Introduction
2. Materials and Methods
2.1. Patients and Clinical Samples
2.2. Antigen Detection
- Detection of Histoplasma GM (EIA test kit)
- Detection of Histoplasma capsulatum by PlateliaTM Aspergillus Ag (Bio-Rad)
2.3. Antibody Detection
2.4. DNA Detection
2.5. Biomarkers Quantification
2.6. Statistical Analysis
3. Results
3.1. Patients and Clinical Samples
3.2. Antigen Detection
3.3. Antibody Detection
3.4. DNA Detection
3.5. Comparative Analysis of Tools Used for Diagnostic
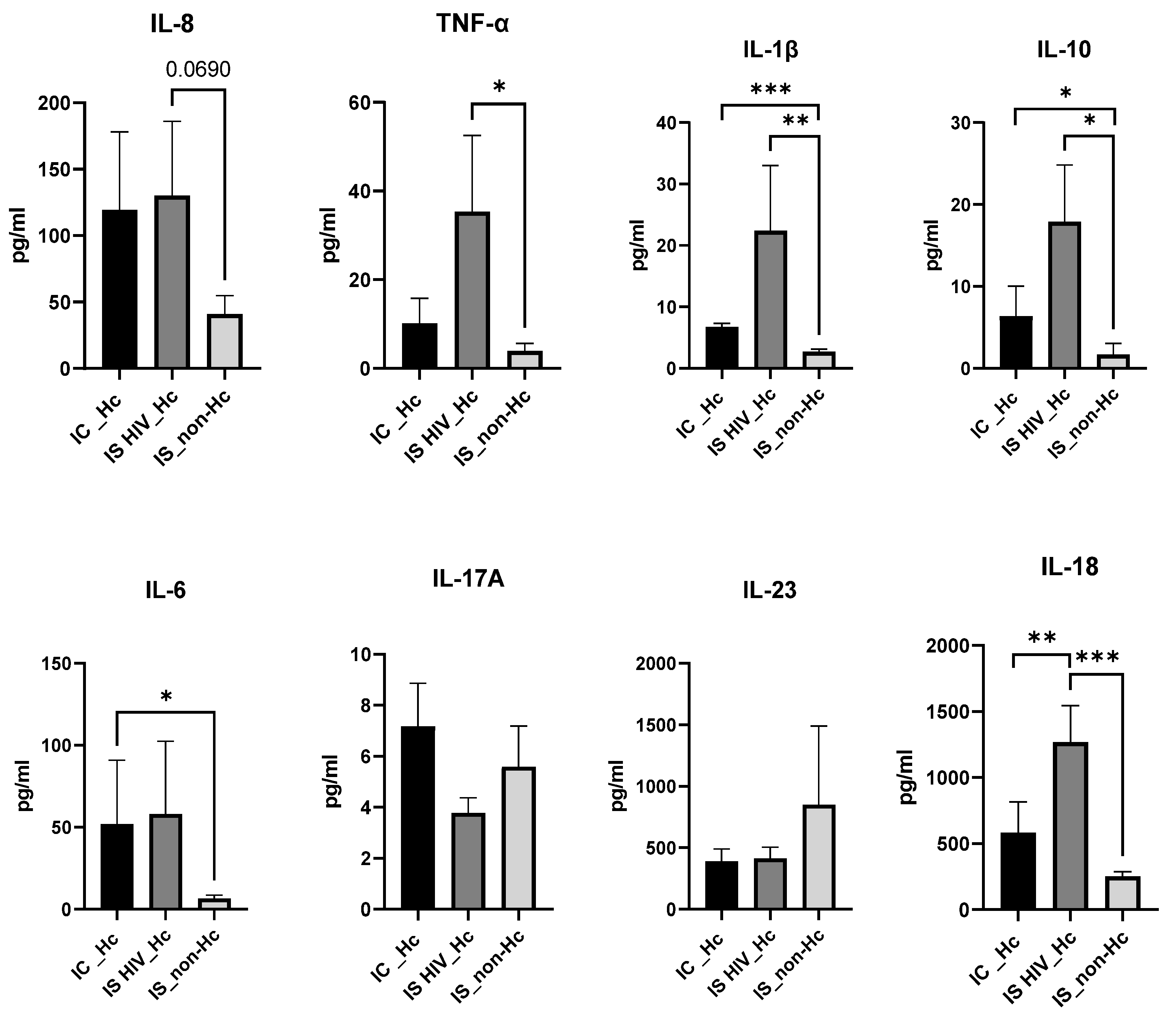
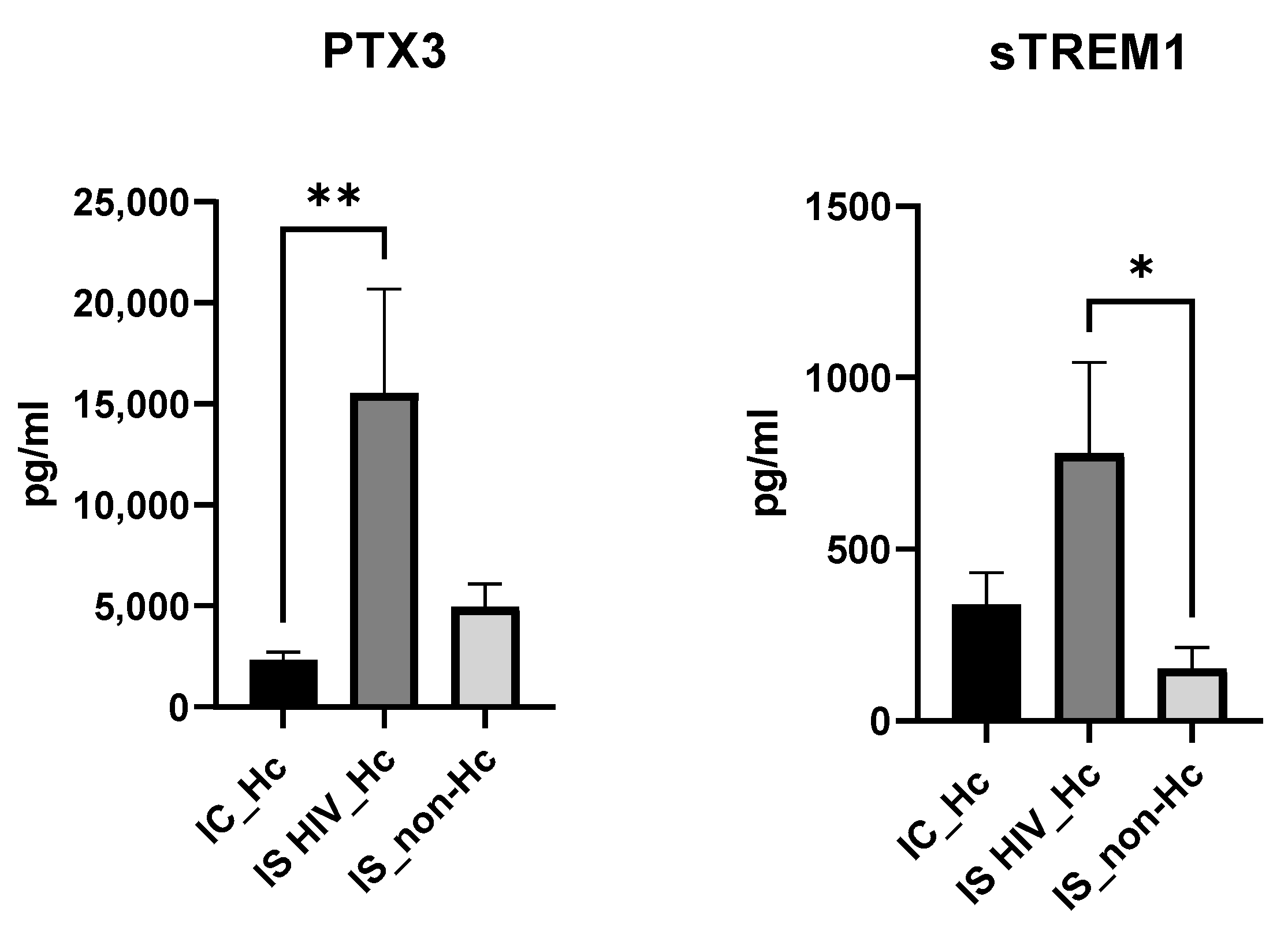
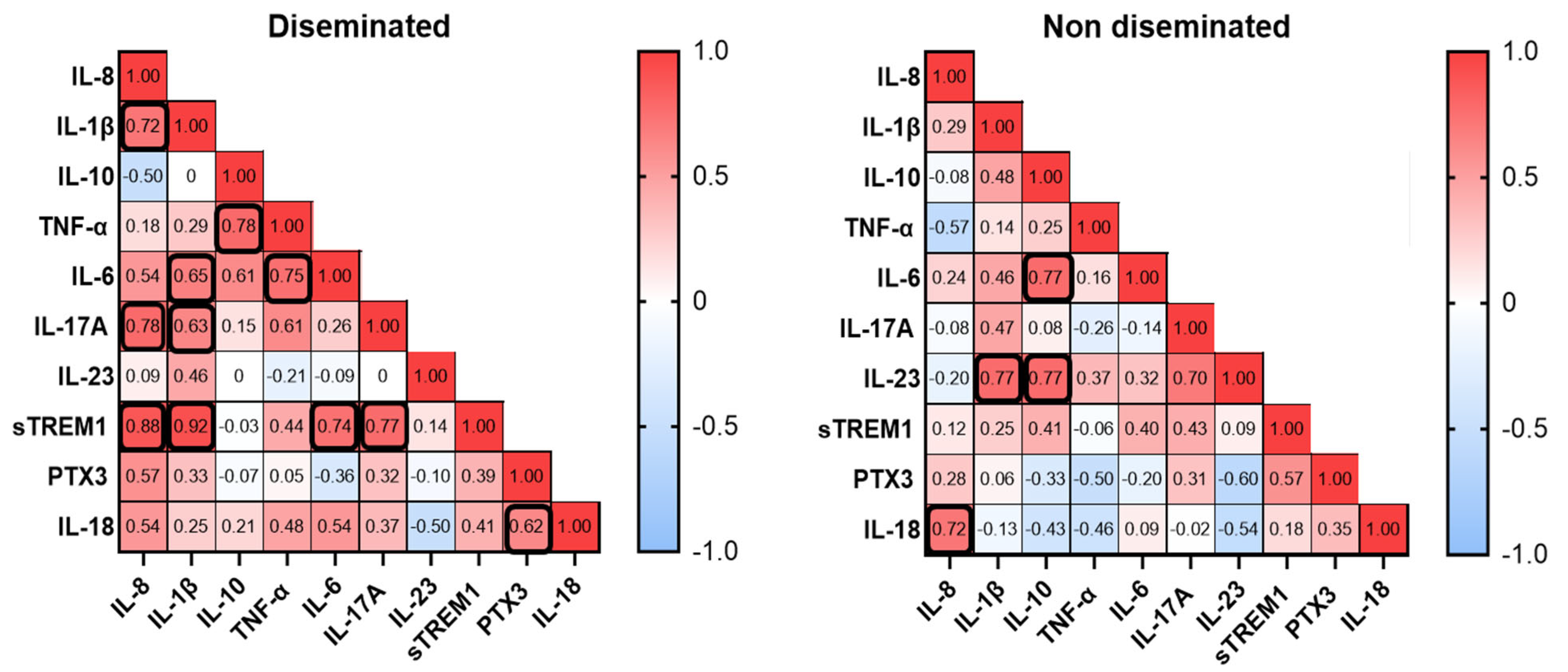
3.6. Biomarker Quantification
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Deepe, G.S., Jr. Immune response to early and late Histoplasma capsulatum infections. Curr. Opin. Microbiol. 2000, 3, 359–362. [Google Scholar] [CrossRef] [PubMed]
- Antinori, S. Histoplasma capsulatum: More widespread than previously thought. Am. J. Trop. Med. Hyg. 2014, 90, 982–983. [Google Scholar] [CrossRef] [PubMed]
- Ashraf, N.; Kubat, R.C.; Poplin, V.; Adenis, A.A.; Denning, D.W.; Wright, L.; McCotter, O.; Schwartz, I.S.; Jackson, B.R.; Chiller, T.; et al. Re-drawing the Maps for Endemic Mycoses. Mycopathologia 2020, 185, 843–865. [Google Scholar] [CrossRef] [PubMed]
- Eichenberger, E.M.; Little, J.S.; Baddley, J.W. Histoplasmosis. Infect. Dis. Clin. N. Am. 2025, 39, 145–161. [Google Scholar] [CrossRef]
- Perez, F.; Caceres, D.H.; Ford, N.; Ravasi, G.; Gomez, B.L.; Pasqualotto, A.C.; Hine, P.; Adenis, A.A.; Nacher, M.; Chiller, T.; et al. Summary of Guidelines for Managing Histoplasmosis among People Living with HIV. J. Fungi 2021, 7, 134. [Google Scholar] [CrossRef]
- Buitrago, M.J.; Bernal-Martinez, L.; Castelli, M.V.; Rodriguez-Tudela, J.L.; Cuenca-Estrella, M. Histoplasmosis and paracoccidioidomycosis in a non-endemic area: A review of cases and diagnosis. J. Travel Med. 2011, 18, 26–33. [Google Scholar] [CrossRef]
