Virulence Characterization of Puccinia striiformis f. sp. tritici in China in 2020 Using Wheat Yr Single-Gene Lines
Abstract
1. Introduction
2. Materials and Methods
2.1. Collection of Wheat Stripe Rust Samples
2.2. Purification and Multiplication of Isolates
2.3. Race Identification
2.4. Data Analysis
3. Results
3.1. Virulence Patterns of Pst Races
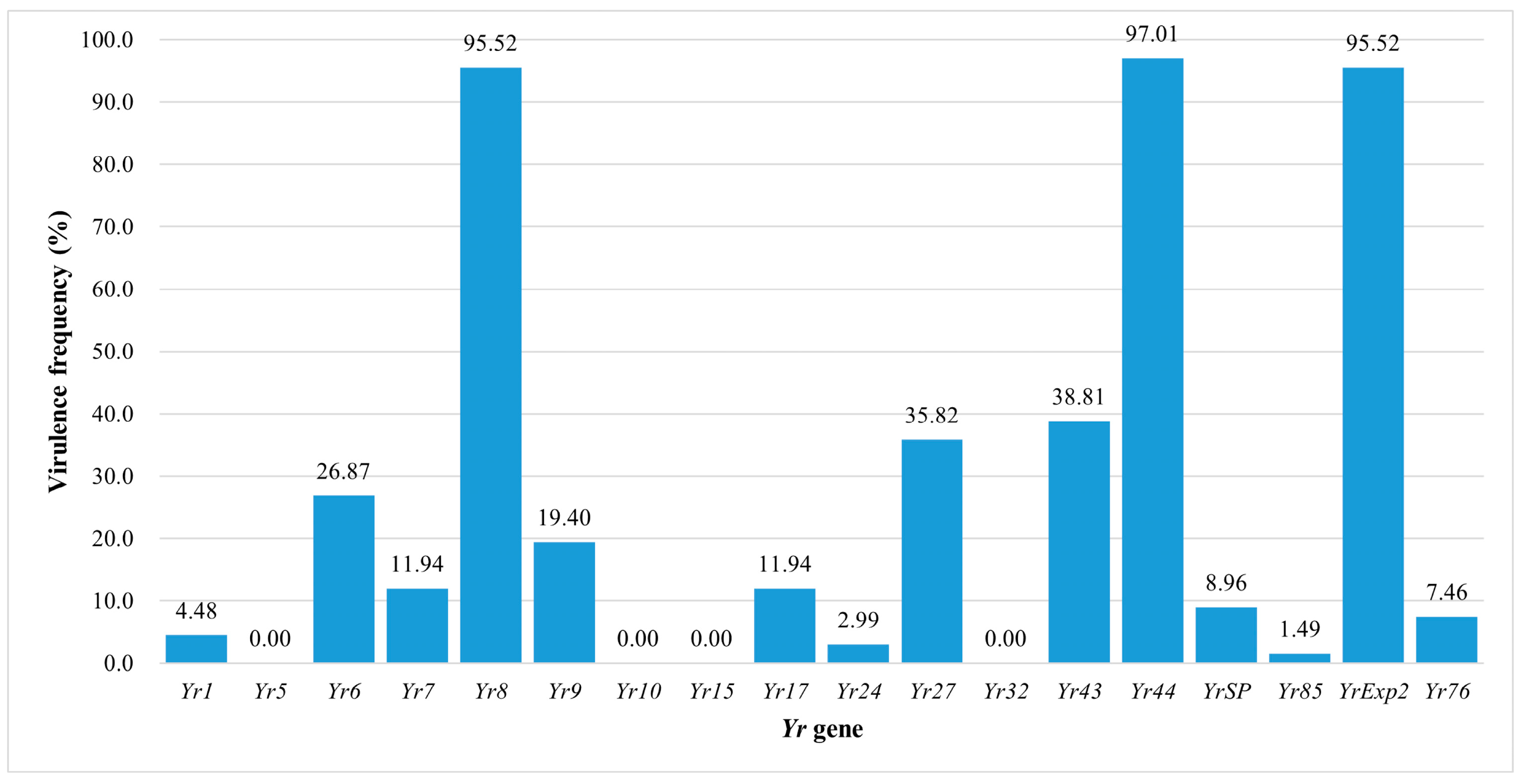
3.2. Frequencies of Virulence Factors
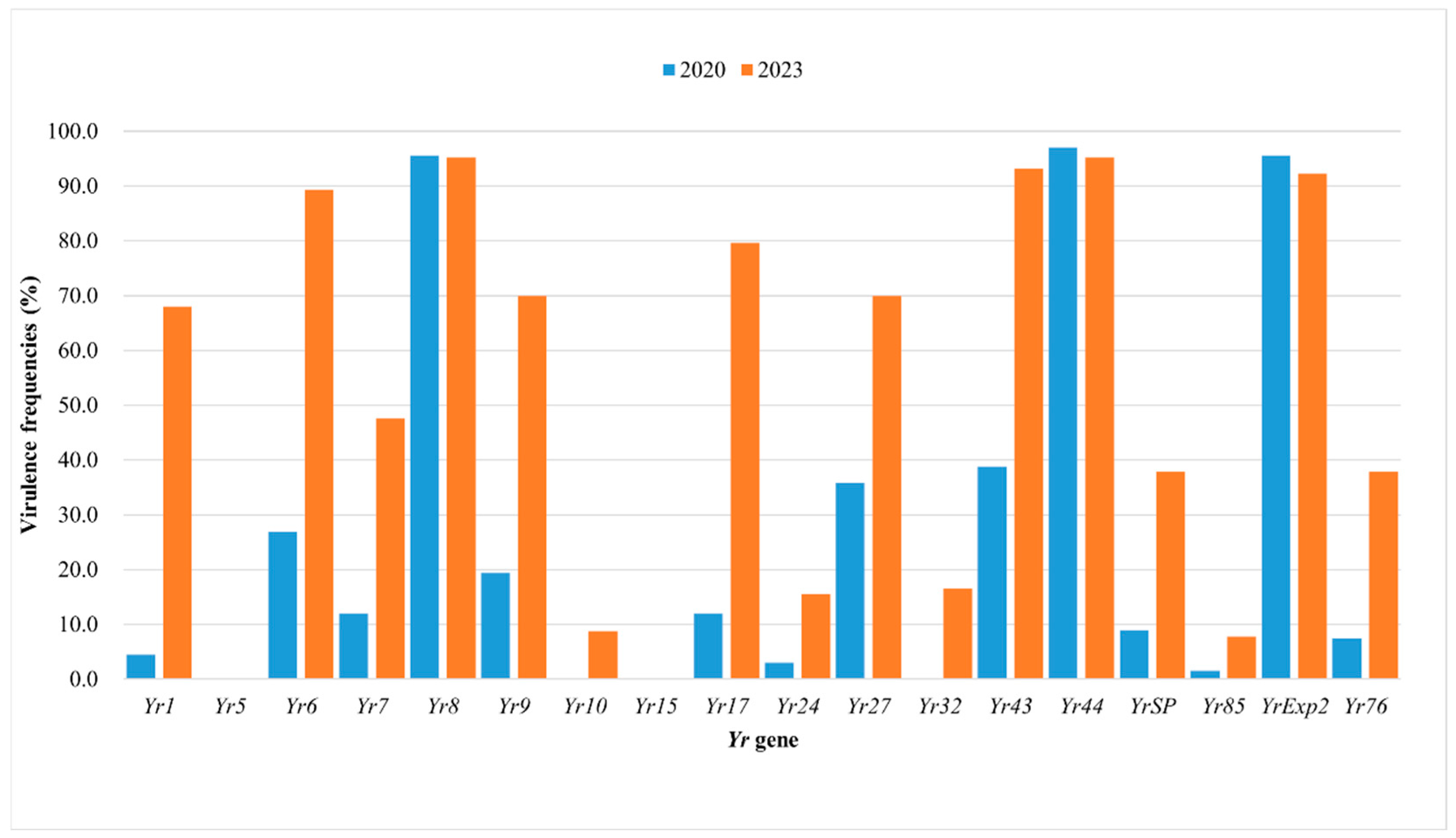
3.3. Comparison of Pst Populations from China in 2020 and 2023
3.4. Correlations Between Virulence Factors
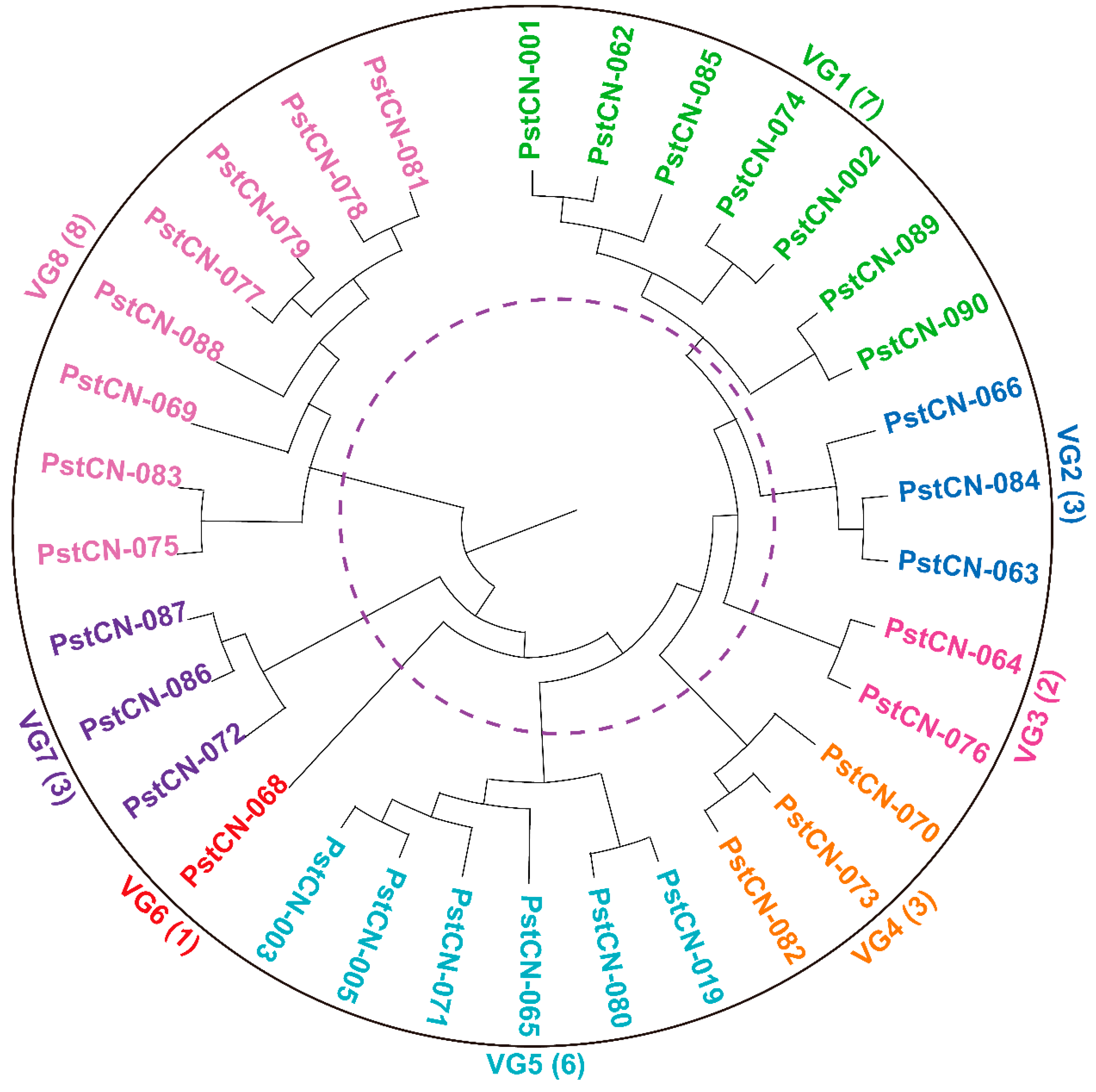
3.5. Pst Race Clustering
4. Discussion
4.1. Races and Epidemics
4.2. Effectiveness of Resistance Genes and Gene Interactions
4.3. Virulence Dynamics of Pst Populations
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Wellings, C.R. Global status of stripe rust: A review of historical and current threats. Euphytica 2011, 179, 129–141. [Google Scholar] [CrossRef]
- Kang, Z.S.; Wang, X.J.; Zhao, J.; Tang, C.L.; Huang, L.L. Advances in research of pathogenicity and virulence variation of the wheat stripe rust fungus Puccinia striiformis f. sp. tritici. Sci. Agric. Sin. 2015, 48, 3439–3453. [Google Scholar]
- Awais, M.; Ma, J.; Chen, W.; Zhang, B.; Turakulov, K.S.; Li, L.; Egamberdieva, D.; Karimjonovich, M.S.; Kang, Z.; Zhao, J. Molecular genotyping revealed the gene flow of Puccinia striiformis f. sp. tritici clonal lineage from Uzbekistan of Central Asia to Xinjiang of China. Phytopathol. Res. 2025, 7, 2. [Google Scholar] [CrossRef]
- Huang, L.; Chen, X.; Dong, Y.; Huang, W.; Ma, H.; Zhang, H.; Xu, Y.; Wang, J. Dynamic Analysis of Regional Wheat Stripe Rust Environmental Suitability in China. Remote Sens. 2023, 15, 2021. [Google Scholar] [CrossRef]
- Zhao, J.; Kang, Z. Fighting wheat rusts in China: A look back and into the future. Phytopathol. Res. 2023, 5, 6. [Google Scholar] [CrossRef]
- Liu, W.C.; Wang, B.T.; Zhao, Z.H.; Li, Y.; Kang, Z.S. Historical review and countermeasures of wheat stripe rust epidemics in China. China Plant Prot. Sin. 2022, 42, 21–27. [Google Scholar]
