Phenotypic and Molecular Characterization of Multidrug-Resistant Clinical Isolates of the Candidozyma haemuli Species Complex (Formerly Candida haemulonii Species Complex) from the Brazilian Amazon Reveals the First Case of Candidozyma pseudohaemuli in Brazil
Abstract
1. Introduction
2. Materials and Methods
2.1. This Study
2.2. Species Identification
2.3. Antifungal Susceptibility Testing
2.4. Evaluation of Virulence Factors
2.4.1. Biofilm Formation and Biomass Quantification
2.4.2. Phospholipase Activity
2.4.3. Lipase Activity
2.4.4. Proteinase Activity
2.5. Statistical Analysis
2.6. Ethics Approval
3. Results
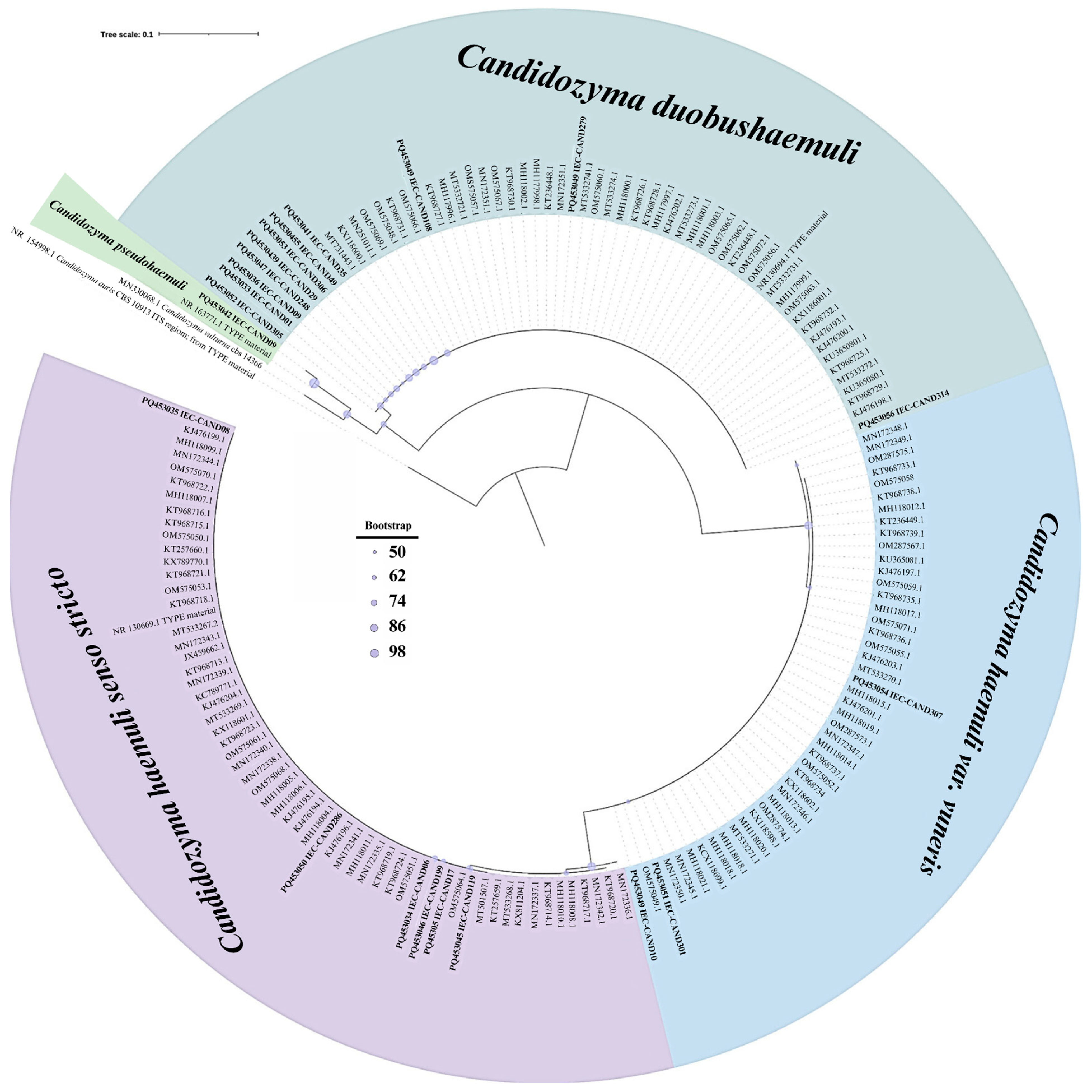
3.1. Patient Characteristics and Isolate Identification
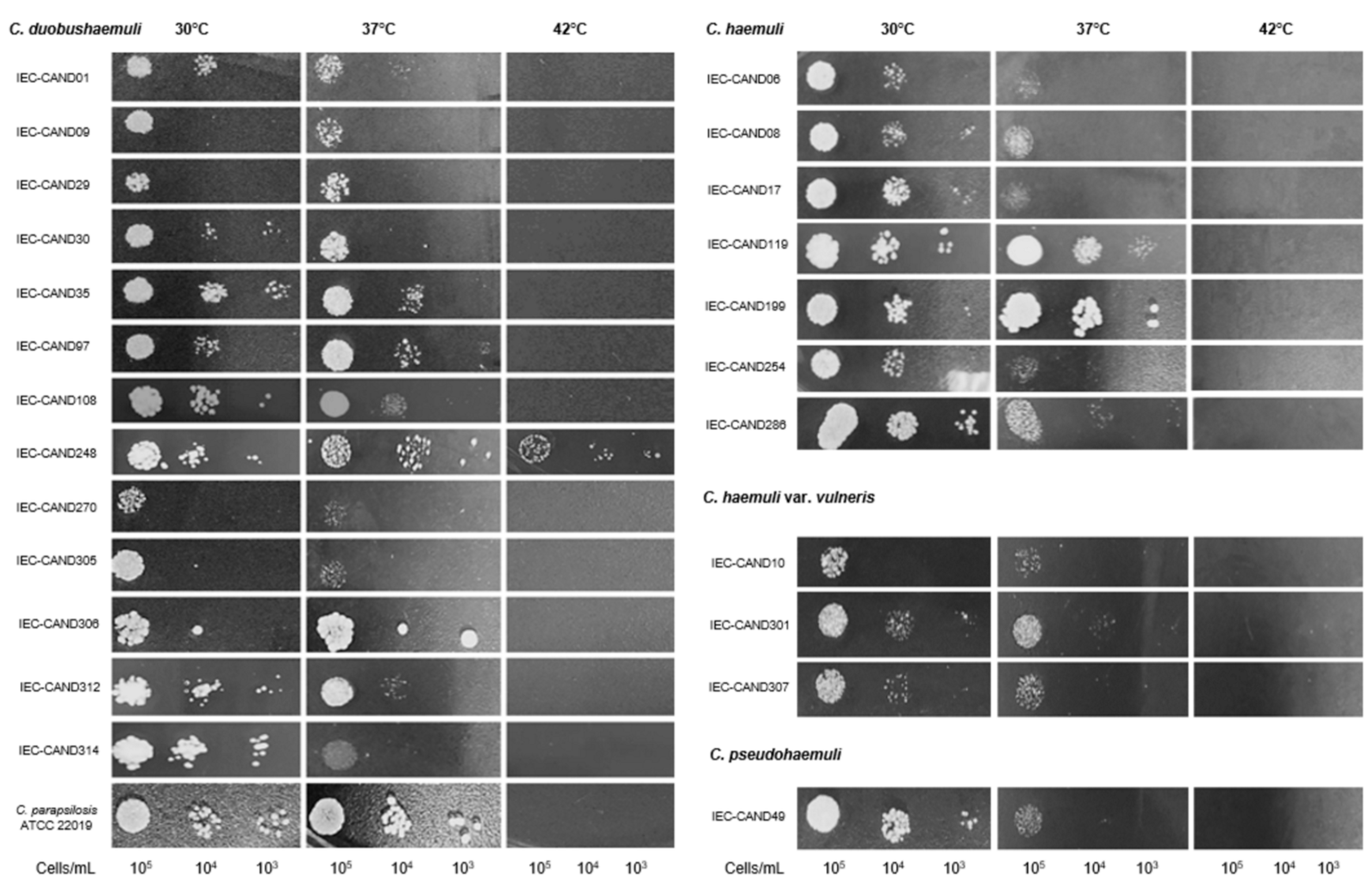
3.2. Phenotypic Virulence Features of the Isolates
3.3. Antifungal Resistance Patterns in Clinical Isolates
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Liu, F.; Hu, Z.-D.; Zhao, X.-M.; Zhao, W.-N.; Feng, Z.-X.; Yurkov, A.; Alwasel, S.; Boekhout, T.; Bensch, K.; Hui, F.-L.; et al. Phylogenomic Analysis of the Candida auris-Candida haemuli Clade and Related Taxa in the Metschnikowiaceae, and Proposal of Thirteen New Genera, Fifty-Five New Combinations and Nine New Species. Persoonia 2024, 52, 22–43. [Google Scholar] [CrossRef] [PubMed]
- Fisher, M.C.; Gurr, S.J.; Cuomo, C.A.; Blehert, D.S.; Jin, H.; Stukenbrock, E.H.; Stajich, J.E.; Kahmann, R.; Boone, C.; Denning, D.W.; et al. Threats Posed by the Fungal Kingdom to Humans, Wildlife, and Agriculture. mBio 2020, 11, 10–1128. [Google Scholar] [CrossRef]