- Molina-Morant, D.; Sanchez-Montalva, A.; Salvador, F.; Sao-Aviles, A.; Molina, I. Imported endemic mycoses in Spain: Evolution of hospitalized cases, clinical characteristics and correlation with migratory movements, 1997–2014. PLoS Negl. Trop. Dis. 2018, 12, e0006245. [Google Scholar] [CrossRef]
- Dao, A.; Kim, H.Y.; Halliday, C.L.; Oladele, R.; Rickerts, V.; Govender, M.N.P.; Shin, J.H.; Heim, J.; Ford, N.P.; Nahrgang, S.A.; et al. Histoplasmosis: A systematic review to inform the World Health Organization of a fungal priority pathogens list. Med. Mycol. 2024, 62, myae039. [Google Scholar] [CrossRef]
- Nacher, M. Histoplasmosis in Persons Living with HIV. J. Fungi 2019, 6, 3. [Google Scholar] [CrossRef]
- Buitrago, M.J.; Martin-Gomez, M.T. Timely Diagnosis of Histoplasmosis in Non-endemic Countries: A Laboratory Challenge. Front. Microbiol. 2020, 11, 467. [Google Scholar] [CrossRef]
- Benedict, K.; Beer, K.D.; Jackson, B.R. Histoplasmosis-related Healthcare Use, Diagnosis, and Treatment in a Commercially Insured Population, United States. Clin. Infect. Dis. 2020, 70, 1003–1010. [Google Scholar] [CrossRef] [PubMed]
- Medina, N.; Alastruey-Izquierdo, A.; Bonilla, O.; Gamboa, O.; Mercado, D.; Perez, J.C.; Salazar, L.R.; Arathoon, E.; Denning, D.W.; Rodriguez-Tudela, J.L. A Rapid Screening Program for Histoplasmosis, Tuberculosis, and Cryptococcosis Reduces Mortality in HIV Patients from Guatemala. J. Fungi 2021, 7, 268. [Google Scholar] [CrossRef] [PubMed]
- Samayoa, B.; Aguirre, L.; Bonilla, O.; Medina, N.; Lau-Bonilla, D.; Mercado, D.; Moller, A.; Perez, J.C.; Alastruey-Izquierdo, A.; Arathoon, E.; et al. The Diagnostic Laboratory Hub: A New Health Care System Reveals the Incidence and Mortality of Tuberculosis, Histoplasmosis, and Cryptococcosis of PWH in Guatemala. Open Forum Infect. Dis. 2020, 7, ofz534. [Google Scholar] [CrossRef]
- Azar, M.M.; Hage, C.A. Laboratory Diagnostics for Histoplasmosis. J. Clin. Microbiol. 2017, 55, 1612–1620. [Google Scholar] [CrossRef]