- Chunlei TANG, X.W.; Cheng, Y.; Liu, M.; Wei, J.; Kang, Z. New insights in the battle between wheat and Puccinia striiformis. Front. Agr. Sci. Eng. 2015, 2, 101–114. [Google Scholar] [CrossRef]
- Esmail, S.M.; Draz, I.S.; Sehsah, M.D.; Saad-El-Din, H.I.; Youssef, W.A.; Komeil, D.A. Evolutionary and dispersal dynamics in invasive races of Puccinia striiformis f. sp. tritici attributed to virulence and SCAR markers. Physiol. Mol. Plant Pathol. 2025, 136, 102524. [Google Scholar] [CrossRef]
- Rehman, S.u.; Qiao, L.; Shen, T.; Hua, L.; Li, H.; Ahmad, Z.; Chen, S. Exploring the Frontier of Wheat Rust Resistance: Latest Approaches, Mechanisms, and Novel Insights. Plants 2024, 13, 2502. [Google Scholar] [CrossRef]
- Wang, L.; Tang, X.; Wu, J.; Shen, C.; Dai, M.; Wang, Q.; Zeng, Q.; Kang, Z.; Wu, Y.; Han, D. Stripe rust resistance to a burgeoning Puccinia striiformis f. sp. tritici race CYR34 in current Chinese wheat cultivars for breeding and research. Euphytica 2019, 215, 68. [Google Scholar] [CrossRef]
- Jiang, B.; Wang, C.; Guo, C.; Lv, X.; Gong, W.; Chang, J.; He, H.; Feng, J.; Chen, X.; Ma, Z. Genetic relationships of Puccinia striiformis f. sp. tritici in southwestern and northwestern China. Microbiol. Spectr. 2022, 10, e01530-22. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Xu, Y.; Peng, Y.; Wang, Y.; Kang, Z.; Zhao, J. A fully haplotype-resolved and nearly gap-free genome assembly of wheat stripe rust fungus. Sci. Data 2024, 11, 508. [Google Scholar] [CrossRef] [PubMed]
- Wang, K.N.; Hong, X.W. Discussion on the causes of stripe rust resistance variation of ‘Bima 1’. Sci. Agric. 1957, 97, 650–652. [Google Scholar]
- Hu, X.P.; Wang, B.T.; Kang, Z.S. Research progress on virulence variation of Puccinia striiformis f. sp. tritici in China. Triticeae Crops 2014, 34, 709–716. [Google Scholar]
- Cao, Z.J.; Jing, J.X.; Wang, M.N.; Xu, Z.B.; Shang, H.S.; Li, Z.Q. Analysis of stripe rust resistance inherence of wheat cultivar Guinong 22. Acta Bot. Boreali-Occident. Sin. 2004, 6, 991–996. [Google Scholar]