- Brown, G.D.; Denning, D.W.; Gow, N.A.R.; Levitz, S.M.; Netea, M.G.; White, T.C. Hidden Killers: Human Fungal Infections. Sci. Transl. Med. 2012, 4, 165rv13. [Google Scholar] [CrossRef]
- Francisco, E.C.; de Jong, A.W.; Colombo, A.L. Candida haemulonii Species Complex: A Mini-Review. Mycopathologia 2023, 188, 909–917. [Google Scholar] [CrossRef]
- Cao, C.; Bing, J.; Liao, G.; Nobile, C.J.; Huang, G. Candida haemulonii Species Complex: Emerging Fungal Pathogens of the Metschnikowiaceae Clade. Zoonoses 2023, 3, 957. [Google Scholar] [CrossRef]
- de Jong, A.W.; Al-Obaid, K.; Mohd Tap, R.; Gerrits van den Ende, B.; Groenewald, M.; Joseph, L.; Ahmad, S.; Hagen, F. Candida khanbhai sp. Nov., a New Clinically Relevant Yeast within the Candida haemulonii Species Complex. Med. Mycol. 2023, 61, myad009. [Google Scholar] [CrossRef] [PubMed]
- Chow, N.A.; Gade, L.; Tsay, S.V.; Forsberg, K.; Greenko, J.A.; Southwick, K.L.; Barrett, P.M.; Kerins, J.L.; Lockhart, S.R.; Chiller, T.M.; et al. Multiple Introductions and Subsequent Transmission of Multidrug-Resistant Candida auris in the USA: A Molecular Epidemiological Survey. Lancet Infect. Dis. 2018, 18, 1377–1384. [Google Scholar] [CrossRef]
- Garcia, J.A. Distribuição Regional da Resistência Antifúngica em Cepas Causadoras de Candidemia no Brasil. Ph.D. Thesis, Secretaria de Estado da Saúde de São Paulo, São Paulo, Brazil, 2024. Available online: https://fi-admin.bvsalud.org/document/view/pgzh2 (accessed on 20 March 2024).