- Caceres, D.H.; Samayoa, B.E.; Medina, N.G.; Tobon, A.M.; Guzman, B.J.; Mercado, D.; Restrepo, A.; Chiller, T.; Arathoon, E.E.; Gomez, B.L. Multicenter Validation of Commercial Antigenuria Reagents To Diagnose Progressive Disseminated Histoplasmosis in People Living with HIV/AIDS in Two Latin American Countries. J. Clin. Microbiol. 2018, 56, 10–1128. [Google Scholar] [CrossRef]
- Caceres, D.H.; Knuth, M.; Derado, G.; Lindsley, M.D. Diagnosis of Progressive Disseminated Histoplasmosis in Advanced HIV: A Meta-Analysis of Assay Analytical Performance. J. Fungi 2019, 5, 76. [Google Scholar] [CrossRef]
- Valero, C.; Martin-Gomez, M.T.; Buitrago, M.J. Molecular Diagnosis of Endemic Mycoses. J. Fungi 2022, 9, 59. [Google Scholar] [CrossRef]
- Kroetz, D.N.; Deepe, G.S. The role of cytokines and chemokines in Histoplasma capsulatum infection. Cytokine 2012, 58, 112–117. [Google Scholar] [CrossRef]
- Carreto-Binaghi, L.E.; Tenorio, E.P.; Morales-Villarreal, F.R.; Aliouat, E.M.; Zenteno, E.; Martinez-Orozco, J.A.; Taylor, M.L. Detection of Cytokines and Collectins in Bronchoalveolar Fluid Samples of Patients Infected with Histoplasma capsulatum and Pneumocystis jirovecii. J. Fungi 2021, 7, 938. [Google Scholar] [CrossRef]
- Peng, Y.Y.; Guo, S.L.; Yan, X.F.; Zhang, L.L.; Wang, J.; Yuan, G.D.; Qing, G.; Xu, L.L.; Zhan, Q. Collective outbreak of severe acute histoplasmosis in immunocompetent Chinese in South America: The clinical characteristics and continuous monitoring of serum cytokines/chemokines. BMC Prim. Care 2022, 23, 197. [Google Scholar] [CrossRef]
- Donnelly, J.P.; Chen, S.C.; Kauffman, C.A.; Steinbach, W.J.; Baddley, J.W.; Verweij, P.E.; Clancy, C.J.; Wingard, J.R.; Lockhart, S.R.; Groll, A.H.; et al. Revision and Update of the Consensus Definitions of Invasive Fungal Disease From the European Organization for Research and Treatment of Cancer and the Mycoses Study Group Education and Research Consortium. Clin. Infect. Dis. 2020, 71, 1367–1376. [Google Scholar] [CrossRef] [PubMed]