- Andrivon, D.; Vallavieille, P.C. Race diversity and complexity in selected populations of fungal biotrophic pathogens of cereals. Phytopathology 1995, 85, 897–905. [Google Scholar] [CrossRef]
- Zhang, G.; Liu, W.; Wang, L.; Cheng, X.; Tian, X.; Du, Z.; Kang, Z.; Zhao, J. Evaluation of the potential risk of the emerging Yr5-virulent races of Puccinia striiformis f. sp. tritici to 165 Chinese wheat cultivars. Plant Dis. 2022, 106, 1867–1874. [Google Scholar] [CrossRef] [PubMed]
- Haider, M.W.; Kaur, J.; Bala, R.; Singh, S.; Srivastava, P.; Sharma, A.; Singh, R.; Kumari, J. Stripe rust resistance gene(s) postulation in wheat germplasm with the help of differentials and tagged molecular markers. Sci. Rep. 2023, 13, 9007. [Google Scholar]
- Ghanbarnia, K.; Gourlie, R.; Amundsen, E.; Aboukhaddour, R. The changing virulence of stripe rust in Canada from 1984 to 2017. Phytopathology 2021, 111, 1840–1850. [Google Scholar] [CrossRef]
- Wan, A.M.; Chen, X.M. Virulence characterization of Puccinia striiformis f. sp. tritici using a new set of Yr single-gene line differentials in the United States in 2010. Plant Dis. 2014, 98, 1534–1542. [Google Scholar] [CrossRef]
- Li, M.J.; Chen, X.M.; Wang, A.M.; Ding, M.L.; Cheng, J.S. Virulence characterization of stripe rust pathogen Puccinia striiformis f. sp. tritici population to 18 near-isogenic lines resistant to wheat yellow rust in Yunnan Province. J. Plant Prot. Sin. 2018, 45, 75–82. [Google Scholar]
- Zhang, X.Z.; Huang, L.; Tang, W.Q.; Qiu, A.G.; Li, X.; Zhou, X.L.; Yang, S.Z.; Wang, M.N.; Chen, X.M.; Chen, W.Q.; et al. Virulence characterization of wheat stripe rust population in China in 2023. Plant Pathol. 2024, 74, 363–377. [Google Scholar] [CrossRef]
- Kosman, E. Measuring diversity: From individuals to populations. Eur. J. Plant Pathol. 2014, 138, 467–486. [Google Scholar] [CrossRef]
- Wellings, C.R. Puccinia striiformis in Australia: A review of the incursion, evolution, and adaptation of stripe rust in the period 1979–2006. Aus. J. Agric. Res. 2007, 58, 567–575. [Google Scholar] [CrossRef]