- Lima, S.L.; Francisco, E.C.; de Almeida Júnior, J.N.; Santos, D.W.d.C.L.; Carlesse, F.; Queiroz-Telles, F.; Melo, A.S.d.A.; Colombo, A.L. Increasing Prevalence of Multidrug-Resistant Candida haemulonii Species Complex among All Yeast Cultures Collected by a Reference Laboratory over the Past 11 Years. J. Fungi 2020, 6, 110. [Google Scholar] [CrossRef] [PubMed]
- Gómez-Gaviria, M.; Martínez-Álvarez, J.A.; Chávez-Santiago, J.O.; Mora-Montes, H.M. Candida haemulonii Complex and Candida auris: Biology, Virulence Factors, Immune Response, and Multidrug Resistance. Infect. Drug Resist. 2023, 16, 1455–1470. [Google Scholar] [CrossRef]
- Chen, X.; Jia, X.; Bing, J.; Zhang, H.; Hong, N.; Liu, Y.; Xi, H.; Wang, W.; Liu, Z.; Zhang, Q.; et al. Clonal Dissemination of Antifungal-Resistant Candida haemulonii, China. Emerg. Infect. Dis. 2023, 29, 576–584. [Google Scholar] [CrossRef]
- Lima, S.L. Caracterização Molecular, Avaliação do Perfil de Susceptibilidade a Antifúngicos e Formação de Biofilme de Isolados Clínicos Pertencentes ao Complexo Candida haemulonii e Candida auris. Master’s Thesis, Universidade Federal de São Paulo (Escola Paulista de Medicina), São Paulo, Brazil, 2017. Available online: https://repositorio.unifesp.br/handle/11600/50775 (accessed on 25 March 2024).
- Subcommittee on Antifungal Susceptibility Testing (AFST) of the ESCMID European Committee for Antimicrobial Susceptibility Testing (EUCAST). EUCAST Definitive Document EDef 7.1: Method for the Determination of Broth Dilution MICs of Antifungal Agents for Fermentative Yeasts. Clin. Microbiol. Infect. 2008, 14, 398–405. [Google Scholar] [CrossRef]
- Centers for Disease Control and Prevention. Antifungal Susceptibility Testing for C. auris. Updated 23 April 2024. Available online: https://www.cdc.gov/candida-auris/hcp/laboratories/antifungal-susceptibility-testing.html (accessed on 27 November 2024).
- Approved Standard M27-A3; Reference Method for Broth Dilution Antifungal Susceptibility Testing of Yeasts. Clinical and Laboratory Standards Institute: Malvern, PA, USA, 2008.
- Ramos, L.S.; Figueiredo-Carvalho, M.H.G.; Silva, L.N.; Siqueira, N.L.M.; Lima, J.C.; Oliveira, S.S.; Almeida-Paes, R.; Zancopé-Oliveira, R.M.; Azevedo, F.S.; Ferreira, A.L.P.; et al. The Threat Called Candida haemulonii Species Complex in Rio de Janeiro State, Brazil: Focus on Antifungal Resistance and Virulence Attributes. J. Fungi 2022, 8, 574. [Google Scholar] [CrossRef] [PubMed]
- Price, M.F.; Wilkinson, I.D.; Gentry, L.O. Plate Method for Detection of Phospholipase Activity in Candida Albicans. Sabouraudia 1982, 20, 7–14. [Google Scholar] [CrossRef]
- Muhsin, T.M.; Aubaid, A.H.; al-Duboon, A.H. Extracellular Enzyme Activities of Dermatophytes and Yeast Isolates on Solid Media. Mycoses 1997, 40, 465–469. [Google Scholar] [CrossRef] [PubMed]
- Rüchel, R.; Tegeler, R.; Trost, M. A Comparison of Secretory Proteinases from Different Strains of Candida Albicans. Sabouraudia 1982, 20, 233–244. [Google Scholar] [CrossRef] [PubMed]
- Ferreira, N.; Belloch, C.; Querol, A.; Manzanares, P.; Vallez, S.; Santos, A. Yeast Microflora Isolated from Brazilian Cassava Roots: Taxonomical Classification Based on Molecular Identification. Curr. Microbiol. 2010, 60, 287–293. [Google Scholar] [CrossRef]
- Almeida, J.N.d.; Motta, A.L.; Rossi, F.; Abdala, E.; Pierrotti, L.C.; Kono, A.S.G.; Diz, M.D.P.E.; Benard, G.; Del Negro, G.M.B. First Report of a Clinical Isolate of Candida haemulonii in Brazil. Clinics 2012, 67, 1229–1231. [Google Scholar] [CrossRef]
- Ramos, L.S.; Figueiredo-Carvalho, M.H.G.; Barbedo, L.S.; Ziccardi, M.; Chaves, A.L.S.; Zancopé-Oliveira, R.M.; Pinto, M.R.; Sgarbi, D.B.G.; Dornelas-Ribeiro, M.; Branquinha, M.H.; et al. Candida haemulonii Complex: Species Identification and Antifungal Susceptibility Profiles of Clinical Isolates from Brazil. J. Antimicrob. Chemother. 2015, 70, 111–115. [Google Scholar] [CrossRef]