- Falci, D.R.; Monteiro, A.A.; Braz Caurio, C.F.; Magalhaes, T.C.O.; Xavier, M.O.; Basso, R.P.; Melo, M.; Schwarzbold, A.V.; Ferreira, P.R.A.; Vidal, J.E.; et al. Histoplasmosis, An Underdiagnosed Disease Affecting People Living With HIV/AIDS in Brazil: Results of a Multicenter Prospective Cohort Study Using Both Classical Mycology Tests and Histoplasma Urine Antigen Detection. Open Forum Infect. Dis. 2019, 6, ofz073. [Google Scholar] [CrossRef] [PubMed]
- Gago, S.; Esteban, C.; Valero, C.; Zaragoza, O.; Puig de la Bellacasa, J.; Buitrago, M.J. A multiplex real-time PCR assay for identification of Pneumocystis jirovecii, Histoplasma capsulatum, and Cryptococcus neoformans/Cryptococcus gattii in samples from AIDS patients with opportunistic pneumonia. J. Clin. Microbiol. 2014, 52, 1168–1176. [Google Scholar] [CrossRef]
- Wheat, L.J.; Azar, M.M.; Bahr, N.C.; Spec, A.; Relich, R.F.; Hage, C. Histoplasmosis. Infect. Dis. Clin. N. Am. 2016, 30, 207–227. [Google Scholar] [CrossRef]
- Powers-Fletcher, M.V.; Hanson, K.E. Nonculture Diagnostics in Fungal Disease. Infect. Dis. Clin. N. Am. 2016, 30, 37–49. [Google Scholar] [CrossRef]
- Toscanini, M.A.; Nusblat, A.D.; Cuestas, M.L. Diagnosis of histoplasmosis: Current status and perspectives. Appl. Microbiol. Biotechnol. 2021, 105, 1837–1859. [Google Scholar] [CrossRef]
- Forno, D.; Samayoa, B.; Medina, N.; Arathoon, E.; Mejia, C.R.; Gordillo, R.; Cedillos, R.; Rodas, J.; Ahlquist Cleveland, A.; Chiller, T.; et al. Diagnosis of fungal opportunistic infections in people living with HIV from Guatemala and El Salvador. Mycoses 2021, 64, 1563–1570. [Google Scholar] [CrossRef]
- Bloch, K.C.; Myint, T.; Raymond-Guillen, L.; Hage, C.A.; Davis, T.E.; Wright, P.W.; Chow, F.C.; Woc-Colburn, L.; Khairy, R.N.; Street, A.C.; et al. Improvement in Diagnosis of Histoplasma Meningitis by Combined Testing for Histoplasma Antigen and Immunoglobulin G and Immunoglobulin M Anti-Histoplasma Antibody in Cerebrospinal Fluid. Clin. Infect. Dis. 2018, 66, 89–94. [Google Scholar] [CrossRef]