- Zhao, Y.; Wang, R.J.; Li, Z.H.; Sun, X.Y.; Wang, H.Y.; Wang, X.Y. Analysis on the effects of wheat planting status and quality evaluation on high quality product rate improvement in Shandong province. Sci. Technol. Cereals Oils Foods. Sin. 2021, 29, 84–90. [Google Scholar]
- Zhang, H.; Ren, Y.; He, Y.J.; Zheng, S.H.; Wu, G.; Zou, F.L.; Lei, J.R.; Du, X.Y.; Tao, J.; Ou, J.M. Molecular detection of disease resistance genes in 153 Sichuan wheat varieties and lines. T. Crops. Sin. 2022, 42, 26–35. [Google Scholar]
- Huang, C.; Jiang, Y.Y.; Li, P.L.; Peng, H.; Cui, Y.; Yang, J.J.; Xie, F.Z. Epidemics analysis of wheat stripe rust in China in 2017. Plant Prot. Sin. 2018, 44, 166–183. [Google Scholar]
- Xi, L.; Wang, Y.-Q.; Zhu, W.; Wang, Y.; Chen, G.-Y.; Pu, Z.-J.; Zhou, Y.-H.; Kang, H.-Y. Identification of resistance to wheat and molecular detection of resistance genes to wheat stripe rust of 78 wheat cultivars (lines) in Sichuan province. Acta Agron. Sin. 2021, 47, 1309–1323. [Google Scholar]
- Gao, P.; Zhou, Y.; Gebrewahid, T.W.; Zhang, P.; Yan, X.; Li, X.; Yao, Z.; Li, Z.; Liu, D. Identification of known leaf rust resistance genes in common wheat cultivars from Sichuan province in China. Crop Prot. 2019, 115, 122–129. [Google Scholar] [CrossRef]
- Huang, L.; Xiao, X.Z.; Liu, B.; Gao, L.; Gong, G.S.; Chen, W.Q.; Zhang, M.; Liu, T.G. Identification of stripe rust resistance genes in common wheat cultivars from the Huang-Huai-Hai region of China. Plant Dis. 2020, 104, 1763–1770. [Google Scholar] [CrossRef]
- Riley, R.; Chapman, V.; Johnson, R. Introduction of yellow rust resistance of Aegilops comosa. Nature 1968, 217, 383–384. [Google Scholar] [CrossRef]
- Liu, T.L.; Wan, A.M.; Liu, D.C.; Chen, X.M. Changes of races and virulence genes in Puccinia striiformis f. sp. tritici, the wheat stripe rust pathogen, in the United States from 1968 to 2009. Plant Dis. 2017, 101, 1522–1532. [Google Scholar] [CrossRef] [PubMed]
- Yuan, C.L.; Wu, J.Z.; Yan, B.Q.; Hao, Q.Q.; Zhang, C.Z.; Lyu, B.; Ni, F.; Caplan, A.; Wu, J.J.; Fu, D.L. Remapping of the stripe rust resistance gene Yr10 in common wheat. Theor. Appl. Genet. 2018, 131, 1253–1262. [Google Scholar] [CrossRef]