- de Souza Ramos, L.; Barbedo, L.S.; Braga-Silva, L.A.; dos Santos, A.L.S.; Pinto, M.R.; da Graça Sgarbi, D.B. Protease and Phospholipase Activities of Candida spp. Isolated from Cutaneous Candidiasis. Revi. Iberoam. Micol. 2015, 32, 122–125. [Google Scholar] [CrossRef]
- Almeida, J.N.d.; Assy, J.G.P.L.; Levin, A.S.; Negro, G.M.B.D.; Giudice, M.C.; Tringoni, M.P.; Thomaz, D.Y.; Motta, A.L.; Abdala, E.; Pierroti, L.C.; et al. Candida haemulonii complex species, Brazil, January 2010–March 2015. Emerg. Infect. Dis. 2016, 22, 561–563. [Google Scholar] [CrossRef]
- Boatto, H.F.; Cavalcanti, S.D.B.; Del Negro, G.M.; Girão, M.J.B.; Francisco, E.C.; Ishida, K.; Gompertz, O.F. Candida Duobushaemulonii: An Emerging Rare Pathogenic Yeast Isolated from Recurrent Vulvovaginal Candidiasis in Brazil. Mem. Inst. Oswaldo Cruz 2016, 111, 407–410. [Google Scholar] [CrossRef]
- Pagani, D.M.; Heidrich, D.; Paulino, G.V.B.; de Oliveira Alves, K.; Dalbem, P.T.; de Oliveira, C.F.; Andrade, Z.M.M.; Silva, C.; Correia, M.D.; Scroferneker, M.L.; et al. Susceptibility to Antifungal Agents and Enzymatic Activity of Candida Haemulonii and Cutaneotrichosporon Dermatis Isolated from Soft Corals on the Brazilian Reefs. Arch. Microbiol. 2016, 198, 963–971. [Google Scholar] [CrossRef]
- Ramos, L.S.; Branquinha, M.H.; Santos, A.L.S. Different Classes of Hydrolytic Enzymes Produced by Multidrug-Resistant Yeasts Comprising the Candida haemulonii Complex. Med. Mycol. 2017, 55, 228–232. [Google Scholar] [CrossRef] [PubMed]
- Ramos, L.S.; Mello, T.P.; Branquinha, M.H.; Santos, A.L.S. Biofilm Formed by Candida Haemulonii Species Complex: Structural Analysis and Extracellular Matrix Composition. J. Fungi 2020, 6, 46. [Google Scholar] [CrossRef]
- de Barros Rodrigues, D.K.; Lockhart, S.R.; Berkow, E.L.; Gade, L.; Bonfietti, L.X.; Mazo Fávero Gimenes, V.; Silva Ruiz, L.; Bronze Macioni, M.; de Souza Carvalho Melhem, M. Whole-Genome Sequencing of Candida haemulonii Species Complex from Brazil and the United States: Genetic Diversity and Antifungal Susceptibility. Med. Mycol. 2023, 61, myad030. [Google Scholar] [CrossRef] [PubMed]
- Françoise, U.; Desnos-Ollivier, M.; Le Govic, Y.; Sitbon, K.; Valentino, R.; Peugny, S.; Chouaki, T.; Mazars, E.; Paugam, A.; Nicolas, M.; et al. Candida haemulonii Complex, an Emerging Threat from Tropical Regions? PLoS Negl. Trop. Dis. 2023, 17, e0011453. [Google Scholar] [CrossRef] [PubMed]
- Poulopoulou, A.; Sidiropoulou, A.; Sarmourli, T.; Zachrou, E.; Michailidou, C.; Zarras, C.; Vagdatli, E.; Massa, E.; Mouloudi, E.; Pyrpasopoulou, A.; et al. Candida auris: Outbreak, Surveillance and Epidemiological Monitoring in Northern Greece. Med. Mycol. 2024, 62, myae062. [Google Scholar] [CrossRef]
- Bilal, H.; Shafiq, M.; Hou, B.; Islam, R.; Khan, M.N.; Khan, R.U.; Zeng, Y. Distribution and Antifungal Susceptibility Pattern of Candida Species from Mainland China: A Systematic Analysis. Virulence 2022, 13, 1573–1589. [Google Scholar] [CrossRef]
- Sugita, T.; Takashima, M.; Poonwan, N.; Mekha, N. Candida pseudohaemulonii sp. Nov., an Amphotericin B-and Azole-Resistant Yeast Species, Isolated from the Blood of a Patient from Thailand. Microbiol. Immunol. 2006, 50, 469–473. [Google Scholar] [CrossRef]
- Kim, M.-N.; Shin, J.H.; Sung, H.; Lee, K.; Kim, E.-C.; Ryoo, N.; Lee, J.-S.; Jung, S.-I.; Park, K.H.; Kee, S.J.; et al. Candida haemulonii and Closely Related Species at 5 University Hospitals in Korea: Identification, Antifungal Susceptibility, and Clinical Features. Clin. Infect. Dis. 2009, 48, e57–e61. [Google Scholar] [CrossRef]
- Gade, L.; Muñoz, J.F.; Sheth, M.; Wagner, D.; Berkow, E.L.; Forsberg, K.; Jackson, B.R.; Ramos-Castro, R.; Escandón, P.; Dolande, M.; et al. Understanding the Emergence of Multidrug-Resistant Candida: Using Whole-Genome Sequencing to Describe the Population Structure of Candida haemulonii Species Complex. Front. Genet. 2020, 11, 554. [Google Scholar] [CrossRef] [PubMed]