- Richer, S.M.; Smedema, M.L.; Durkin, M.M.; Herman, K.M.; Hage, C.A.; Fuller, D.; Wheat, L.J. Improved Diagnosis of Acute Pulmonary Histoplasmosis by Combining Antigen and Antibody Detection. Clin. Infect. Dis. 2016, 62, 896–902. [Google Scholar] [CrossRef]
- Vidal, J.E.; Werlang, P.C.; Muniz, B.M.; Rego, C.M.; Barbalho, R.E.; Baptista, A.M.; Telles, J.P.; da Cruz, A.B.; Pereira, I.S.; Gava, R.; et al. Combining urine antigen and blood polymerase chain reaction for the diagnosis of disseminated histoplasmosis in hospitalized patients with advanced HIV disease. Med. Mycol. 2021, 59, 916–922. [Google Scholar] [CrossRef]
- Alanio, A.; Gits-Muselli, M.; Lanternier, F.; Sturny-Leclere, A.; Benazra, M.; Hamane, S.; Rodrigues, A.M.; Garcia-Hermoso, D.; Lortholary, O.; Dromer, F.; et al. Evaluation of a New Histoplasma spp. Quantitative RT-PCR Assay. J. Mol. Diagn. 2021, 23, 698–709. [Google Scholar] [CrossRef] [PubMed]
- Wilmes, D.; Hagen, F.; Verissimo, C.; Alanio, A.; Rickerts, V.; Buitrago, M.J. A multicentre external quality assessment: A first step to standardise PCR protocols for the diagnosis of histoplasmosis and coccidioidomycosis. Mycoses 2023, 66, 774–786. [Google Scholar] [CrossRef] [PubMed]
- Muraosa, Y.; Toyotome, T.; Yahiro, M.; Watanabe, A.; Shikanai-Yasuda, M.A.; Kamei, K. Detection of Histoplasma capsulatum from clinical specimens by cycling probe-based real-time PCR and nested real-time PCR. Med. Mycol. 2016, 54, 433–438. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Dantas, K.C.; Freitas, R.S.; da Silva, M.V.; Criado, P.R.; Luiz, O.D.C.; Vicentini, A.P. Comparison of diagnostic methods to detect Histoplasma capsulatum in serum and blood samples from AIDS patients. PLoS ONE 2018, 13, e0190408. [Google Scholar] [CrossRef]