- Zhang, G.S.; Zhao, Y.Y.; Kang, Z.S.; Zhao, J. First report of a Puccinia striiformis f. sp. tritici race virulent to wheat stripe rust resistance gene Yr5 in China. Plant Dis. 2020, 104, 284–285. [Google Scholar] [CrossRef]
- Tekin, M.; Cat, A.; Akan, K.; Catal, M.; Akar, T. A new virulent race of wheat stripe rust pathogen (Puccinia striiformis f. sp. tritici) on the resistance gene Yr5 in Turkey. Plant Dis. 2021, 105, 3292. [Google Scholar] [CrossRef]
- Wang, M.N.; Chen, X.M. Stripe rust epidemics of wheat and barley and races of Puccinia striiformis identified in the United States in 2020. Phytopathology 2021, 111, S2.147. [Google Scholar]
- de Vallavieille-Pope, C.; Ali, S.; Leconte, M.; Enjalbert, J.; Delos, M.; Rouzet, J. Virulence dynamics and regional structuring of Puccinia striformis f. sp triitici in france between 1984 and 2009. Plant Dis. 2012, 96, 131–140. [Google Scholar] [CrossRef]
- Hovmller, M.S.; Justesen, A.F. Rates of evolution of avirulence phenotypes and DNA markers in a northwest European population of Puccinia striiformis f. sp. tritici. Mol. Ecol. 2007, 16, 637–4647. [Google Scholar] [CrossRef]
- Sthapit Kandel, J.; Krishnan, V.; Jiwan, D.; Chen, X.; Skinner, D.Z.; See, D.R. Mapping genes for resistance to stripe rust in spring wheat landrace PI 480035. PLoS ONE 2017, 12, e0177898. [Google Scholar] [CrossRef]
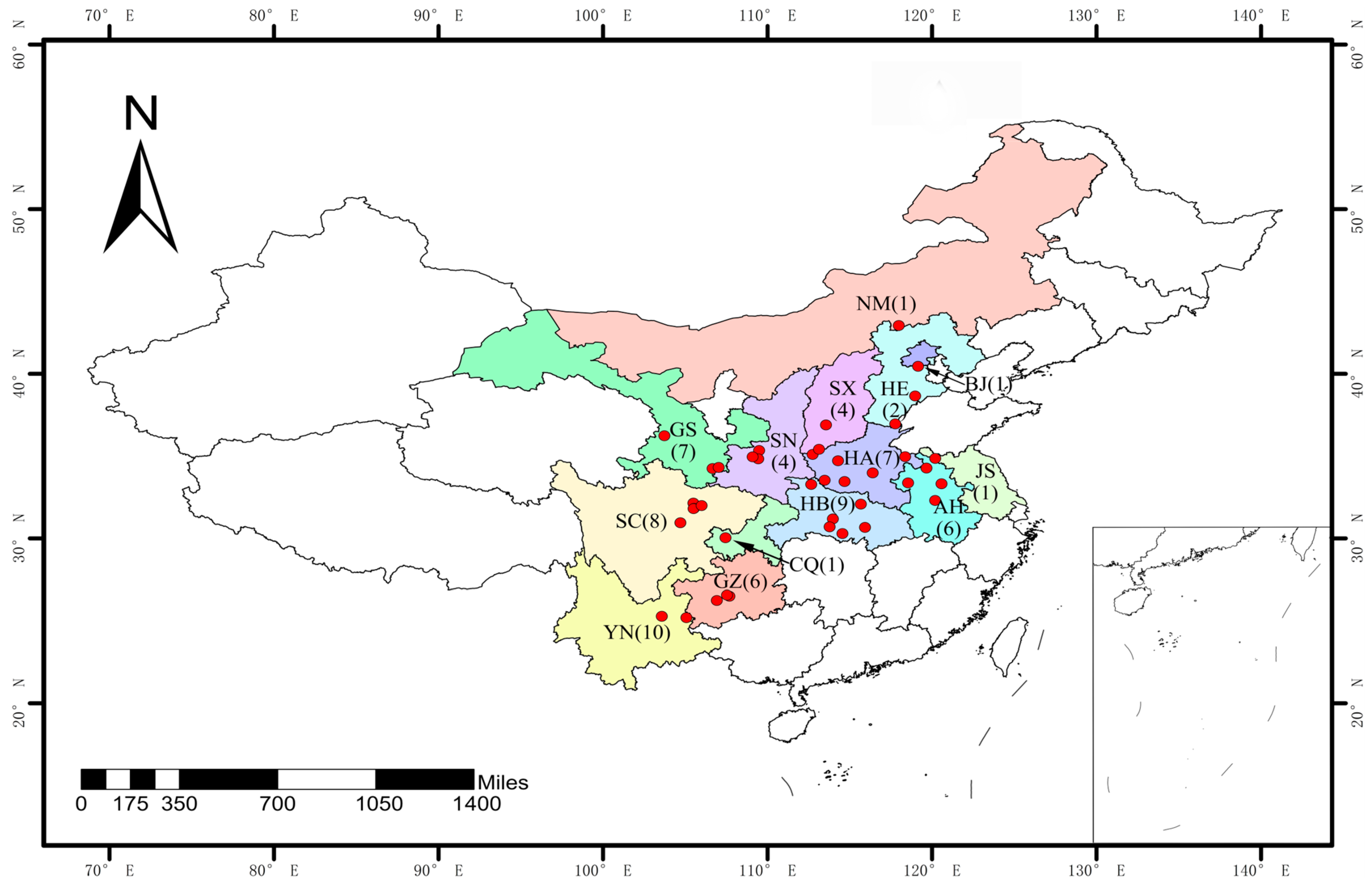

| Province 1 | No. of Cities | No. of Samples | No. of Isolates | Races | Isolate/Race Ratio |
|---|---|---|---|---|---|
| HE | 7 | 8 | 3 | 3 | 1.00 |
| HA | 7 | 13 | 7 | 5 | 1.40 |
| SX | 3 | 6 | 4 | 4 | 1.00 |
| SN | 3 | 7 | 4 | 4 | 1.00 |
| SC | 3 | 11 | 8 | 6 | 1.33 |
| GS | 2 | 11 | 7 | 5 | 1.40 |
| AH | 4 | 6 | 6 | 4 | 1.50 |
| JS | 5 | 2 | 1 | 1 | 1.00 |
| GZ | 2 | 8 | 6 | 5 | 1.20 |
| HB | 9 | 13 | 9 | 8 | 1.12 |
| YN | 3 | 13 | 10 | 8 | 1.25 |
| CQ | 1 | 2 | 1 | 1 | 1.00 |
| NM | 1 | 2 | 1 | 1 | 1.00 |