- Excoffier, L.; Foll, M.; Petit, R.J. Genetic Consequences of Range Expansions. Annu. Rev. Ecol. Evol. Syst 2009, 40, 481–501. [Google Scholar] [CrossRef]
- Merseguel, K.B.; Nishikaku, A.S.; Rodrigues, A.M.; Padovan, A.C.; e Ferreira, R.C.; de Azevedo Melo, A.S.; Briones, M.R.d.S.; Colombo, A.L. Genetic Diversity of Medically Important and Emerging Candida Species Causing Invasive Infection. BMC Infect. Dis. 2015, 15, 57. [Google Scholar] [CrossRef]
- Cendejas-Bueno, E.; Kolecka, A.; Alastruey-Izquierdo, A.; Theelen, B.; Groenewald, M.; Kostrzewa, M.; Cuenca-Estrella, M.; Gómez-López, A.; Boekhout, T. Reclassification of the Candida Haemulonii Complex as Candida haemulonii (C. haemulonii Group I), C. duobushaemulonii sp. Nov. (C. haemulonii Group II), and C. Haemulonii Var. Vulnera Var. Nov.: Three Multiresistant Human Pathogenic Yeasts. J. Clin. Microbiol. 2012, 50, 3641–3651. [Google Scholar] [CrossRef] [PubMed]
- Kathuria, S.; Singh, P.K.; Sharma, C.; Prakash, A.; Masih, A.; Kumar, A.; Meis, J.F.; Chowdhary, A. Multidrug-Resistant Candida auris Misidentified as Candida haemulonii: Characterization by Matrix-Assisted Laser Desorption Ionization—Time of Flight Mass Spectrometry and DNA Sequencing and Its Antifungal Susceptibility Profile Variability by Vitek 2, CLSI Broth Microdilution, and Etest Method. J. Clin. Microbiol. 2015, 53, 1823–1830. [Google Scholar] [CrossRef]
- Kumar, A.; Sachu, A.; Mohan, K.; Vinod, V.; Dinesh, K.; Karim, S. Simple Low Cost Differentiation of Candida auris from Candida haemulonii Complex Using CHROMagar Candida Medium Supplemented with Pal’s Medium. Rev. Iberoam. Micol. 2017, 34, 109–111. [Google Scholar] [CrossRef] [PubMed]
- Jurado-Martín, I.; Marcos-Arias, C.; Tamayo, E.; Guridi, A.; de Groot, P.W.J.; Quindós, G.; Eraso, E. Candida Duobushaemulonii: An Old But Unreported Pathogen. J. Fungi 2020, 6, 374. [Google Scholar] [CrossRef]
- Pharkjaksu, S.; Boonmee, N.; Mitrpant, C.; Ngamskulrungroj, P. Immunopathogenesis of Emerging Candida auris and Candida haemulonii Strains. J. Fungi 2021, 7, 725. [Google Scholar] [CrossRef]
- Tamura, T.; Alshahni, M.M.; Makimura, K. Evaluation of CHROMagarTM Candida Plus Chromogenic Agar for the Presumptive Identification of Candida auris. Microbiol. Immunol. 2022, 66, 292–298. [Google Scholar] [CrossRef]
- Saluja, P.; Prasad, G.S. Candida ruelliae sp. Nov., a Novel Yeast Species Isolated from Flowers of Ruellia sp. (Acanthaceae). FEMS Yeast Res. 2008, 8, 660–666. [Google Scholar] [CrossRef]
- Satoh, K.; Makimura, K.; Hasumi, Y.; Nishiyama, Y.; Uchida, K.; Yamaguchi, H. Candida auris sp. Nov., a Novel Ascomycetous Yeast Isolated from the External Ear Canal of an Inpatient in a Japanese Hospital. Microbiol. Immunol. 2009, 53, 41–44. [Google Scholar] [CrossRef]
- Ruv Lemes, M.; Sampaio, G.; Fisch, G.; Alves, L.M.; Maksic, J.; Guatura, M.; Shimizu, M. Impacts of Atmospheric CO2 Increase and Amazon Deforestation on the Regional Climate: A Water Budget Modelling Study. Int. J. Climatol. 2023, 43, 1497–1513. [Google Scholar] [CrossRef]
- Butt, E.W.; Baker, J.C.A.; Bezerra, F.G.S.; von Randow, C.; Aguiar, A.P.D.; Spracklen, D.V. Amazon Deforestation Causes Strong Regional Warming. Proc. Natl. Acad. Sci. USA 2023, 120, e2309123120. [Google Scholar] [CrossRef]
- Bottino, M.J.; Nobre, P.; Giarolla, E.; da Silva Junior, M.B.; Capistrano, V.B.; Malagutti, M.; Tamaoki, J.N.; de Oliveira, B.F.A.; Nobre, C.A. Amazon Savannization and Climate Change Are Projected to Increase Dry Season Length and Temperature Extremes over Brazil. Sci. Rep. 2024, 14, 5131. [Google Scholar] [CrossRef]
- Alves, V.; de Andrade, I.B.; Corrêa-Junior, D.; Avellar-Moura, I.; Passos, K.; Soares, J.; Pontes, B.; Almeida, M.A.; Almeida-Paes, R.; Frases, S. Revealing the Impact of Rapamycin on the Virulence Factors of the Candida haemulonii Complex. Curr. Res. Microb. Sci. 2024, 7, 100247. [Google Scholar] [CrossRef]