- Moussiegt, A.; Donald, S.M.; Bougnoux, M.E.; Van Eer, M.; Vreden, S.; Chiller, T.; Caceres, D.H.; Gomez, B.L.; Nacher, M.; Lortholary, O.; et al. Fungal biomarkers in HIV-associated disseminated histoplasmosis: A multicenter diagnostic accuracy study on the Guiana shield. Int. J. Infect. Dis. 2025, 153, 107360. [Google Scholar] [CrossRef]
- Ranque, S.; Pelletier, R.; Michel-Nguyen, A.; Dromer, F. Platelia Aspergillus assay for diagnosis of disseminated histoplasmosis. Eur. J. Clin. Microbiol. Infect. Dis. 2007, 26, 941–943. [Google Scholar] [CrossRef]
- Caceres, D.H.; Gomez, B.L.; Tobon, A.M.; Chiller, T.M.; Lindsley, M.D. Evaluation of OIDx Histoplasma Urinary Antigen EIA. Mycopathologia 2022, 187, 129–131. [Google Scholar] [CrossRef]
- Horwath, M.C.; Fecher, R.A.; Deepe, G.S., Jr. Histoplasma capsulatum, lung infection and immunity. Future Microbiol. 2015, 10, 967–975. [Google Scholar] [CrossRef]
- Alcantara, C.; Almeida, B.R.; Barros, B.; Orikaza, C.M.; Toledo, M.S.; Suzuki, E. Histoplasma capsulatum chemotypes I and II induce IL-8 secretion in lung epithelial cells in distinct manners. Med. Mycol. 2020, 58, 1169–1177. [Google Scholar] [CrossRef]
- Sahaza, J.H.; Suarez-Alvarez, R.; Estrada-Barcenas, D.A.; Perez-Torres, A.; Taylor, M.L. Profile of cytokines in the lungs of BALB/c mice after intra-nasal infection with Histoplasma capsulatum mycelial propagules. Comp. Immunol. Microbiol. Infect. Dis. 2015, 41, 1–9. [Google Scholar] [CrossRef]
- Bernal-Martinez, L.; Goncalves, S.M.; de Andres, B.; Cunha, C.; Gonzalez Jimenez, I.; Lagrou, K.; Mellado, E.; Gaspar, M.L.; Maertens, J.A.; Carvalho, A.; et al. TREM1 regulates antifungal immune responses in invasive pulmonary aspergillosis. Virulence 2021, 12, 570–583. [Google Scholar] [CrossRef]
| Characteristics | Number |
|---|---|
| Age, mean, years | 40 |
| Gender: | |
| Male (%) | 62 |
| Female (%) | 38 |
| Originating from an endemic region (Nicaragua, Colombia, Guatemala, Costa Rica, Bolivia-via, Uruguay, Ecuador, Peru, Nigeria, Senegal, Venezuela, Brazil, Paraguay, Dominican Republic, Pana-ma, Argentina, El Salvador, Equatorial Guinea, Mexico, and Ghana) | 32 |