| Race | PSTv 1 | Octal Caode | Virulence to Yr Gene | No. of Virulence Factors | No. of Isolates | Frequency (%) (n = 67) | Province 2 (Number of Isolates) |
|---|---|---|---|---|---|---|---|
| PstCN-001 | PSTv-136 | 020062 | 8, 43, 44, Exp2 | 4 | 8 | 11.94 | YN(2), SC(3), SN(2), HE(1) |
| PstCN-002 | \ | 020262 | 8, 27, 43, 44, Exp2 | 5 | 6 | 8.98 | HB(1), GS(2), HA(1), YN(1), SC(1) |
| PstCN-003 | \ | 120062 | 6, 8, 43, 44, Exp2 | 5 | 1 | 1.49 | AH(1) |
| PstCN-005 | \ | 120262 | 6, 8, 27, 43, 44, Exp2 | 6 | 1 | 1.49 | SC(1) |
| PstCN-019 | \ | 420062 | 1, 8, 43, 44, Exp2 | 5 | 1 | 1.49 | GZ(1) |
| PstCN-062 | PSTv-137 | 020022 | 8, 44, Exp2 | 3 | 19 | 28.36 | AH(3), GS(2), GZ(2), HA(3), HB(2), JS(1), SX(1), SN(1), SC(2), YN(2) |
| PstCN-063 | \ | 120022 | 6, 8, 44, Exp2 | 4 | 4 | 5.97 | GS(1), SN(1), HE(1), HB(1) |
| PstCN-064 | \ | 060002 | 7, 8, Exp2 | 3 | 1 | 1.49 | HB(1) |
| PstCN-065 | \ | 100062 | 6, 43, 44, Exp2 | 4 | 1 | 1.49 | YN(1) |
| PstCN-066 | \ | 030022 | 8, 9, 44, Exp2 | 4 | 1 | 1.49 | HB(1) |
| PstCN-068 | \ | 020243 | 8, 27, 43, Exp2, 76 | 5 | 1 | 1.49 | HA(1) |
| PstCN-069 | \ | 061262 | 7, 8, 17, 27, 43, 44, Exp2 | 7 | 1 | 1.49 | HB(1) |
| PstCN-070 | \ | 530222 | 1, 6, 8, 9, 27, 44, Exp2 | 7 | 1 | 1.49 | GZ(1) |
| PstCN-071 | \ | 120662 | 6, 8, 24, 27, 43, 44, Exp2 | 7 | 1 | 1.49 | YN(1) |
| PstCN-072 | \ | 030626 | 8, 9, 24, 27, 44, 85, Exp2 | 7 | 1 | 1.49 | SX(1) |
| PstCN-073 | \ | 120222 | 6, 8, 27, 44, Exp2 | 5 | 1 | 1.49 | GZ(1) |
| PstCN-074 | \ | 020222 | 8, 27, 44, Exp2 | 4 | 1 | 1.49 | SC(1) |
| PstCN-075 | \ | 031233 | 8, 9, 17, 27, 44, SP, Exp2, 76 | 8 | 1 | 1.49 | HB(1) |
| PstCN-076 | \ | 060022 | 7, 8, 44, Exp2 | 4 | 1 | 1.49 | YN(1) |
| PstCN-077 | \ | 171233 | 6, 7, 8, 9, 17, 27, 44, SP, Exp2, 76 | 10 | 1 | 1.49 | HA(1) |
| PstCN-078 | PSTv-323 | 171272 | 6, 7, 8, 9, 17, 27, 43, 44, SP, Exp2 | 10 | 1 | 1.49 | SX(1) |
| PstCN-079 | \ | 170273 | 6, 7, 8, 9, 27, 43, 44, SP, Exp2, 76 | 10 | 1 | 1.49 | NM(1) |
| PstCN-080 | \ | 530062 | 1, 6, 8, 9, 43, 44, Exp2 | 7 | 1 | 1.49 | HA(1) |
| PstCN-081 | PSTv-052 | 171262 | 6, 7, 8, 9, 17, 27, 43, 44, Exp2 | 9 | 1 | 1.49 | HB(1) |