- Ambaraghassi, G.; Dufresne, P.J.; Dufresne, S.F.; Vallières, É.; Muñoz, J.F.; Cuomo, C.A.; Berkow, E.L.; Lockhart, S.R.; Luong, M.-L. Identification of Candida auris by Use of the Updated Vitek 2 Yeast Identification System, Version 8.01: A Multilaboratory Evaluation Study. J. Clin. Microbiol. 2019, 57, e00884-19. [Google Scholar] [CrossRef]
- Novak, A.R.; Bradley, M.E.; Kiser, T.H.; Mueller, S.W. Azole-Resistant Aspergillus and Echinocandin-Resistant Candida—What Are the Treatment Options? Curr. Fungal Infect. Rep. 2020, 14, 141–152. [Google Scholar] [CrossRef]
- Ben-Ami, R.; Berman, J.; Novikov, A.; Bash, E.; Shachor-Meyouhas, Y.; Zakin, S.; Maor, Y.; Tarabia, J.; Schechner, V.; Adler, A.; et al. Multidrug-Resistant Candida haemulonii and C. auris, Tel Aviv, Israel. Emerg. Infect. Dis. 2017, 23, 195–203. [Google Scholar] [CrossRef]
- Nobrega de Almeida, J.; Brandão, I.B.; Francisco, E.C.; de Almeida, S.L.R.; de Oliveira Dias, P.; Pereira, F.M.; Santos Ferreira, F.; de Andrade, T.S.; de Miranda Costa, M.M.; de Souza Jordão, R.T.; et al. Axillary Digital Thermometers Uplifted a Multidrug-Susceptible Candida auris Outbreak among COVID-19 Patients in Brazil. Mycoses 2021, 64, 1062–1072. [Google Scholar] [CrossRef]
- de Almeida, J.N.; Francisco, E.C.; Hagen, F.; Brandão, I.B.; Pereira, F.M.; Presta Dias, P.H.; de Miranda Costa, M.M.; de Souza Jordão, R.T.; de Groot, T.; Colombo, A.L. Emergence of Candida auris in Brazil in a COVID-19 Intensive Care Unit. J. Fungi 2021, 7, 220. [Google Scholar] [CrossRef]
- Spruijtenburg, B.; Nobrega de Almeida Júnior, J.; Ribeiro, F.d.C.; Kemmerich, K.K.; Baeta, K.; Meijer, E.F.J.; de Groot, T.; Meis, J.F.; Colombo, A.L.; Brazilian Candida auris collaborative network. Multicenter Candida auris Outbreak Caused by Azole-Susceptible Clade IV in Pernambuco, Brazil. Mycoses 2024, 67, e13752. [Google Scholar] [CrossRef] [PubMed]
- Falci, D.R.; Pasqualotto, A.C. Clinical Mycology in Latin America and the Caribbean: A Snapshot of Diagnostic and Therapeutic Capabilities. Mycoses 2019, 62, 368–373. [Google Scholar] [CrossRef] [PubMed]


| Variables | No. (%) |
|---|---|
| Demographic variables | |
| Age range | |
| 13–18 | 1 (4.2) |
| 19–30 | 3 (12.5) |
| 31–45 | 5 (20.8) |
| 46–60 | 7 (29.2) |
| 61–75 | 4 (16.7) |
| 76+ | 4 (16.7) |
| Male gender | 16 (66.6) |
| Female gender | 8 (33.3) |
| Sites of isolation | |
| Bronchoalveolar lavage | 14 (58.3) |
| Sputum | 4 (16.7) |
| Tracheal secretion | 2 (8.3) |
| Urine | 1 (4.2) |
| Skin lesion | 1 (4.2) |
| Blood | 1 (4.2) |
| Pleural fluid | 1 (4.2) |
| Main clinical condition | |
| Tuberculosis | 5 (20.8) |
| Chronic pneumopathy † | 5 (20.8) |
| Neoplasia | 5 (20.8) |
| PLHIV + tuberculosis | 3 (12.5) |
| PLHIV + cryptococcosis | 1 (4.2) |
| PLHIV + histoplasmosis | 1 (4.2) |
| Erysipelas | 2 (8.3) |
| Paracoccidioidomycosis | 1 (4.2) |
| Pneumonia | 1 (4.2) |
| Clinical history | |
| Hospitalization | 15 (62.5) |
| Intensive care | 5 (20.8) |
| Prior antifungal therapy (within 60 days) | 8 (33.3) |
| Prior antibiotic therapy (within 60 days) | 17 (70.83) |
| Isolated species | |
| C. duobushaemuli | 13 (54.2) |
| C. haemuli sensu stricto | 7 (29.2) |
| C. haemuli var. vulneris | 3 (12.5) |
| C. pseudohaemuli | 1 (4.2) |
| Isolate No. | Source | Year | Age/ Gender | Identification | |||
|---|---|---|---|---|---|---|---|
| Previous Identification (VITEK® 2) ** | MALDI-TOF MS (Biotyper® Sirius System) ** | ITS Region Sequencing | GenBank Accession No. | ||||
| IEC-CAND01 | BAL | 2021 | 46/M | C. duobushaemulonii | NA | C. duobushaemuli | PQ453033 |
| IEC-CAND09 | BAL | 2021 | 33/M | C. famata | C. duobushaemulonii | C. duobushaemuli | PQ453036 |
| IEC-CAND29 | Tracheal secretion | 2021 | 84/F | NA | C. duobushaemulonii | C. duobushaemuli | PQ453039 |