| Travelers | 8 |
| Underliying condition: | |
| HIV/AIDS | 20 |
| Inmunosupressed non-HIV | 1 |
| None | 19 |
| Patient nº | Immunosuppression | RT-PCR | ID | Platelia Asp | GM EIA | Clinical Data | Classification |
|---|---|---|---|---|---|---|---|
| 1 | No | + | + | − | + | APH | Proven |
| 2 | No | + | + | − | + | APH | Proven |
| 3 | No | − | + | + | + | APH | Probable |
| 7 | No | − | + | − | − | APH | Proven |
| 7 * | No | − | + | − | + | APH | Proven |
| 28 | No | − | − | − | − | APH | Proven |
| 29 | No | − | + | − | − | APH | Proven |
| 33 | No | − | + | − | − | CPH | Probable |
| 35 | No | − | + | − | − | APH | Probable |
| 37 | No | − | + | − | + | ACPH | Probable |
| 38 | No | + | + | − | + | ND | Probable |
| 4 | Yes | + | − | − | + | DH | Proven |
| 8 | Yes | + | + | + | + | DH | Proven |
| 9 | Yes | − | − | − | + | DH | Proven |
| 10 | Yes | + | − | + | + | DH | Proven |
| 11 | Yes | − | − | − | − | GIH | Proven |
| 13 | Yes | − | + | − | + | DH | Proven |
| 16 | Yes | + | − | + | + | DH | Proven |
| 17 | Yes | − | + | − | + | DH | Proven |
| 18 | Yes | + | − | + | + | DH | Proven |
| 19 | Yes | − | + | − | + | GIH | Proven |
| 20 | Yes | + | + | + | + | DH | Proven |
| 30 | Yes | + | + | − | + | DH | Proven |
| 32 | Yes | − | + | + | + | DH | Proven |
| 34 | Yes | − | + | − | + | DH | Proven |
| 36 | Yes | − | + | − | − | APH | Proven |
| 40 | Yes | − | + | − | + | SAPH | Probable |
| Immunosuppression | RT-PCR | ID | Platelia Asp | GM EIA |
|---|---|---|---|---|
| YES 16/27 (59%) | 7/16 (44%) | 10/16 (62.5%) | 6/16 (37.5%) | 14/16 (87.5%) |
| NO 11/27 (41%) | 3/11 (27%) | 10/11 (90.9%) | 1/11 (9%) | 6/11 (54.5%) |
| TOTAL 27 | 10/27 (37%) | 20/27 (74%) | 7/27 (26%) | 20/27 (74%) |
| Immunosuppression | RT-PCR +ID | RT-PCR + Platelia Asp | RT-PCR + GM EIA | ID + Platelia Asp | ID + EIA GM | Platelia Asp + GM EIA |
|---|---|---|---|---|---|---|