| PstCN-082 | \ | 160222 | 6, 7, 8, 27, 44, Exp2 | 6 | 1 | 1.49 | YN(1) |
| PstCN-083 | \ | 021033 | 8, 17, 44, SP, Exp2, 76 | 6 | 1 | 1.49 | AH(1) |
| PstCN-084 | \ | 130022 | 6, 8, 9, 44, Exp2 | 5 | 1 | 1.49 | AH(1) |
| PstCN-085 | PSTv-130 | 000022 | 44, Exp2 | 2 | 1 | 1.49 | YN(1) |
| PstCN-086 | \ | 131222 | 6, 8, 9, 17, 27, 44, Exp2 | 7 | 1 | 1.49 | GS(1) |
| PstCN-087 | \ | 011222 | 9, 17, 27, 44, Exp2 | 5 | 1 | 1.49 | SX(1) |
| PstCN-088 | \ | 030272 | 8, 9, 27, 43, 44, SP, Exp2 | 7 | 1 | 1.49 | GS(1) |
| PstCN-089 | \ | 020220 | 8, 27, 44 | 3 | 1 | 1.49 | HE(1) |
| PstCN-090 | \ | 020020 | 8, 44 | 2 | 2 | 2.99 | GZ(1), CQ(1) |
| Race | Octal | Virulence to Yr Gene | 2020 (N = 67) | 2023 (N = 103) | ||||
|---|---|---|---|---|---|---|---|---|
| No. | Fre. (%) | Distribution | No. | Fre. (%) | Distribution | |||
| PstCN-001 | 020062 | 8, 43, 44, Exp2 | 8 | 11.94 | YN(2), SC(3), SN(2), HE(1) | 5 | 4.85 | SC(4), GZ(1) |
| PstCN-002 | 020262 | 8, 27, 43, 44, Exp2 | 6 | 8.96 | HB, GS(2), HA, YN, SC | 1 | 0.97 | QH(1) |
| PstCN-003 | 120062 | 6, 8, Exp2, 44, 43 | 1 | 1.49 | AH | 3 | 2.91 | HB(1), GS(1), AH(1) |
| PstCN-005 | 120262 | 8, 6, 27, Exp2, 44, 43 | 1 | 1.49 | SC | 3 | 2.91 | AH(2), SC(1) |
| PstCN-019 | 420062 | 1, 8, 43, Exp2, 44 | 1 | 1.49 | GZ | 1 | 0.97 | HE(1) |
| Virulence to Yr Gene | |||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Vir to | Yr5 | Yr6 | Yr7 | Yr8 | Yr9 | Yr10 | Yr15 | Yr17 | Yr24 | Yr27 | Yr32 | Yr43 | Yr44 | YrSP | Yr85 | YrExp2 | Yr76 |
| Yr1 | 0.00 | 0.21 | −0.08 | 0.05 | 0.26 | 0.00 | −0.03 | −0.08 | −0.04 | −0.02 | 0.00 | 0.13 | 0.04 | −0.07 | −0.03 | 0.05 | −0.06 |
| Yr5 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | |
| Yr6 | 0.32 | −0.04 | 0.32 | 0.00 | −0.07 | 0.21 | 0.10 | 0.26 | 0.00 | 0.11 | 0.10 | 0.18 | −0.07 | 0.12 | 0.10 | ||
| Yr7 | 0.08 | 0.29 | 0.00 | −0.05 | 0.43 | −0.06 | 0.29 | 0.00 | 0.09 | −0.21 | 0.37 | −0.05 | 0.08 | 0.25 | |||
| Yr8 | −0.08 | 0.00 | 0.03 | −0.14 | 0.04 | 0.02 | 0.00 | 0.02 | −0.04 | 0.07 | 0.03 | −0.05 | 0.06 | ||||