| IEC-CAND30 | Urine | 2021 | 60/F | C. famata | C. duobushaemulonii | C. duobushaemuli | PQ453040 |
| IEC-CAND35 | BAL | 2021 | 31/M | NA | C. duobushaemulonii | C. duobushaemuli | PQ453041 |
| IEC-CAND97 | Blood | 2020 | 77/M | C. duobushaemulonii | C. duobushaemulonii | C. duobushaemuli | PQ453043 |
| IEC-CAND108 | BAL | 2021 | 26/M | C. duobushaemulonii | C. duobushaemulonii | C. duobushaemuli | PQ453044 |
| IEC-CAND248 | BAL | 2022 | 43/M | NA | C. duobushaemulonii | C. duobushaemuli | PQ453047 |
| IEC-CAND270 | Sputum | 2022 | 65/F | NA | C. duobushaemulonii | C. duobushaemuli | PQ453049 |
| IEC-CAND305 | BAL | 2022 | 82/F | C. duobushaemulonii | C. duobushaemulonii | C. duobushaemuli | PQ453052 |
| IEC-CAND306 | BAL | 2022 | 57/M | C. spherica/Saccharomyces cerevisiae † | NA | C. duobushaemuli | PQ453053 |
| IEC-CAND312 | BAL | 2022 | 40/M |
C. auris/
C. duobushaemulonii † | C. duobushaemulonii | C. duobushaemuli | PQ453055 |
| IEC-CAND314 | BAL | 2024 | 18/M | C. haemulonii | NA | C. duobushaemuli | PQ453056 |
| IEC-CAND06 | BAL | 2021 | 30/M | C. haemulonii | NA | C. haemuli | PQ453034 |
| IEC-CAND08 | Tracheal secretion | 2021 | 73/F | C. parapsilosis | NA | C. haemuli | PQ453035 |
| IEC-CAND17 | Sputum | 2021 | 20/M | NA | C. haemulonii | C. haemuli | PQ453038 |
| IEC-CAND119 | BAL | 2021 | 72/F | NA | C. haemulonii | C. haemuli | PQ453045 |
| IEC-CAND199 | Sputum | 2021 | 42/M | NA | NA | C. haemuli | PQ453046 |
| IEC-CAND254 | Skin lesion | 2022 | 51/F | C. duobushaemulonii | NA | C. haemuli | PQ453048 |
| IEC-CAND286 | sputum | 2022 | 53/M | NA | C. haemulonii | C. haemuli | PQ453050 |
| IEC-CAND10 | BAL | 2021 | 72/M | C. famata | C. duobushaemulonii | C. haemuli var. vulneris | PQ453037 |
| IEC-CAND301 | BAL | 2022 | 49/M | C. haemulonii/C. haemulonii var. vulneris † | C. haemulonii var. vulneris | C. haemuli var. vulneris | PQ453051 |
| IEC-CAND307 | Pleural fluid | 2022 | 50/F | C. haemulonii var. vulneris | C. haemulonii var. vulneris | C. haemuli var. vulneris | PQ453054 |
| IEC-CAND49 | BAL | 2021 | 81/M | NA | C. haemulonii | C. pseudohaemuli | PQ453042 |
| Isolates | Proteinase | Lipase | Phospholipase | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Pz Index | Activity | Median Pz (IQR) | Pz Index | Activity | Median Pz (IQR) | Pz Index | Activity | Median Pz (IQR) | |
| C. duobushaemuli (n = 13) | |||||||||
| IEC-CAND09 | 0.69 | Strong | 0.82 (0.78–1.00) † | 1.00 | None | 1.0 (1.0–1.10) † | 1.00 | None | 1.0 (1.0–1.10) |
| IEC-CAND30 | 0.79 | Moderate | 1.00 | None | 1.00 | None | |||
| IEC-CAND01 | 0.75 | Moderate | 1.00 | None | 1.00 | None | |||
| IEC-CAND248 | 0.82 | Moderate | 1.00 | None | 1.00 | None | |||
| IEC-CAND270 | 0.73 | Moderate | 1.00 | None | 1.00 | None | |||
| IEC-CAND305 | 0.82 | Moderate | 1.00 | None | 1.00 | None | |||
| IEC-CAND312 | 0.78 | Moderate | 1.00 | None | 1.00 | None | |||
| IEC-CAND314 | 0.78 | Moderate | 1.00 | None | 1.00 | None | |||
| IEC-CAND29 | 0.90 | Weak | 1.00 | None | 0.85 | Moderate | |||
| IEC-CAND35 | 1.00 | None | 1.00 | None | 1.00 | None | |||
| IEC-CAND97 | 1.00 | None | 1.00 | None | 1.00 | None | |||
| IEC-CAND108 | 1.00 | None | 1.00 | None | 1.00 | None | |||
| IEC-CAND306 | 1.00 | None | 1.00 | None | 1.00 | None | |||
| C. haemuli (n = 7) | |||||||||
| IEC-CAND06 | 0.52 | Strong | 0.57 (0.54–0.61) † | 0.53 | Strong | 0.57 (0.55–0.79) † | 1.00 | None | 1.0 (1.0–1.10) |
| IEC-CAND17 | 0.57 | Strong | 0.56 | Strong | 1.00 | None | |||
| IEC-CAND119 | 0.55 | Strong | 0.57 | Strong | 1.00 | None | |||
| IEC-CAND286 | 0.50 | Strong | 0.37 | Strong | 1.00 | None | |||
| IEC-CAND08 | 0.65 | Strong | 1.00 | None | 1.00 | None | |||
| IEC-CAND254 | 0.57 | Strong | 1.00 | None | 0.83 | Moderate | |||
| IEC-CAND199 | 0.89 | Moderate | 0.57 | Strong | 1.00 | None | |||
| Candida haemuloni var. vulneris (n = 3) | |||||||||
| IEC-CAND301 | 0.54 | Strong | 0.75 (0.65–0.79) † | 0.39 | Strong | 0.51 (0.45–0.76) † | 1.00 | None | 1.0 (1.0–1.10) |