| YES 16/27 (59%) | 13/16 (81.25%) | 8/16 (50%) | 13/16 (81.25%) | 12/16 (75%) | 15/16 (93.7%) | 14/16 (87.5%) |
| NO 11/27 (41%) | 10/11 (90.9%) | 3/11 (27.2%) | 6/11 (54.5%) | 10/11 (90.9%) | 10/11 (90.9%) | 6/11 (54.5%) |
| TOTAL 27 | 23/27 (85.1%) | 11/27 (40.7%) | 19/27 (70.3%) | 12/27 (81.4%) | 25/27 (92.6%) | 20/27 (74%) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bernal-Martínez, L.; De la Cruz-Ríos, P.; Viedma, R.; Gago, S.; Ortega-Madueño, S.; Alcazar-Fuoli, L.; Buitrago, M.J. Usefulness of Serum as a Non-Invasive Sample for the Detection of Histoplasma capsulatum Infections: Retrospective Comparative Analysis of Different Diagnostic Techniques and Quantification of Host Biomarkers. J. Fungi 2025, 11, 448. https://doi.org/10.3390/jof11060448
Bernal-Martínez L, De la Cruz-Ríos P, Viedma R, Gago S, Ortega-Madueño S, Alcazar-Fuoli L, Buitrago MJ. Usefulness of Serum as a Non-Invasive Sample for the Detection of Histoplasma capsulatum Infections: Retrospective Comparative Analysis of Different Diagnostic Techniques and Quantification of Host Biomarkers. Journal of Fungi. 2025; 11(6):448. https://doi.org/10.3390/jof11060448
Chicago/Turabian StyleBernal-Martínez, L., P. De la Cruz-Ríos, R. Viedma, S. Gago, S. Ortega-Madueño, L. Alcazar-Fuoli, and M. J. Buitrago. 2025. "Usefulness of Serum as a Non-Invasive Sample for the Detection of Histoplasma capsulatum Infections: Retrospective Comparative Analysis of Different Diagnostic Techniques and Quantification of Host Biomarkers" Journal of Fungi 11, no. 6: 448. https://doi.org/10.3390/jof11060448
APA StyleBernal-Martínez, L., De la Cruz-Ríos, P., Viedma, R., Gago, S., Ortega-Madueño, S., Alcazar-Fuoli, L., & Buitrago, M. J. (2025). Usefulness of Serum as a Non-Invasive Sample for the Detection of Histoplasma capsulatum Infections: Retrospective Comparative Analysis of Different Diagnostic Techniques and Quantification of Host Biomarkers. Journal of Fungi, 11(6), 448. https://doi.org/10.3390/jof11060448