| Yr9 | 0.00 | −0.06 | 0.52 | 0.14 | 0.41 | 0.00 | 0.00 | 0.09 | 0.51 | 0.25 | 0.10 | 0.29 | |||||
| Yr10 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | ||||||
| Yr15 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | |||||||
| Yr17 | −0.06 | 0.38 | 0.00 | −0.01 | 0.06 | 0.53 | −0.05 | 0.08 | 0.42 | ||||||||
| Yr24 | 0.23 | 0.00 | 0.04 | 0.03 | −0.05 | 0.70 | 0.04 | −0.05 | |||||||||
| Yr27 | 0.00 | 0.28 | −0.05 | 0.30 | 0.16 | 0.02 | 0.25 | ||||||||||
| Yr32 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | |||||||||||
| Yr43 | −0.04 | 0.08 | −0.10 | 0.17 | 0.01 | ||||||||||||
| Yr44 | 0.05 | 0.02 | −0.04 | −0.28 | |||||||||||||
| YrSP | −0.04 | 0.07 | 0.71 | ||||||||||||||
| Yr85 | 0.03 | −0.03 | |||||||||||||||
| YrExp2 | 0.06 | ||||||||||||||||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Huang, J.; Zhang, X.; Tan, W.; Wu, Y.; Xu, H.; Wang, S.; Mehmood, S.; Zhou, X.; Yang, S.; Wang, M.; et al. Virulence Characterization of Puccinia striiformis f. sp. tritici in China in 2020 Using Wheat Yr Single-Gene Lines. J. Fungi 2025, 11, 447. https://doi.org/10.3390/jof11060447
Huang J, Zhang X, Tan W, Wu Y, Xu H, Wang S, Mehmood S, Zhou X, Yang S, Wang M, et al. Virulence Characterization of Puccinia striiformis f. sp. tritici in China in 2020 Using Wheat Yr Single-Gene Lines. Journal of Fungi. 2025; 11(6):447. https://doi.org/10.3390/jof11060447
Chicago/Turabian StyleHuang, Jie, Xingzong Zhang, Wenjing Tan, Yi Wu, Hai Xu, Shuwaner Wang, Sajid Mehmood, Xinli Zhou, Suizhuang Yang, Meinan Wang, and et al. 2025. "Virulence Characterization of Puccinia striiformis f. sp. tritici in China in 2020 Using Wheat Yr Single-Gene Lines" Journal of Fungi 11, no. 6: 447. https://doi.org/10.3390/jof11060447
APA StyleHuang, J., Zhang, X., Tan, W., Wu, Y., Xu, H., Wang, S., Mehmood, S., Zhou, X., Yang, S., Wang, M., Chen, X., Chen, W., Liu, T., Li, X., & Xia, C. (2025). Virulence Characterization of Puccinia striiformis f. sp. tritici in China in 2020 Using Wheat Yr Single-Gene Lines. Journal of Fungi, 11(6), 447. https://doi.org/10.3390/jof11060447