| IEC-CAND307 | 0.75 | Moderate | 0.51 | Strong | 1.00 | None | |||
| IEC-CAND10 | 0.83 | Moderate | 1.00 | None | 1.00 | None | |||
| C. pseudohaemuli (n = 1) | |||||||||
| IEC-CAND49 | 0.69 | Strong | 0.69 (0.69–0.69) † | 1.00 | None | 1.0 (1.0–1.10) † | 1.00 | None | 1.0 (1.0–1.10) |
| MIC, μg/mL | ||||
|---|---|---|---|---|
| Clinical Isolates | FLU | ITC | AB | FC |
| C. duobushaemuli (n = 13) | ||||
| IEC-CAND01 | >64 | 0.5 | 8 | 0.12 |
| IEC-CAND09 | 4 | >8 | 2 | 0.12 |
| IEC-CAND29 | 16 | 0.5 | 1 | 0.12 |
| IEC-CAND30 | 64 | >8 | ≥8 | 0.25 |
| IEC-CAND35 | 16 | >8 | 1 | 0.25 |
| IEC-CAND97 | 64 | 1 | 8 | >64 |
| IEC-CAND108 | >64 | 8 | >8 | 0.25 |
| IEC-CAND248 | 64 | 0.5 | 4 | 0.25 |
| IEC-CAND270 | >64 | >8 | 4 | 0.12 |
| IEC-CAND305 | >64 | >8 | >8 | 0.25 |
| IEC-CAND306 | >64 | 8 | 0.5 | 0.25 |
| IEC-CAND312 | 4 | 0.25 | >8 | 0.25 |
| IEC-CAND314 | >64 | 8 | >8 | 0.12 |
| C. haemuli (n = 7) | ||||
| IEC-CAND06 | 64 | 0.25 | >8 | 0.12 |
| IEC-CAND08 | 2 | 0.25 | 2 | 0.12 |
| IEC-CAND17 | 8 | 0.25 | >8 | 0.12 |
| IEC-CAND119 | 2 | 0.5 | 1 | 0.12 |
| IEC-CAND199 | 2 | 0.125 | 1 | 0.12 |
| IEC-CAND254 | 64 | 1 | 1 | 0.12 |
| IEC-CAND286 | 4 | 0.25 | 1 | 0.12 |
| C. haemuli var. vulneris (n = 3) | ||||
| IEC-CAND10 | 2 | 1 | 1 | 0.12 |
| IEC-CAND301 | 1 | 0.125 | 2 | 0.25 |
| IEC-CAND307 | 16 | 2 | 2 | 0.12 |
| C. pseudohaemuli (n = 1) | ||||
| IEC-CAND49 | 64 | 0.5 | 2 | 0.25 |
| % Resistance | |||||
|---|---|---|---|---|---|
| Clinical Isolates | FLU | ITC | AB | FC | %MDR |
| C. duobushaemuli (n = 13) | 69.2 | 69.2 | 69.2 | 7.7 | 69.2 |
| C. haemuli sensu stricto (n = 7) | 28.6 | 14.3 | 42.9 | 0.0 | 14.3 |
| C. haemuli var. vulneris (n = 3) | 0.0 | 66.7 | 66.7 | 0.0 | 33.3 |
| C. pseudohaemuli (n = 1) | 100.0 | 0.0 | 100.0 | 0.0 | 100.0 |
| Total (n = 24) | 50.0 | 50.0 | 66.7 | 4.2 | 50.0 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
França, S.L.; Oliveira, R.S.d.; Sousa, G.S.M.; Sá, S.R.d.; Nogueira, W.d.S.; Santo, E.P.T.d.E.; Caldas, D.d.S.; Silva, S.H.M.d. Phenotypic and Molecular Characterization of Multidrug-Resistant Clinical Isolates of the Candidozyma haemuli Species Complex (Formerly Candida haemulonii Species Complex) from the Brazilian Amazon Reveals the First Case of Candidozyma pseudohaemuli in Brazil. J. Fungi 2025, 11, 394. https://doi.org/10.3390/jof11050394
França SL, Oliveira RSd, Sousa GSM, Sá SRd, Nogueira WdS, Santo EPTdE, Caldas DdS, Silva SHMd. Phenotypic and Molecular Characterization of Multidrug-Resistant Clinical Isolates of the Candidozyma haemuli Species Complex (Formerly Candida haemulonii Species Complex) from the Brazilian Amazon Reveals the First Case of Candidozyma pseudohaemuli in Brazil. Journal of Fungi. 2025; 11(5):394. https://doi.org/10.3390/jof11050394
Chicago/Turabian StyleFrança, Sérgio Lobato, Rodrigo Santos de Oliveira, Gabriel Silas Marinho Sousa, Sarah Rodrigues de Sá, Walber da Silva Nogueira, Elaine Patrícia Tavares do Espírito Santo, Daniel dos Santos Caldas, and Silvia Helena Marques da Silva. 2025. "Phenotypic and Molecular Characterization of Multidrug-Resistant Clinical Isolates of the Candidozyma haemuli Species Complex (Formerly Candida haemulonii Species Complex) from the Brazilian Amazon Reveals the First Case of Candidozyma pseudohaemuli in Brazil" Journal of Fungi 11, no. 5: 394. https://doi.org/10.3390/jof11050394
APA StyleFrança, S. L., Oliveira, R. S. d., Sousa, G. S. M., Sá, S. R. d., Nogueira, W. d. S., Santo, E. P. T. d. E., Caldas, D. d. S., & Silva, S. H. M. d. (2025). Phenotypic and Molecular Characterization of Multidrug-Resistant Clinical Isolates of the Candidozyma haemuli Species Complex (Formerly Candida haemulonii Species Complex) from the Brazilian Amazon Reveals the First Case of Candidozyma pseudohaemuli in Brazil. Journal of Fungi, 11(5), 394. https://doi.org/10.3390/jof11050394






